Abstract
Despite their rigidity, microtubules in living cells bend significantly during polymerization resulting in greater curvature than can be explained by thermal forces alone. However, the source of the non-thermal forces that bend growing microtubules remains obscure. We analyzed the motion of microtubule tips in NIH-3T3 fibroblasts expressing EGFP-EB1, a fluorescent +TIP protein that specifically binds to the growing ends of microtubules. We found that dynein inhibition significantly reduced the deviation of the growing tip from its initial trajectory. Inhibiting myosin modestly reduced tip fluctuations, while simultaneous myosin and dynein inhibition caused no further decrease in fluctuations compared to dynein inhibition alone. Our results can be interpreted with a model in which dynein linkages play a key role in generating and transmitting fluctuating forces that bend growing microtubules.
Key Terms: Microtubule bending, dynein, myosin
Introduction
Microtubules play a central role in cell polarization28, migration8, intracellular trafficking6, cell division22 and nuclear positioning26. In vitro experiments show that microtubules subjected to thermal forces have a persistence length on the order of millimeters10, yet in vivo they exhibit bends on micron length scales. These bends develop primarily by deflections from linear tip trajectories rather than subsequent bending of intact microtubules5, although initial bends may be amplified by compressive forces once the tip reaches the cell periphery4 or by the activity of cytoplasmic molecular motors32.
Thermal forces are too small to explain the deflections of growing microtubules in living cells5, and the causes of microtubule bending before the tips reach the cell periphery remains obscure. Growing microtubules can bend due to compressive stresses generated by polymerization against a barrier2,4,7. Furthermore, the activity of myosin motors drives fluctuations in the cytomatrix, causing bending of existing microtubules in vivo3; myosin forces could bend growing microtubules by a similar mechanism. Processive microtubule motors such as dynein or kinesin can exert forces on the microtubule if they are anchored to the cytomatrix at the cargo-binding end2,12,25,32. As the motor walks along the microtubule it pulls on the anchor point, resulting in a tensile force between the anchor and the point of attachment to the microtubule. Odde and coworkers2 have suggested that the anterograde motion of buckling microtubules (moving towards the cell periphery) can be explained by microtubule-bound motor activity. In laser severing experiments with single microtubules in endothelial cells, newly created minus-ended microtubules bend (instead of straightening) in a dynein dependent manner32, consistent with a model for dynein pulling forces along the entire length of the microtubule. This model also explains amplified buckles at the cell periphery and how the centrosome can be centered by an astral array of microtubules32.
Given the athermal nature of forces that deflect growing microtubule tips5, we reasoned that motor activity may also be responsible for bending growing microtubules. Using in vivo experiments, we show that bending of the trajectories of growing microtubule tips is significantly decreased in dynein-inhibited cells. Myosin-inhibition also decreases bends while kinesin-inhibition has no effect. Simultaneous inhibition of myosin and dynein in cells does not have any additional reduction in bending beyond that by inhibition of dynein alone. We interpret these results with a mathematical model in which dynein linkages generate and transmit fluctuating forces that bend growing microtubules.
Materials and Methods
Cell culture, transfection and inhibition experiments
NIH-3T3 fibroblasts were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Mediatech, Manassas, VA) with 10% Donor Bovine Serum (DBS) (Gibco, Grand Island, NY). The cells were maintained at 37 °C in humidified 5% CO2. For microscopy the cells were plated on 35 mm glass-bottomed dishes (WPI, Sarasota, FL) and allowed to spread overnight at 37° C and 5% CO2. The glass-bottomed dishes were coated with 5 μg/ml fibronection (BD Biocoat™, Franklin Lakes, NJ) and kept at 4° C overnight before cell seeding.
In control experiments, cells were transiently co-transfected with pGFP-EB1 (Addgene plasmid 17234) and DsRed and were incubated for 18–24 hours prior to plating. DsRed was expressed to allow comparisons with cells expressing fluorescently labeled proteins such as DsRed-CC1. In dynein inhibition experiments, cells were co-transfected with DsRed-CC1 (Fig. S1) and with mCherry-KHC in kinesin-1 inhibition experiments (Fig. S1). DsRed-CC1 renders dynein inactive by competitively binding to it and preventing dynein interactions with dynactin31,32 while mCherry-KHC can inhibit kinesin-1 by multiple mechanisms23. Transient transfection of plasmids into cells was performed with Lipofectamine™ 2000 transfection reagent (Life Technologies, Invitrogen, Carlsbad, CA). Some cells were treated for 30 minutes with 10 μM Y27632, which is a Rho-kinase (ROCK) inhibitor that causes substantial inhibition of non-muscle myosin16,18.
