In the decade following the publication of the Human Genome, noncoding RNAs (ncRNAs) have reshaped our understanding of the broad landscape of genome regulation. During this period, natural antisense transcripts (NATs), which are transcribed from the opposite strand of either protein or non-protein coding genes, have vaulted to prominence. Recent findings have shown that NATs can exert their regulatory functions by acting as epigenetic regulators of gene expression and chromatin remodeling. Here, we review recent work on the mechanisms of epigenetic modifications by NATs and their emerging role as master regulators of chromatin states. Unlike other long ncRNAs, antisense RNAs usually regulate their counterpart sense mRNA by modulating chromatin structure in cis and by bridging epigenetic effectors and regulatory complexes at specific genomic loci. Understanding the broad range of effects of NATs will shed light on the complex mechanisms that regulate chromatin remodeling and gene expression in development and disease.
Chromatin and ncRNAs: coupling structure and dynamic information
Histone octamer proteins and their tightly associated 146 bp of DNA form the nucleosome, the structural and functional core of eukaryotic chromatin. Specific combinations of DNA and histone post-translational modification patterns lead to diverse changes in chromatin states and distinct functional genomic outputs [1, 2]. DNA methylation is perhaps the best-characterized chemical modification of DNA that impacts chromatin structure and function. In mammalian cells, DNA methylation occurs on cytosine residues in CpG dinucleotides and correlates with transcriptional repression. Promoter regions have a high density of CpG dinucleotides, whose methylation state dictates the transcriptional activity of the gene. Chromatin structure and function are also regulated by post-translational modifications of histone proteins. Histone-modifying enzymes are protein complexes that dynamically recognize (read), add (write), remove (erase) or replace various chromatin modifications. Examples of writers include EZH2, the catalytic subunit of the polycomb repressive complex 2 (PRC2), which is responsible for the trimethylation of histone H3 at lysine 27 (H3K27me3), and G9a, the histone methyltransferase (HMT) that catalyzes the di- or trimethylation of histone H3 at lysine 9 (H3K9me2/3) [2, 3]. “Erasers”, such as the demethylase LSD1, specifically remove certain histone marks [4]. “Readers” function as interpreters and include effector proteins that recognize specific histone marks and transduce this information into a genomic response [5–7]. Writers, erasers and readers have to work in concert, with their action tightly coordinated to produce an integrated regulatory effect. Recent discoveries of frequent interactions between ncRNAs and chromatin strongly suggest pivotal roles for ncRNAs in orchestrating the function of these protein complexes. How chromatin-modifying enzymes specifically recognize and bind to their target loci still remains mysterious. One tempting hypothesis is that local transcription of low abundance ncRNAs might be the key event in the locus-specific recruitment of different reader, eraser and writer complexes.
Dynamic transcriptional regulation at the level of chromatin
The classic division of chromatin into two opposing states, gene rich euchromatin versus the silenced, tightly packed heterochromatin, has been challenged by recent discoveries suggesting the existence of different chromatin states in various organisms, including humans [8–13]. The two-state chromatin model assumed that the chromatin structure was essentially an on/off switch whereby a gene was either active or repressed, without any intermediate states. By contrast, a dynamic chromatin state varies between these extremes and represents an integration of information derived from an intricate network of histone-modifying enzymes, chromatin binding proteins, transcription factors and chromatin-associated RNA transcripts [14, 15].
Globally, RNA, which is an integral structural component of chromatin, is required for the maintenance of compact chromatin fibers [16]. RNA has also been shown to be involved in the maintenance of higher-order chromatin structure at pericentric heterochromatin in mouse cells [17], highlighting the important contribution of RNA to the regulation of chromatin structure and function. Recently, a genome-wide next-generation RNA sequencing approach was used to identify the RNA content of chromatin in human fibroblasts [18]. Surprisingly, more than 70% of the sequencing reads aligned with intergenic and intronic regions of the human genome. Although this result could be an artifact of incompletely processed mRNAs or DNA contamination, functional experiments on a small number of chromatin-RNA transcripts imply an interaction with chromatin-modifying enzymes, which raises the possibility of a functional role of these transcripts in chromatin regulation [18].
Further support for the notion that RNA regulates chromatin comes from a small but growing number of antisense transcripts [19, 20] and other long ncRNAs [21–24] that interact with epigenetic effectors to orchestrate chromatin remodeling and epigenetic changes during development and disease. Cell-type specific ncRNAs interact with ubiquitously expressed regulatory proteins to form RNA-protein complexes that can interact with histones, DNA, other RNAs and other chromatin-modifying complexes to dynamically coordinate changes in gene expression programs (reviewed in [25]). RNA motifs composed of primary sequence information coupled to highly diverse secondary structure elements underlie the complexity and dynamic nature of these interactions. The combination of structural and regulatory elements of the chromatin contributes to the acquisition of a specific chromatin state and is key to understanding the mechanisms governing the organization of the human genome and the regulation of gene expression.