Confocal Microscopy
The cells were imaged on a Leica SP5 DM6000 confocal microscope equipped with a 63X oil immersion objective. During microscopy, cells were maintained at 37 °C in a temperature, CO2 and humidity controlled environmental chamber. In order to image EGFP-EB1, a 488 nm laser with 10% power and an appropriate GFP bandpass filter was used. Images were taken at a resolution of 1024×1024 and with a speed of 400 Hz, at a rate of 3 seconds/frame. The images were further analyzed using LAS AF Lite (Leica Systems) software. For dynein and kinesin-1 inhibition studies, expression of DsRed-CC1 and mCherry-KHC was confirmed using epifluorescence microscopy.
Trajectory Analysis
Microtubule trajectories were constructed from an analysis of EB1 movies using plusTipTracker, a MATLAB based open source software package that combines automated tracking, data analysis and visualization tools for analysis of movies of fluorescently labeled microtubule plus end binding proteins (+TIPs)1. The software detects EB1 comets by application of locally optimal thresholds using a watershed-based technique. The track reconstruction is described in more detail elsewhere14.
To test the accuracy of the software for our experiments, we first measured the positional error in the measurements. Fixed NIH-3T3 cells expressing EGFP-EB1 were imaged for two minutes at three-second intervals. Since the position of the tips is fixed, the variation in the +TIP detection by the software provides an estimate of the positional error. The deviation from the mean was calculated for about eight tips. Fig. S2 shows the plot of the frequency versus the deviation from the mean. Fitting a normal distribution to the data yielded a standard deviation of 0.06 μm while the average distance travelled by the tips in between successive frames was 1.1 μm. Thus there is a 6% error in the measurements, which was deemed to be small enough to enable reliable tracking of the tips.
To reduce uncertainty due to positioning error in the trajectory analysis, the initial direction of the trajectory was determined by fitting a line to the first four points in the trajectory. Each trajectory was first translated to the origin and rotated onto the positive x-axis by an angle that was determined by the slope of the fitted line. The rotated trajectories thus had the same initial direction. To quantitatively investigate microtubule curvature, we next performed a Fourier decomposition of the reconstructed trajectories10, with a trajectory length of 8 μm. The tangent angle, θ = tan−1(dy/dx), was decomposed as a sum of sine waves,
| 1) |
We assume that the microtubules are clamped at the centrosome (i.e. a fixed orientation), so that the boundary condition at the origin, θ(0)= 0 is automatically imposed by the basis functions. From a set of N coordinates (xk,yk) making up a trajectory, the length Δsk and tangent angles θk of each of the (N−1) segments that connect the coordinates were calculated:
| 2) |
| 3) |
The amplitudes were determined by taking an approximate Fourier transform of Eq. 1)
| 4) |
where
| 5) |
and
| 6) |
Model for dynein force generation
Motor forces are included as linkages distributed along the length of the microtubule, as illustrated in Fig. 3a. We imagine that a particular segment of the microtubule is occasionally captured by a cytoskeletal bound dynein motor and bound for times of the order of , where we take the dynein off rate to be of the order of 1 s−1 32. A segment of length h will capture a dynein motor in a time step δt with a probability p = nhkoffδt where n is the (linear) density of dynein motors per unit length of the microtubule, estimated at about two cytoskeletal-bound motors per micron32. On average the binding and unbinding rates will balance at the desired motor density n if each bound motor unbinds with a probability koffδt. Note that these formulas assume that the binding and unbinding probabilities are small, koffδt ≪ 1; more general formulas can be derived in cases where koffδt is not small. The motor model resembles that proposed by Nedelec and Foethke24; the main difference is that in our implementation the motors only exist while bound to the microtubule instead of being permanently bound to random positions in the cytomatrix. It seems unlikely that this will cause any substantial difference in the simulations.
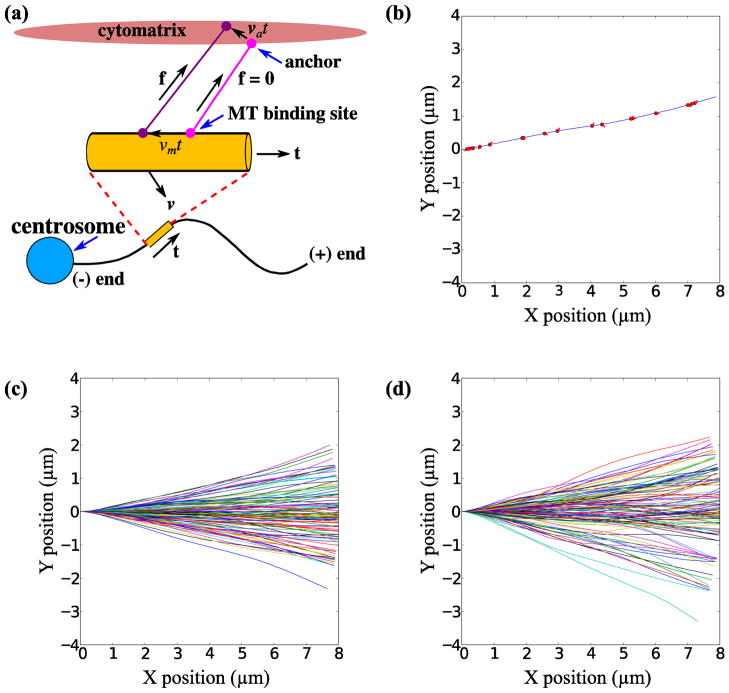
FIGURE 3.
Numerical simulations of microtubule bending by dynein and myosin. The model for dynein and myosin force generation is illustrated in (a). A dynein-cytomatrix linkage is formed in a force-free state at time t = 0; the light mauve circles indicate the points of attachment to the microtubule and cytomatrix. As the motor walks along the microtubule, shown by the dark mauve circle, the linkage is extended and a force is exerted in the direction shown by the black arrow. In addition the anchor point in the cytomatrix may also be moved by myosin activity. The total force in the linkage is a combination of dynein and myosin activity. A simulation of filament growth is illustrated in (b), which shows the configuration of a microtubule polymerized to a contour length of 8 μm. The red circles are the positions of the dynein motors and the red lines indicate the dynein-cytomatrix linkage. (c) and (d) show the trajectories of 100 different samples (in each case) under the action of dynein forces (stationary anchor points) and dynein plus myosin forces (moving anchors) respectively.
A model for dynein force generation was constructed based on the following simplifying assumptions that are nevertheless consistent with what is known about dynein-force generation: 1) The dynein-cytoskeletal linkage is formed in a force-free state, with a vector bi linking the point of attachment to the microtubule, xi with the anchor point in the cytomatrix, ai 2) The dynein motor walks towards the microtubule minus end, with a force-dependent speed δt until the tangential force reaches the stall force fmax or the motor unbinds. The motor is displaced from its initial position both by walking along the microtubule and from the motion of the microtubule itself,
| 7) |
where v is the velocity of the microtubule segment. The motor walks with a speed vm,i in the direction of the local tangent to the microtubule t, which is formed by assuming the filament is made up of straight segments connecting the nodes. We assume a linear force velocity relation varying from a maximum speed in the force-free state, vmax, to zero at the stall force, fmax. The coordinate si, which measures the position of the motor along the contour length of the microtubule, is incremented by vm,iδt at each time step, and the motor position is then determined by interpolating from the neighboring nodes. The velocity of the segment is found by interpolating the nodal velocities. 3) The initial distribution of directions of the microtubule-cytomatrix linkage, b, is isotropic with the anchor points initialized as ai(0) = xi(0)+bi. To model the action of the myosin network, the anchor points ai are moved with random velocities, which represents the motor-induced motions of the cytomatrix. Each anchor point is assumed to move with a constant velocity
| 8) |
for as long as the motor remains bound.
Under these assumptions, both the displacement of the motor from its binding and the motion of the anchor point can cause an extension of the dynein linkage, ri(t) = ai(t)−xi(t), with a linear force along the linkage (Fig. 3A)
| 9) |
It is possible for the motor to exert a compressive force on the dynein-cytomatrix linkage if b·t < 0. However this is always transient since eventually the motor will walk to a position such that b·t > 0.
Equation of motion
The strain energy in a bent elastic filament of length L (free from external couples) can be written as a line integral of the mean-square curvature17,
| 10) |
where s is the distance along the filament arc, and B is the bending modulus. If we discretize the filament into N segments of uniform length h = L/N, then a second-order approximation to the strain energy is given by20,21
| 11) |
where the curvature Cn is related to the angle between adjacent segments, . The discrete approximation to H can then be differentiated to find the elastic force on each node of the filament. In addition to the elastic forces, there is also a constraint force that is required to maintain the length of the individual segments, rn,n−1·rn,n−1 = h2 20, 21.
In the overdamped limit, the nodal velocities vn are obtained from the force balance:
| 12) |
where fn is the total force on node n from the motors on neighboring segments. The friction coefficient γ is taken to represent the background friction of the cellular matrix; we take value of 10Pa s, or 10 N m−2 s, based on measurements of microtubule relaxation in dynein-inhibited cells32.
Results
Fluctuations in growing microtubule tips
We tracked the position of EGFP-EB1 labeled microtubule tips in cells using plusTipTracker and microtubule trajectories were reconstructed from the measured tip positions. The tips were tracked in a region that encircled the centrosome, but not closer than 5 μm to the cell periphery. This was done to exclude events where the growing tip might experience compressive loading as it grew against the membrane. To avoid spurious tracks, each trajectory was visually confirmed to be correctly detected and trajectories that weren’t reconstructions of the growth of a single tip were discarded. A minimum of 12–13 cells were imaged and 300–600 trajectories per condition were quantified.