Natural antisense transcripts (NATs)
A substantial fraction of the mammalian genome is transcribed in the form of non-protein coding RNAs [26–29] that have important regulatory functions in development, differentiation [30–32] and human diseases [19, 33–35]. Although there is no unequivocal classification of non-protein coding transcripts found in the mammalian genome, ncRNAs can be roughly divided on the basis of size into short ncRNAs (less than 200 nt in length) and long ncRNAs (lncRNAs) that are more than 200 nt long [36, 37]. Short ncRNAs include miRNAs, piRNA, endogenous siRNAs and snoRNAs, which have been extensively reviewed elsewhere [38–40] and therefore will not be discussed here. lncRNAs are a heterogeneous group of RNAs transcribed from intergenic [41] or intragenic regions [42], which vary in length from 200 nt to over 100 kb [37]. NATs are a conserved class of lncRNA molecules [43] that are transcribed from the opposite DNA strand of other RNA transcripts with which they share sequence complementarity [26, 44–46]. Antisense RNAs could potentially exert a regulatory function on their corresponding sense mRNA at different levels [47]. NAT regulatory mechanisms fall into four main categories: mechanisms related to transcription (including epigenetic interactions), RNA–DNA interactions, RNA–RNA interactions in the nucleus and RNA– RNA interactions in the cytoplasm [48]. Among these four mechanisms, RNA-mediated epigenetic modification has received an increasing amount of experimental support. Antisense transcripts can provide a scaffold for effector proteins to interact with DNA and chromatin in a locus specific way.
NATs: cis-acting epigenetic silencers
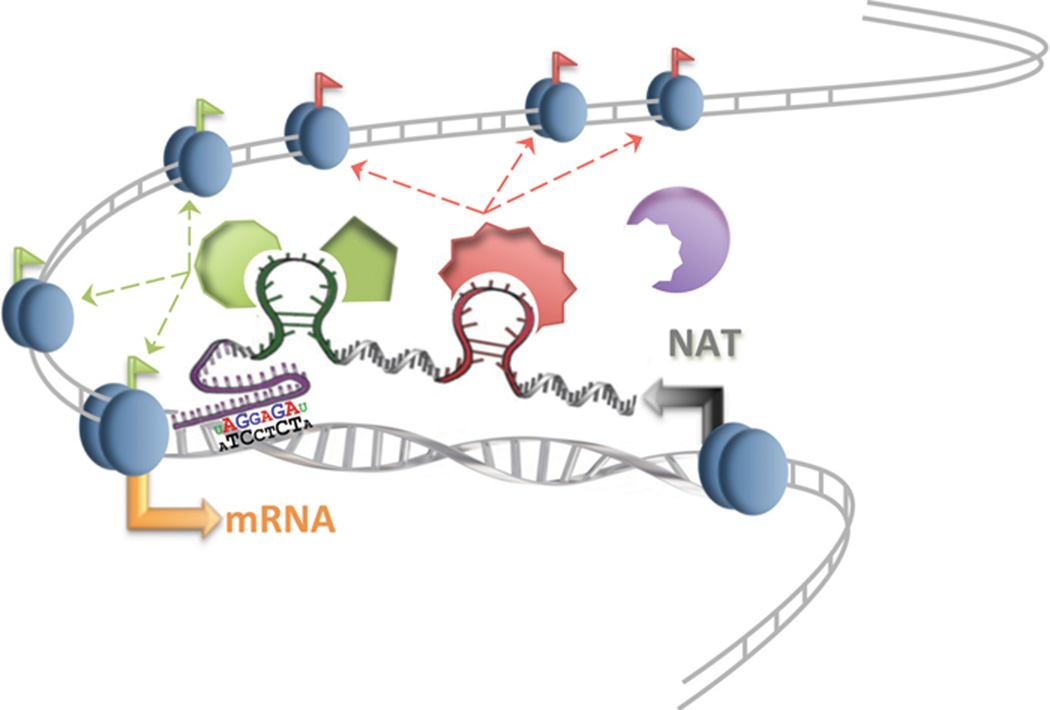
Unlike transcription factors, many histone-modifying enzymes lack specific DNA-binding domains [15]. Based on this important observation, it has been postulated that ncRNAs might interact with ubiquitously expressed histone-modifying enzymes providing the required level of binding specificity (Figure 1).
Figure 1. Epigenetic regulation induced by NATs.
NATs regulate the epigenetic landscape of genomic loci from which they are transcribed (cis regulation). A specific secondary structure permits the NAT to interact with different chromatin-modifying enzymes (green and red shapes), thereby coordinating their action and directing specific epigenetic modifications of the nearby chromatin (green and red flags). Locus specificity may be achieved through sequence-specific interactions between the NAT and the DNA.
In mammalian cells, dosage compensation offered the first characterized examples of antisense lncRNA-mediated chromatin remodeling and gene silencing [49]. One of the two mammalian female X chromosomes is inactivated via an RNA-based mechanism in which the antisense ncRNA Xist, expressed from the X chromosome, mediates the recruitment of polycomb repressive complex 2 (PRC2) that in turn catalyzes the heterochromatinization of the entire X chromosome [21,49].