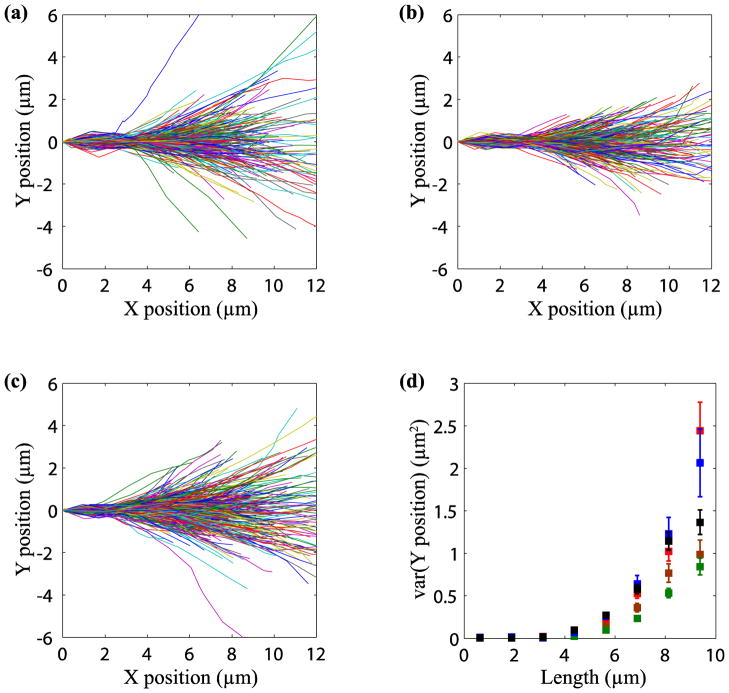
Figure 1a shows the trajectory plots for growing microtubules in normal cells. All trajectories were translated to the origin and rotated so that the initial direction was along the positive x-axis, as described in materials and methods. In control cells that expressed DsRed the trajectories were observed to spread out significantly over tens of seconds.
FIGURE 1.
Motor activity causes bending during microtubule growth. Trajectories of growing microtubules under different conditions were reconstructed using plusTipTracker. Plots for growing microtubule trajectories are shown for (a) control cells, (b) dynein-inhibited cells and (c) myosin-inhibited cells. In comparison to control cells, inhibition of dynein and myosin reduces the spread of the trajectories. Panel (d) shows a comparison of the length dependent variance under different conditions: control (red), kinesin inhibited (blue), myosin inhibited (black), dynein inhibited (green) and dynein and myosin inhibited (brown). Errors bars indicate the standard error in the mean (SEM). About 12 cells and 300–600 trajectories were analyzed for each case.
Motor activity bends growing microtubules
We next over-expressed DsRed-CC1 in NIH-3T3 fibroblasts to competitively inhibit dynein31,32. The spread in the trajectories was less in dynein-inhibited cells compared to control cells (compare Figs. 1a and b). When kinesin was inhibited by expressing mCherry-KHC 23, the spread was similar to the control cells (Fig. S3).
We next attempted to inhibit non-muscle myosin activity with blebbistatin, a specific myosin II inhibitor15. However, at doses that inhibited myosin activity, blebbistatin treatment caused a significant rounding of cells making it difficult to track EB1 tips reliably in these cells. We therefore treated cells with Y27632, a ROCK inhibitor that has been shown to cause a large decrease in non-muscle myosin activity without causing cell rounding16,18. The spread of the trajectories on Y27632 treatment is shown in Fig. 1c and appears to be decreased when compared to control cells, but to a lesser extent than in dynein-inhibited cells (Fig. 1b).
As all trajectories start at the origin and are initially oriented along the positive x-axis, any deviation of the trajectories in the y-direction is a measure of the spread. One method of quantifying the effects of motor inhibition on trajectory spread is to compare the variance of the y positions of the tips at different times. However, modest but statistically significant differences in tip speed were observed when dynein, kinesin, or ROCK was inhibited (Fig. S4). Owing to the differences in the polymerization velocity, we instead chose to examine the length-dependence of the variance at each condition (Fig. 1d). The variance in dynein-inhibited cells is decreased by a factor of two in comparison with control cells. There is a smaller decrease in variance in ROCK inhibited cells, but there is no statistically significant difference between kinesin-1 inhibited cells and control cells.