A similar mechanism of RNA-based epigenetic regulation of gene expression was found to silence various imprinted mammalian alleles. Most imprinted mammalian genes associate in clusters [50], and the presence of NATs is a common feature of these loci [26,51,52]. For example, Air is an imprinted, paternally expressed lncRNA transcribed from the second intron of the mouse insulin-like growth factor 2 receptor (Igf2r) gene [53]. In the mouse placenta, expression of Air induces the epigenetic silencing of both the paternal allele of Igf2r, from which Air is expressed, and neighboring upstream genes. Although the transcription unit of Air only overlaps with Igf2r, Air recognizes and binds to the promoter regions of its neighboring genes. The molecular mechanisms underlying these interactions have not been clarified and might rely on specific secondary structure adopted by Air or on the involvement of mediator proteins. The Air ncRNA interaction with the promoter of upstream genes in the cluster results in the recruitment of the HMT G9a, which generates a repressive chromatin state [56]. The ability of Air to silence non-overlapping genes in cis is reminiscent of Xist-induced X-chromosome inactivation. In the case of Xist, epigenetic silencing spreads through the entire X chromosome, in contrast to the case of imprinted genes, epigenetic silencing spread only to a significant portion of the locus. The extent of the spread of epigenetic silencing may be related to the presence of insulator elements in the DNA sequence and their association with the CCCTC-binding factor (CTCF) [54], a multifunctional protein that enables insulator function and facilitates higher-order chromatin interactions [55].
Another interesting example of imprinting regulation is the antisense ncRNA transcript Kcnq1ot1, which is transcribed from intron 10 of the imprinted gene Kcnq1 [57]. This paternally expressed NAT silences Kcnq1 in cis, as well as neighboring genes on the paternal chromosome, by controlling chromatin and DNA modifications at that locus [58]. Kcnq1ot1 mediates the allele-specific deposition of the repressive histone marks H3K27me3 and H3K9me3 by direct interaction with the PRC2 components Ezh2, Suz12 and the H3K9-specific HMT, G9a [58, 59]. Similar to the situation with Air, the epigenetic changes caused by Kcnq1ot1 occur outside the sequence bound by this lncRNA, emanating bidirectionally from the Kcnq1 locus. Some of the imprinted genes in this cluster, although silenced, lack Kcnq1ot1 enrichment [60].
Based on these examples, cis-acting NATs may remain linked to their transcription loci but exert their regulatory function on the neighboring genes via the recruitment of different proteins and the organization of higher-order chromatin structures. The presence or absence of insulator elements may influence the extension of chromatin alterations in each locus [61]. In this hypothetical scenario, the antisense transcript acts as a scaffold for recruitment of chromatin-modifying enzymes, initiating events that expand in both directions to the entire chromosome, as in the case of X-chromosome inactivation, or to the entire imprinted cluster. In this model, the recruitment of chromatin-modifying complexes is dependent on antisense RNA expression, while the expansion of these effects depends on the subsequent involvement of DNA insulator elements.
Taken together, these imprinting studies imply that a large portion of NATs could exert their regulatory role by binding chromatin enzymes and recruiting them in cis to their targets. In favor of this hypothesis, RNA immunoprecipitation (RIP) experiments targeting Ezh2, coupled with directional RNA sequencing (RIP-seq), revealed that the PRC2 complex associates with almost 10 000 RNAs in mouse embryonic stem cells (mESCs) [62]. Almost 3000 of these RNAs are NATs, and around 1000 are bidirectional transcripts. Interestingly, some NATs linked to disease loci were found to immunoprecipitate with Ezh2, such as Hspa1a–AS, Bgn-AS, Foxn2-AS and Malat1-AS [62], suggesting that ncRNAs target the PRC2 complex to chromatin. Unfortunately, in this study RIP-sequencing data were not integrated with ChIP-sequencing data, and the authors did not investigate the possible overlap between the genomic localization of PRC2 and the immunoprecipitated RNA transcripts. Nevertheless, the presence of NATs associated with PRC2 suggests the importance of these RNA transcripts in mediating the recruitment of chromatin-modifying complexes.
Accumulating evidence implies that the interaction of NATs with EZH2 and other HMTs is more common than previously believed, contributing to the epigenetic regulation of many autosomal loci. In addition to the finding that lncRNAs interact with histone-modifying enzymes, they have also been shown to play a role in DNA methylation. ANRIL is a NAT that overlaps with the INK4b/ARF/INK4a locus [63]. This locus encodes two cyclin-dependent kinase inhibitors, p15INK4b and p16INK4a, and a regulator of the p53 pathway, ARF [64]. The ANRIL transcript also overlaps with several polymorphisms discovered in genome-wide association studies (GWAS) that correspond to increased risk for cardiovascular disease and diabetes [65]. An initial study showed that ANRIL expression inversely correlates with p15INK4b expression in acute lymphoblastic leukemia and acute myeloid leukemia. It was demonstrated that ANRIL mediates the silencing of the tumor suppressor gene p15INK4b via DNA methylation and heterochromatin formation in a Dicer-independent manner, thus excluding the involvement of endogenous small RNAs in the process [20]. Later, it was shown that ANRIL, EZH2 and the PRC1 component CBX7 are upregulated in several prostate cancer tissue specimens with an inverse correlation to the expression of p16INK4a [19]. Moreover, ANRIL physically associates with CBX7 and colocalized with EZH2 and CBX7 to the promoter region of p16INK4a in prostate cancer cells. Thus, the NAT ANRIL participates in the silencing of two very important tumor suppressor genes via two distinct mechanisms, and the alteration of these regulatory circuits has been found in different types of cancer.
Evidence of a functional interaction between NATs and PRC2 comes from a study on the cyclin dependent kinase inhibitor p21, another important tumor suppressor gene. Bidirectional transcription at the p21 locus generates an antisense transcript and p21 mRNA. The p21 NAT represses p21 mRNA in a process involving the deposition of the repressive histone mark H3K27me3 [66]. This mechanism is AGO1-independent, further excluding involvement of endogenous small RNA mediators in the process. Thus, depending on the cellular context, an imbalanced expression of NATs can result in the silencing or activation of partner protein coding genes, providing an interesting potential mechanism to explain the aberrant upregulation or silencing of cancer-related genes.
Among the different body tissues, the brain expresses a high abundance of ncRNAs [67]. Discovered in the developing mouse forebrain, the NAT Evf2 is transcribed from the ultra-conserved Dlx5/6 region encoding the homeodomain transcription factors DLX5 and DLX6 [68]. Evf2 forms a complex with the Dlx-2 homeodomain protein to function as a transcriptional coactivator that increases Dlx5/6 enhancer activity [68]. Recently, studies of an Evf2 loss-of-function mouse revealed more complex regulatory functions of this NAT in the development of GABAergic interneurons [69]. Through antisense interference, Evf2 negatively regulates the expression of Dlx6 mRNA. Moreover, Evf2 exerts a silencing effect on Dlx5 by recruiting DLX and the methyl CpG binding protein 2 (MECP2) to the enhancer region [69]. Mutant Evf2 mice have reduced numbers of GABAergic interneurons in the dentate gyrus of the early postnatal hippocampus and reduced synaptic inhibition in the adult hippocampus [69]. This study highlights the importance of NATs in regulating gene expression during neuronal maturation and raises the possibility of a more extended role of antisense transcripts in central nervous system development.
In recent studies, repeat expansion diseases have often been characterized by bidirectional transcription overlapping the repeat region [70]. Spinocerebellar ataxia type 7 (SCA7) is a neurological disorder associated with a polyglutamine repeat (CAG) expansion in the ataxin−7 gene [71]. SCAANT1 is a 1.4 kb long NAT overlapping the ataxin−7 gene that is actively transcribed upon CTCF binding to target sites flanking the CAG repeat region [72]. SCAANT1 expression is associated with an increased level of the repressive H3K27me3 mark and a decreased level of the activating histone H3 acetylation mark at the ataxin−7 gene promoter. The pathological increase of CAG expansion is accompanied by reduced expression of SCAANT1 ncRNA and increased expression of ataxin−7 mRNA, showing an inverse relationship between the NAT and its partner sense transcript [72]. This study reveals an interesting NAT-based mechanism that is potentially involved in SCA7 pathogenesis.
NATs can silence gene expression in cis, making them attractive therapeutic targets to achieve specific upregulation of gene expression. It has recently been shown that brain-derived neurotrophic factor (BDNF) is under the epigenetic control of an antisense transcript, BDNF-AS [73]. Depletion of BDNF-AS can alter chromatin marks at the BDNF locus and upregulate locus-specific gene expression. This study also described NAT-mediated endogenous gene suppression of glia-derived neurotrophic factor (GDNF) and ephrin B2 receptor (EPHB2), suggesting that antisense RNA-mediated transcriptional suppression is a frequent phenomenon [73]. Considering the frequency with which NATs are transcribed, these examples may represent only the tip of the iceberg, with the regulatory role of NATs in epigenetic modifications representing a more common event than previously imagined.
NATs: cis-acting epigenetic activators
The first observation that lncRNAs are involved in epigenetic gene activation stems from dosage compensation studies in Drosophila, where the imbalanced presence of X chromosomes in the sexes necessitates compensation by a twofold upregulation of all the genes on the single male X chromosome [74]. Two lncRNAs, roX1 and roX2, play a fundamental role in the correct targeting of the Dosage Compensation Complex to many different binding sites on the male X chromosome, which results in transcriptional upregulation. These and other examples provide accumulating evidence of a central role for NATs in the epigenetic activation of specific loci on a genome-wide basis, providing insight into the biological language of lncRNAs [75].
Following these initial findings in Drosophila, several other examples of ncRNAs in vertebrates have been reported. Among these, a ncRNA-expression profile study of mESC differentiation identified several ncRNAs associated with important mESC protein coding genes [30]. Among these ncRNAs, two concordantly upregulated NATs colocalized with their sense mRNA partners during a specific step of mESC differentiation. The NATs, named Evx1as and Hoxb5/6as, are transcribed from the opposite DNA strand of Evx1 and Hoxb5/6, respectively [30]. Using RNA-ChIP, the authors found that these NATs immunoprecipitate with H3K4me3, demonstrating a physical interaction with a transcriptional activation mark [30]. Furthermore, RNA-IP experiments showed direct interaction between Evx1as and Hoxb5/6as with MLL1, the mammalian trithorax protein responsible for H3K4me3 in the promoter region of several developmental genes [30]. This finding raise the possibility that these NATs are involved in the epigenetic activation of their mRNA partners during differentiation.