Fourier mode analysis of tip trajectories
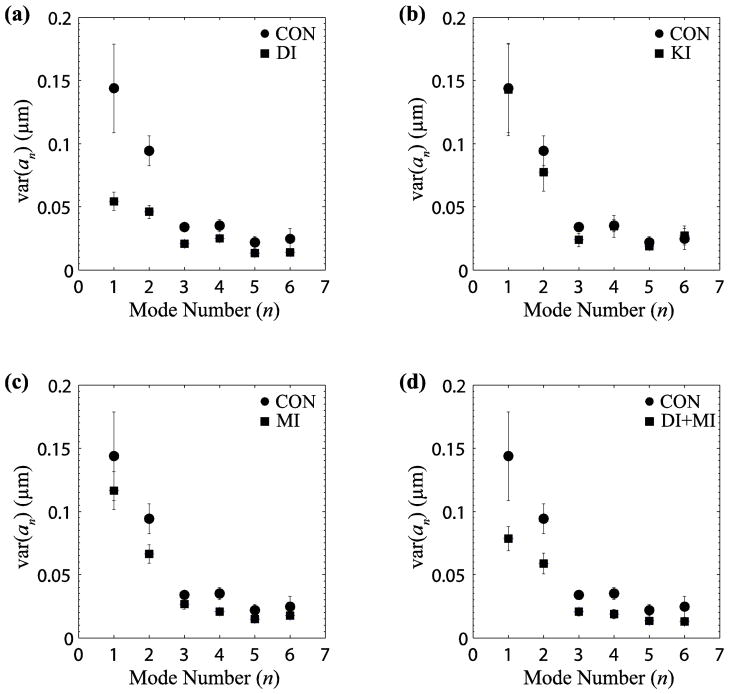
To further characterize the deflections of polymerizing microtubules, we performed a Fourier mode analysis (described in the materials and methods) on the measured trajectories. A comparison of the Fourier mode amplitudes for different experimental conditions is shown in Fig 2. Shape fluctuations in control cells were significantly higher than those in dynein-inhibited cells at almost all mode numbers (p < 0.05 for mode 1 by Student’s T-test), indicating that dynein linkages contribute significantly to the bending of microtubule trajectories (Fig. 2a). In contrast, trajectories in cells where only kinesin was inhibited showed no discernible differences from the control cells (Fig. 2b). Inhibition of myosin only reduced the mean-square amplitudes at some wavelengths, and to a lesser extent than inhibition of dynein (Fig. 2c). Inhibition of both myosin and dynein showed a similar reduction in mode amplitudes as inhibition of dynein alone (Fig. 2d).
FIGURE 2.
Fourier mode analysis of microtubule growth trajectories. The variance of Fourier amplitudes was calculated for each mode10. Plots (a)–(d) show amplitude variances for different conditions (squares) in comparison to control cells (circles). Amplitude variances of microtubules in dynein inhibited (DI) cells are less than half those of control cells (a), are unchanged in kinesin inhibited (KI) cells (b), and decreased in myosin inhibited (MI) cells (c). Simultaneous myosin and dynein inhibition (MI+DI) showed a similar reduction to dynein inhibition alone (d). The results suggest that dynein and myosin, but not kinesin contribute to microtubule bending. Error bars indicate SEM.
These results suggest that both myosin and dynein contribute to bending the trajectories of microtubule tips. One possibility, consistent with these results, is that myosin motor-induced force fluctuations are transmitted primarily through dynein linkages between the microtubule and the actin cytoskeleton; blocking these linkages by CC1 would then be sufficient to block both myosin-induced forces and dynein-induced forces on the polymerizing microtubules.
Simulations of fluctuations in growing microtubules
In order to better understand the results of the motor-inhibition experiments, we used numerical simulations of growing microtubules to investigate possible mechanisms whereby microtubule trajectories are deflected by motor forces, either dynein or myosin generated. The cartoon in Fig. 3a illustrates the model for the motor forces. We assume that the dynein-cytomatrix linkage is formed in a stress-free state; for simplicity we further assume that the microtubules grow in the basal plane of the cell and the linkage is also in the same plane, oriented randomly with respect to the tangent to the microtubule at the point of attachment. As the motor begins to walk along the microtubule it exerts a force along the line between the attachment point on the microtubule and the anchor point in the cytomatrix. This force has components parallel to and perpendicular to the local tangent direction that can deflect the microtubule as illustrated in Fig. 3A. The parameters for the microtubules and dynein motors (Table 1) are the same as in our previous simulations 32.
Table 1.