In another example of epigenetic activation, the chromatin-associated ncRNA transcript termed Intergenic10, located in the region 3’ to FANK1 in the opposite orientation, overlaps with the protein-coding gene ADAM12 [18]. The expression of Intergenic10 positively correlates with the expression of the neighboring protein coding genes. siRNA depletion of Intergenic10 resulted in the concordant downregulation of ADAM12 and FANK1 and a decrease in the levels of the active chromatin mark H3K4me2 in the promoter regions of the downregulated genes [18]. NATs may bind and recruit in cis chromatin-modifying enzymes to establish a locus-specific transcriptionally active chromatin state.
Taken together, these observations show that a chromatin-associated ncRNA can act as a chromatin remodeler in cis to positively or negatively regulate the expression of neighboring genes.
LncRNAs: trans-acting chromatin remodelers
Controversy still exists regarding the functional significance of many long and short ncRNA transcripts that are pervasively transcribed in the human genome and particularly those originating in the proximity of the transcriptional start sites (TSSs) of many active genes. However, cell-, tissue- and developmental-specific transcription of lncRNAs argues against the simplistic assumption that these arise from transcriptional noise. Moreover, removal of these ncRNAs often correlates with functional consequences. Aside from NATs, the human genome produces many other classes of lncRNAs. For example, the analysis of chromatin signatures revealed a family of over one thousand highly conserved lncRNAs, termed large intergenic non-coding RNAs (lincRNAs), that contain sense and antisense members with many potential regulatory functions [41]. RNA-IP experiments of the PRC2 complex component EZH2 followed by hybridization to a custom exon-tiling array for 900 human lincRNAs showed that almost 30% of expressed lincRNAs physically interact with PRC2 [76]. Immunoprecipitation of lncRNAs with EZH2 is highly suggestive of functional roles of these transcripts through the PRC2 pathway. The catalog of lincRNAs encoded in the human genome as well as the understanding of their roles in mediating the function of chromatin modifying complexes is rapidly expanding.
Unlike most NATs, lincRNAs exert their regulatory roles in trans to alter chromatin shape and gene expression at distant loci. HOTAIR is a lincRNA encoded in antisense orientation in the HOX-C cluster on chromosome 12 that is necessary for the correct expression of the HOX-D cluster of genes on chromosome 2 [23]. HOTAIR associates with the PRC2 complex to silence and maintain a large domain of heterochromatin in the HOX-D gene cluster. Genomic regions flanking HOX-D contain high levels of H3K27me3 and low levels of H3K4me2/3 [77]. It was shown in several cellular systems that HOTAIR acts as a modular scaffold for the recruitment of both PRC2 and LSD1, the catalytic subunit of the repressor complex CoREST/REST, which in turn coordinate the methylation of H3K27me3 and demethylation of H3K4me2/3, respectively, in trans at many different target genomic regions [78]. Interestingly, altered HOTAIR expression in primary breast tumors is a powerful predictor of metastasis and poor prognosis [35]. Inhibition of HOTAIR expression in cancer cells reduces invasiveness and metastatic potential, consistent with its physiological function in dictating chromatin states of fibroblast during development [35].
A loss-of-function study in mESCs produced a functional characterization of a large number of lincRNAs. [32]. It was shown that lincRNAs maintain the pluripotent state and repress lineage programs in mESCs by trans-acting mechanisms of global gene expression regulation. mESCs lincRNAs associate with 12 different chromatin complexes involved in different aspects of epigenetic regulation, such as writers (Tip60/P400, Prc2, Setd8, Eset, Suv39), readers (Prc1, Cbx1, Cbx3) and erasers (Jarid1b, Jarid1c, Hdac1) [32]. Seventy-four lincRNAs associate with at least one of these complexes and several lincRNAs associate with functionally related chromatin complexes [32]. Because lincRNAs physically associate with multiple chromatin regulatory proteins, they may serve as scaffolds to bridge together similar complexes into larger functional units.
Like NATs, trans-acting lncRNAs can be involved in the epigenetic activation of specific loci. HOTTIP is a spliced, polyadenylated lncRNA transcribed in the opposite orientation from the 5’ end of the HOXA locus [79]. HOTTIP knockdown in fibroblasts and chick embryos resulted in decreased HOXA expression, affecting a region 40kb downstream of the 5’ end of the HOXA locus. This repressive effect depends on the distance from the HOTTIP gene; genes in close proximity exhibit a greater decrease in expression levels [79]. These changes in gene expression correlated with a global loss of H3K4me3 and H3K4me2 across the affected region. RIP experiments demonstrated direct binding of HOTTIP with WDR5, a component of the core complex responsible for H3K4 methylation [79]. Ectopically expressed HOTTIP does not induce the expression of 5’ HOXA genes in fibroblast cells, implying a cis mechanism of action for HOTTIP. Artificial recruitment of HOTTIP RNA upstream of a silent GAL4 promoter can boost transcription in the presence of WDR5, confirming the cis effect of the HOTTIP transcript in the proximity of the target genes [79].
Mechanisms of lncRNA interactions with chromatin and chromatin-modifying enzymes
The ability of lncRNAs to function as scaffolds for the recruitment of different yet functionally related enzymes and to confer locus specificity to these enzymes raises two immediate questions:what mediates the interactions between ncRNAs and specific chromatin enzymes, and what is the language of molecular rules governing them? One of the first hints of a mechanism governing ncRNA-enzyme interactions came from studies of the X-chromosome inactivation phenomenon. It was shown that a novel ncRNA called Repeat A (RepA) directly binds to EZH2 and functions in the recruitment of PRC2 to the X chromosome [21]. RepA is a 1.6 kb ncRNA transcribed within Xist and is composed of 7.5 tandem repeat sequences that fold into two conserved stem-loop structures crucial for EZH2 binding [21]. These initial findings were subsequently confirmed by an independent study showing that short RNAs of 50 to 200nts in length are transcribed from the 5’end of polycomb target genes [80]. Interestingly, these short RNAs have stem-loop structures similar to RepA and are able to bind the PRC2 component SUZ12 [80]. Similarly, the antisense Kcnq1ot1 has a conserved RNA repeat that was shown to be necessary for the epigenetic silencing of imprinted genes [60]. These studies imply that lncRNAs assume specific secondary structures offering different docking sites for different enzymes.
In large part, how NATs bind to target genes to guide chromatin-modifying enzymes to specific loci still remains unexplained (Figure 2). Two recently developed methods for profiling the genome-wide occupancy of lncRNAs revealed the high-throughput identification of RNA-DNA and RNA-protein interactions [81, 82]. The application of these new techniques may represent a promising tool to explore the mechanisms governing ncRNA-chromatin interactions, as shown by the informative analysis performed on a few known lncRNAs (roX2, TERC and HOTAIR) [82]. Interestingly, among the discovered DNA binding sites of both rox2 and TERC, specific consensus DNA sequences have been observed, thus suggesting that specific DNA motifs might be important for the recruitment of these and other lncRNAs to their target genomic loci. HOTAIR binding sites contain a GA-rich polypurine motif, reminiscent of mammalian Polycomb response elements. It is notable that although the HOTAIR binding sites overlap with PRC2 and H3K27me3 chromatin regions, they are restricted to small regions of a few hundred bp, raising the possibility that HOTAIR nucleates PRC2 binding and H3K27me3 spreading [82]. Together these data, and the discovery that HOTAIR binding to its genomic targets does not require EZH2, demonstrate that ncRNAs are required for specific recognition of DNA sequences as well as recruitment of polycomb proteins, which in turn modify the neighboring chromatin. This study demonstrates that locus-specific interaction between ncRNAs and chromatin takes place independently from ncRNA-enzyme interaction and pointed out the existence of specific RNA-targeting motif among ncRNA target sites. These motifs may represent binding sites for structural elements within the ncRNA, in case of direct RNA-DNA interaction, or may function as the binding site for mediator proteins that may induce HOTAIR recruitment.
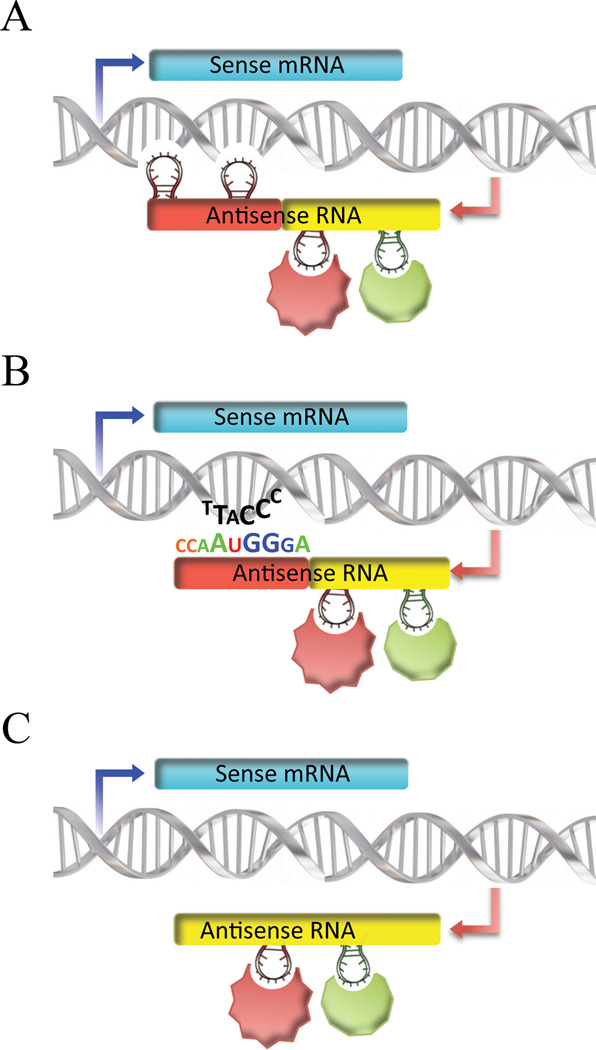
Figure 2. Molecular mechanisms of NATs and chromatin interactions.