Model Parameters
| Symbol | Parameter | Range | Source | Value Used |
|---|---|---|---|---|
| fmax | Maximum dynein force | 5–8 pN | 9 | 8 pN |
| vm | Dynein speed (no force) | 0.8 μm s−1 | 29 | 0.8 μm s−1 |
| κ | Dynein spring constant | 0.1–1 pN nm−1 | 13, 19 | 1 pN nm−1 |
| koff | Dynein-nucleus off-rate | No value | 32 | 1 s−1 |
| ρ | Dynein density | No value | 32 | 2 μm−1 |
| N | Number of microtubules | 200–500 | 11 | 100 |
| vpol | MT polymerization speed | 0.1–0.2 μm s−1 | 11, 27 | 0.1 μm s−1 |
| ξ | Effective friction | 32 | 10 Pa s | |
| va | Myosin speed | 0.1–0.2 μm s−1 | 30 | 0.1 μm s−1 |
| B | Bending modulus | 24–30 pN nm−2 | 13 | 25 pN nm−2 |
A typical simulation of a growing microtubule is illustrated in Movie 1 and Fig. 3b for the case where dynein is anchored to a stationary cytomatrix (i.e. va,i = 0). The dynein motors pull along the direction of the motor-anchor vector, indicated by the red lines, and so exert lateral as well as tangential forces. The microtubule can be significantly diverted from a straight-line path as shown in Fig. 3c, which shows the final configurations of 100 fully-grown (8 μm) microtubules. The variance of the tip position calculated from the simulated trajectories in Fig. 3C is about 0.8 μm2 at 8 μm length, which is somewhat less than that observed in experiments (1.3 μm2 at 8 μm, Fig. 1d). However in dynein-inhibited cells, there is still a significant variance in the tip position (0.47 μm2 at 8 μm, Fig. 1d) which is much larger than can be accounted for by thermal forces alone. Since there are no such background fluctuations in the simulations and microtubules grow straight in the absence of motor forces, it is to be expected that the fluctuations in the tip positions are less than those observed in the experiments. The simulated variance in the tip positions closely matches the observed difference between control cells and dynein-inhibited cells.
We next performed a Fourier decomposition on the simulated trajectories in Fig. 3c. The variance of the first bending mode, var(a1) = 0.09 μm, is due entirely to dynein activity (no other effects are included in the simulation). This variance matches exactly to the reduction in variance of the first bending mode following dynein inhibition, from 0.144 μm in control cells to 0.054 μm in dynein inhibited cells (Fig. 2a), i.e. . However, the observed effect of dynein on the second mode is not well reproduced by the simulation result, var(a2) = 0.01 μm.
Since myosin inhibition produced a measureable decrease in the variance of tip positions, we next investigated the influence of myosin forces on the microtubule tip trajectories. We used a simple model of myosin activity, where the anchor points of dynein on the cytomatrix move with a fixed velocity; here we choose va = 0.05 μm s−1 which is consistent with measurements of myosin motor speeds30. A simulation showing the growth of a typical microtubule with moving anchor points is shown in Movie 2. It can be seen that there is more curvature in the growing filament, which is reflected in larger values for the variance of the first bending mode var(a1) = 0.15 μm. The larger deflections and curvatures can also be seen in the trajectory plots of 100 sample microtubules (Fig. 3d). The amplitude of the second mode, var(a2) = 0.02 μm is larger than in the absence of cytoskeletal flow but still smaller than the experimental observations. Thus our models only account for the large scale fluctuations in microtubule shape, but not the detailed Fourier spectrum.
We found that the shapes of the microtubules were insensitive to variations in the dynein motor parameters. It is the mechanism of dynein pulling that tends to lead to deflected filaments rather than the exact motor parameters. On the other hand the shapes are sensitive to the motion of the anchor points, which we used to represent myosin-driven fluctuations in the cytomatrix The speed of the anchor points (0.05 μm s−1) was less than typical myosin motor speeds (0.05–0.2 μm s−1), based on the assumption that the cytomatrix moves more slowly than the motors pulling it.. In future the compliance of the cytomatrix could be taken into account with a force balance on the myosin-actin network instead of a prescribed motion of the anchor points.
Discussion
Despite their rigidity, microtubules in living cells contain both short-wavelength bends at the periphery and long-wavelength bends in the cell interior2,5,10,32. The action of minus-end directed motors, specifically dynein, provides an explanation for both the short wavelengths and the localization of these buckles near the periphery32. While growth of microtubules along curved paths has been proposed as the reason for long wavelength bends in the cell interior5, the forces that cause growing microtubule tips to deflect from a straight path have remained unclear. In this paper, we suggest that forces bending the growing microtubule are transmitted to it by dynein. These forces may be generated by dynein activity itself, or by myosin forces that cause motion of dynein anchors in the cytomatrix.
Our results show that dynein and myosin inhibition significantly decrease the spread in microtubule trajectories, but kinesin inhibition does not have any noticeable effect. However, even when both dynein and myosin activity is suppressed, microtubules still spread considerably more than would be expected from thermal forces alone. Thus, there appears to be no single cause for the observed microtubule shapes; both dynein, myosin, and some, as yet undetected, causes (organelles or other motors) all contribute to the observed deflections.