Two types of interactions are necessary for any ncRNA-induced chromatin modification to happen: between an antisense RNA molecule and a chromatin-modifying enzyme (CME) and between either a CME and DNA or antisense RNA and DNA. The second type of interaction is necessary to confer sequence specificity to the chromatin modifications. Each one of these interactions (RNA-protein, RNA-DNA or DNA-protein) can either happen through sequence motifs (digital Watson-Crick base pairing) or by RNA secondary structure. NATs function as intermediates that target CMEs to locus-specific regions of the genome. The molecular mechanisms governing the interaction between NATs and chromatin remain poorly characterized. Here, we propose three different possible scenarios by which this interaction occurs:
(a) Specific binding of antisense RNA to a CME as well as to a DNA region by forming a unique secondary structure.
(b) The sequence motif dictates the interaction between the antisense RNA molecule and the target DNA. In this model, antisense RNA binds specifically to CMEs and to a particular DNA region
(c) Nonspecific binding of antisense RNA to a DNA sequence. In this model, local antisense transcription leads to a specific chromatin modification. The specificity in this model comes from the promoter of antisense RNA and the fact that transcription will lead to particular modifications. NATs do not physically associate with the chromatin. In this case, locus-specificity is achieved by nascent NATs that are recognized by chromatin-modifying enzymes.
Concluding remarks
While the examples of NAT and lncRNA mechanisms described above suggest a broad continuum of function for ncRNAs in epigenetic regulation, the exact roles and mechanisms of most of these molecules remain largely unknown. NATs have emerged as powerful transducers of biological information, primarily due to their ability to bridge the interaction between proteins and DNA [83]. The information content and structural features of these ncRNAs collectively establish a dynamic interface with other macromolecules, [83] thus facilitating the formation and modulation of ribonucleoprotein complexes critical for epigenetic signaling. These unique features permit NATs and other lncRNAs to function as scaffolds to regulate epigenetic mechanisms within the cell. The key to future studies of lncRNAs will be to successfully integrate the layers of knowledge gained from multiple genomic, transcriptomic, proteomic and epigenomic approaches in order to create a multidimensional understanding of NATs within the existing cellular framework [84].
Acknowledgments
The authors would like to thank Dr. Chiara Pastori and Roya Pedram Fatemi for helpful discussions and critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chi P, et al. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nature reviews. Cancer. 2010;10:457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daniel JA, et al. Effector proteins for methylated histones: an expanding family. Cell Cycle. 2005;4:919–926. doi: 10.4161/cc.4.7.1824. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 4.Shi Y, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 5.Maurer-Stroh S, et al. The Tudor domain 'Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends in biochemical sciences. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- 6.Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. doi: 10.1016/j.cell.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 8.van Steensel B. Chromatin: constructing the big picture. Embo J. 2011;30:1885–1895. doi: 10.1038/emboj.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schubeler D. Chromatin in multicolor. Cell. 2010;143:183–184. doi: 10.1016/j.cell.2010.09.045. [DOI] [PubMed] [Google Scholar]
- 10.Filion GJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kharchenko PV, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471:480–485. doi: 10.1038/nature09725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstein MB, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonasio R, et al. Molecular signals of epigenetic states. Science. 2010;330:612–616. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E, Allis CD. RNA meets chromatin. Genes Dev. 2005;19:1635–1655. doi: 10.1101/gad.1324305. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Campos A, Azorin F. RNA is an integral component of chromatin that contributes to its structural organization. PLoS One. 2007;2 doi: 10.1371/journal.pone.0001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maison C, et al. Higher-order structure in pericentric heterochromatin involves a distinct pattern of histone modification and an RNA component. Nat Genet. 2002;30:329–334. doi: 10.1038/ng843. [DOI] [PubMed] [Google Scholar]
- 18.Mondal T, et al. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yap KL, et al. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–674. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu W, et al. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao J, et al. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martianov I, et al. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 23.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bierhoff H, et al. Noncoding transcripts in sense and antisense orientation regulate the epigenetic state of ribosomal RNA genes. Cold Spring Harb Symp Quant Biol. 2010;75:357–364. doi: 10.1101/sqb.2010.75.060. [DOI] [PubMed] [Google Scholar]
- 25.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katayama S, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 27.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 28.Birney E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clark MB, et al. The reality of pervasive transcription. PLoS Biol. 2011;9:e1000625. doi: 10.1371/journal.pbio.1000625. discussion e1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinger ME, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahfeldt T, et al. Programming human pluripotent stem cells into white and brown adipocytes. Nat Cell Biol. 2012;14:209–219. doi: 10.1038/ncb2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guttman M, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ji P, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 34.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright MW, Bruford EA. Naming 'junk': human non-protein coding RNA (ncRNA) gene nomenclature. Hum Genomics. 2011;5:90–98. doi: 10.1186/1479-7364-5-2-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapranov P, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 38.