The dynein-cytomatrix linkage plays both an active and a passive role in determining microtubule shapes. Dynein generates bends by its own motor activity (active) and it transmits myosin forces via the motion of the cytomatrix in the region of its anchor point (passive). In this view, inhibiting the dynein–cytomatrix linkage by DsRed-CC1 prevents dynein from pulling on the microtubule but also inhibits transmission of myosin forces to the microtubule. Assuming that the contributions of dynein and myosin are additive, a comparison of the variance of the first mode amplitudes suggests that about 40% of the stored bending energy comes from as yet unidentified sources, 15% from myosin activity5 transmitted by stationary dynein linkages, and 45% from dynein motor activity as if pinned to a static cytomatrix.
An alternative explanation of the data is that myosin does not play a significant role in influencing filament shapes and that dynein accounts for about 60% of the filament bending energy. Myosin inhibition could reduce the effectiveness of dynein molecules that are pulling on the microtubules because the F-actin network is less dense in the absence of myosin crosslinking. Inhibiting myosin in cells where dynein activity is also inhibited would then be expected to have no further effect on the spread, while inhibiting myosin or dynein alone would decrease the deviation from straight paths.
Both these mechanisms are consistent with the experimental observations. To assess the plausibility of these different mechanisms, we carried out simulations of microtubule growth under the action of dynein and myosin motor forces. First, we investigated the possibility that cytoskeletal-bound dynein motors can produce bends in growing microtubules. We found that dynein motors produced significant deflections in the microtubule tips, as measured by the variance in the tip position for 100 microtubules (Fig. 3c). The variance in position for 8 μm segments, ~0.8 μm2 correlates well with the difference in variance in control cells (~1.2 μm2) and dynein-inhibited cells (~0.5 μm2). Moreover, simulations of the variance in the amplitude of the first mode match the experimentally measured change in mode amplitudes when dynein is inhibited. We next implemented a very simple model where the anchor points of the dynein motors move with randomly chosen velocities, mimicking the effect of myosin driven flow in the cytomatrix. Larger bends were observed when the fluctuations in cytomatrix were included, but these are too large to be consistent with the difference between control cells and those with dynein-cytomatrix linkages inhibited. Thus the simulations support the hypothesis that dynein is the key driver of fluctuations in microtubule shape, with myosin generated forces playing at most a secondary role.
Supplementary Material
Acknowledgments
The following grant supported this work: NSF CMMI 0954302, NSF CMMI 0927945, NIH GM102486 and NSF CTS-0505929. DsRed-CC1 and mCherry-KHC were gifts from Prof. Trina Schroer (Johns Hopkins University) and Prof. Kristen Verhey (University of Michigan). We thank the anonymous referee for pointing out the appropriateness of sine functions in the curvature analysis rather than a cosine basis.
References
- 1.Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: Quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bicek AD, Tuzel E, Demtchouk A, Uppalapati M, Hancock WO, Kroll DM, Odde DJ. Anterograde microtubule transport drives microtubule bending in LLC-PK1 epithelial cells. Mol Biol Cell. 2009;20:2943–2953. doi: 10.1091/mbc.E08-09-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brangwynne CP, Koenderink GH, MacKintosh FC, Weitz DA. Nonequilibrium microtubule fluctuations in a model cytoskeleton. Phys Rev Lett. 2008;100:118104–118104. doi: 10.1103/PhysRevLett.100.118104. [DOI] [PubMed] [Google Scholar]
- 4.Brangwynne CP, MacKintosh FC, Kumar S, Geisse NA, Talbot J, Mahadevan L, Parker KK, Ingber DE, Weitz DA. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brangwynne CP, MacKintosh FC, Weitz DA. Force fluctuations and polymerization dynamics of intracellular microtubules. Proc Natl Acad Sci U S A. 2007;104:16128–16133. doi: 10.1073/pnas.0703094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caviston JP, Holzbaur EL. Microtubule motors at the intersection of trafficking and transport. Trends Cell Biol. 2006;16:530–537. doi: 10.1016/j.tcb.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 7.Dogterom M, Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- 8.Etienne-Manneville S. Actin and microtubules in cell motility: which one is in control? Traffic. 2004;5:470–477. doi: 10.1111/j.1600-0854.2004.00196.x. [DOI] [PubMed] [Google Scholar]