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttman M, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakaya HI, et al. Genome mapping and expression analyses of human intronic noncoding RNAs reveal tissue-specific patterns and enrichment in genes related to regulation of transcription. Genome Biol. 2007;8:R43. doi: 10.1186/gb-2007-8-3-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun M, et al. Evidence for variation in abundance of antisense transcripts between multicellular animals but no relationship between antisense transcriptionand organismic complexity. Genome Res. 2006;16:922–933. doi: 10.1101/gr.5210006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiyosawa H, et al. Antisense transcripts with FANTOM2 clone set and their implications for gene regulation. Genome Res. 2003;13:1324–1334. doi: 10.1101/gr.982903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen J, et al. Human antisense genes have unusually short introns: evidence for selection for rapid transcription. Trends Genet. 2005;21:203–207. doi: 10.1016/j.tig.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 46.Chen J, et al. Genome-wide analysis of coordinate expression and evolution of human cis-encoded sense-antisense transcripts. Trends Genet. 2005;21:326–329. doi: 10.1016/j.tig.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Lapidot M, Pilpel Y. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep. 2006;7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faghihi MA, Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee JT, et al. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat Genet. 1999;21:400–404. doi: 10.1038/7734. [DOI] [PubMed] [Google Scholar]
- 50.Verona RI, et al. Genomic imprinting: intricacies of epigenetic regulation in clusters. Annu Rev Cell Dev Biol. 2003;19:237–259. doi: 10.1146/annurev.cellbio.19.111401.092717. [DOI] [PubMed] [Google Scholar]
- 51.Mohammad F, et al. Epigenetics of imprinted long noncoding RNAs. Epigenetics. 2009;4:277–286. [PubMed] [Google Scholar]
- 52.Wan LB, Bartolomei MS. Regulation of imprinting in clusters: noncoding RNAs versus insulators. Adv Genet. 2008;61:207–223. doi: 10.1016/S0065-2660(07)00007-7. [DOI] [PubMed] [Google Scholar]
- 53.Sleutels F, et al. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 54.Kim TH, et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 56.Nagano T, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- 57.Smilinich NJ, et al. A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith-Wiedemann syndrome. Proc Natl Acad Sci U S A. 1999;96:8064–8069. doi: 10.1073/pnas.96.14.8064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 59.Terranova R, et al. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Developmental cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 60.Kanduri C. Kcnq1ot1: a chromatin regulatory RNA. Semin Cell Dev Biol. 2011;22:343–350. doi: 10.1016/j.semcdb.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 61.Ghirlando R, et al. Chromatin domains, insulators, and the regulation of gene expression. Biochim Biophys Acta. 2012 doi: 10.1016/j.bbagrm.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao J, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40:939–953. doi: 10.1016/j.molcel.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pasmant E, et al. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007;67:3963–3969. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 64.Popov N, Gil J. Epigenetic regulation of the INK4b-ARF-INK4a locus: in sickness and in health. Epigenetics. 2010;5:685–690. doi: 10.4161/epi.5.8.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pasmant E, et al. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2011;25:444–448. doi: 10.1096/fj.10-172452. [DOI] [PubMed] [Google Scholar]
- 66.Morris KV, et al. Bidirectional transcription directs both transcriptional gene activation and suppression in human cells. PLoS Genet. 2008;4:e1000258. doi: 10.1371/journal.pgen.1000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qureshi IA, et al. Long non-coding RNAs in nervous system function and disease. Brain Res. 2010;1338:20–35. doi: 10.1016/j.brainres.2010.03.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Feng J, et al. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bond AM, et al. Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci. 2009;12:1020–1027. doi: 10.1038/nn.2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Batra R, et al. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum Mol Genet. 2010;19:R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Martin JJ. Vinken PJ, Bruyn GW, editors. Spinocerebellar ataxia type 7. Handbook of clinical neurology / edited by P.J. Vinken and G.W. Bruyn. 2012;103:475–491. doi: 10.1016/B978-0-444-51892-7.00030-9. [DOI] [PubMed] [Google Scholar]
- 72.Sopher BL, et al. CTCF regulates ataxin-7 expression through promotion of a convergently transcribed, antisense noncoding RNA. Neuron. 2011;70:1071–1084. doi: 10.1016/j.neuron.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Modarresi F, et al. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat Biotechnol. 2012 doi: 10.1038/nbt.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Straub T, Becker PB. Transcription modulation chromosome-wide: universal features and principles of dosage compensation in worms and flies. Curr Opin Genet Dev. 2011;21:147–153. doi: 10.1016/j.gde.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 75.Ilik I, Akhtar A. roX RNAs: non-coding regulators of the male X chromosome in flies. RNA biology. 2009;6:113–121. doi: 10.4161/rna.6.2.8060. [DOI] [PubMed] [Google Scholar]
- 76.Khalil AM, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fanti L, et al. The trithorax group and Pc group proteins are differentially involved in heterochromatin formation in Drosophila. Chromosoma. 2008;117:25–39. doi: 10.1007/s00412-007-0123-7. [DOI] [PubMed] [Google Scholar]
- 78.Tsai MC, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kanhere A, et al. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675–688. doi: 10.1016/j.molcel.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Simon MD, et al. The genomic binding sites of a noncoding RNA. Proc Natl Acad Sci U S A. 2011;108:20497–20502. doi: 10.1073/pnas.1113536108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chu C, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44:667–678. doi: 10.1016/j.molcel.2011.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.St Laurent G, 3rd, Wahlestedt C. Noncoding RNAs: couplers of analog and digital information in nervous system function? Trends Neurosci. 2007;30:612–621. doi: 10.1016/j.tins.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 84.Hawkins RD, et al. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11:476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]