- 9.Gennerich A, Reck-Peterson SL. Probing the Force Generation and Stepping Behavior of Cytoplasmic Dynein. Single Molecule Analysis: Methods and Protocols. 2011;783 doi: 10.1007/978-1-61779-282-3_4. [DOI] [PubMed] [Google Scholar]
- 10.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120:923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gliksman NR, Skibbens RV, Salmon ED. How the transition frequencies of microtubule dynamic instability (nucleation, catastrophe, and rescue) regulate microtubule dynamics in interphase and mitosis: analysis using a Monte Carlo computer simulation. Mol Biol Cell. 1993;4:1035–1050. doi: 10.1091/mbc.4.10.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamaguchi MS, Hiramoto Y. Analysis of the Role of Astral Rays in Pronuclear Migration in Sand Dollar Eggs by the Colcemid-UV Method. Dev Growth Differ. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- 13.Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer; 2001. [Google Scholar]
- 14.Jaqaman K, Loerke D, Mettlen M, Kuwata H, Grinstein S, Schmid SL, Danuser G. Robust single-particle tracking in live-cell time-lapse sequences. Nat Methods. 2008;5:695–702. doi: 10.1038/nmeth.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovacs M, Toth J, Hetenyi C, Malnasi-Csizmadia A, Sellers JR. Mechanism of Blebbistatin Inhibition of Myosin II. Journal of Biological Chemistry. 2004;279:35557–35563. doi: 10.1074/jbc.M405319200. [DOI] [PubMed] [Google Scholar]
- 16.Kumar S, I, Maxwell Z, Heisterkamp A, Polte TR, Lele TP, Salanga M, Mazur E, Ingber DE. Viscoelastic retraction of single living stress fibers and its impact on cell shape, cytoskeletal organization, and extracellular matrix mechanics. Biophys J. 2006;90:3762–3773. doi: 10.1529/biophysj.105.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Landau LD, Lifshitz EM. Theory of Elasticity. Pergamon Press; 1981. [Google Scholar]
- 18.Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 19.Lindemann CB, Hunt AJ. Does axonemal dynein push, pull, or oscillate? Cell Motil Cytoskeleton. 2003;56:237–244. doi: 10.1002/cm.10148. [DOI] [PubMed] [Google Scholar]
- 20.Llopis I, Lagomarsino MC, Pagonabarraga I, Lowe CP. Cooperativity and hydrodynamic interactions in externally driven semiflexible filaments. Comput Phys Commun. 2008;179:150–154. [Google Scholar]
- 21.Llopis I, Pagonabarraga I, Lagomarsino MC, Lowe CPC. Sedimentation of pairs of hydrodynamically interacting semiflexible filaments. Phys Rev E. 2007;76:061901. doi: 10.1103/PhysRevE.76.061901. [DOI] [PubMed] [Google Scholar]
- 22.Lodish HF, Berk A, Kaiser CA, Krieger M, Scott MP, Bretscher A, Ploegh H, Matsudaira P. Molecular Cell Biology. New York: W.H. Freeman; 2008. [Google Scholar]
- 23.Navone F, Niclas J, Hombooher N, Sparks L, Bernstein HD, McCaffrey G, Vale RD. Cloning and expression of a human kinesin heavy chain gene: interaction of the COOH-terminal domain with cytoplasmic microtubules in transfected CV-1 cells. J Cell Biol. 1992;117:1263–1275. doi: 10.1083/jcb.117.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nedelec F, Foethke DC. Collective Langevin dynamics of flexible cytoskeletal fibers. New J Phys. 2007;9:427. [Google Scholar]
- 25.O’Connell CB, Wang YL. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol Biol Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reinsch S, Gonczy P. Mechanisms of Nuclear Positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- 27.Shelden E, Wadsworth P. Observation and quantification of individual microtubule behavior in vivo: microtubule dynamics are cell-type specific. J Cell Biol. 1993;120:935–945. doi: 10.1083/jcb.120.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siegrist SE, Doe CQ. Microtubule-induced cortical cell polarity. Genes Dev. 2007;21:483–496. doi: 10.1101/gad.1511207. [DOI] [PubMed] [Google Scholar]
- 29.Toba S, Watanabe TM, Yamaguchi-Okimoto L, Toyoshima YY, Higuchi H. Overlapping hand-over-hand mechanism of single molecular motility of cytoplasmic dynein. Proc Natl Acad Sci U S A. 2006;103:5741–5745. doi: 10.1073/pnas.0508511103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Umemoto S, Sellers JR. Characterization of in vitro motility assays using smooth muscle and cytoplasmic myosins. J Biol Chem. 1990;265:14864–14869. [PubMed] [Google Scholar]
- 31.Wu J, Lee KC, Dickinson RB, Lele TP. How dynein and microtubules rotate the nucleus. J Cell Physiol. 2010;226:2666–2674. doi: 10.1002/jcp.22616. [DOI] [PubMed] [Google Scholar]
- 32.Wu J, Misra G, Russell RJ, Ladd AJC, Lele TP, Dickinson RB. Effects of Dynein on Microtubule Mechanics and Centrosome Positioning. Mol Biol Cell. 2011;22:4834–4841. doi: 10.1091/mbc.E11-07-0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.