Abstract
Listeria monocytogenes is a cause of concern to food industries, mainly for those producing ready-to-eat (RTE) products. This microorganism can survive processing steps such as curing and cold smoking and is capable of growing under refrigeration temperatures. Its presence in RTE fish products with extended shelf life may be a risk to the susceptible population. One example of such a product is gravlax salmon; a refrigerated fish product not exposed to listericidal processes and was the subject of this study. In order to evaluate the incidence and dissemination of L. monocytogenes 415 samples were collected at different steps of a gravlax salmon processing line in São Paulo state, Brazil. L. monocytogenes was confirmed in salmon samples (41%), food contact surfaces (32%), non-food contact surfaces (43%) and of food handlers’ samples (34%), but could not be detected in any ingredient. 179 L. monocytogenes isolates randomly selected were serogrouped and typed by PFGE. Most of L. monocytogenes strains belonged to serogroup 1 (73%). 61 combined pulsotypes were found and a dendrogram identified six clusters: most of the strains (120) belonged to cluster A. It was suggested that strains arriving into the plant via raw material could establish themselves in the processing environment contaminating the final product. The wide dissemination of L. monocytogenes in this plant indicates that a great effort has to be taken to eliminate the microorganism from these premises, even though it was not observed multiplication of the microorganism in the final product stored at 4°C up to 90 days.
Keywords: Listeria monocytogenes, gravlax salmon, PFGE
INTRODUCTION
Seafood is known as a vehicle for the microorganism and has been linked to episodes of listeriosis (9,18,20,41). Several reports have shown that the prevalence of L. monocytogenes varies in different seafood, seafood products and its processing plant environments (3,22,27,46,50). No information on the dissemination of L. monocytogenes in gravlax salmon processing line could be found.
Since the 90’s the consumption of salmon in Brazil has increased due to better price and accessibility. The salmon consumed and processed is imported mainly from Chile, an important producer of farmed salmon Gravlax (gravad lax or gravad) salmon is a ready-to-eat product made from salmon fillets cured with salt and sugar and seasoned with pepper and dill. It does not undergo any listericidal treatment and in Brazil, it is vacuum-package and stored at 7°C to 8°C or kept frozen.
Restriction endonuclease analysis by pulsed-field gel electrophoresis (PFGE) has proved to be a highly discriminatory and reproducible method, especially if combined with serotyping (7,8,10) and it has been useful for subtyping L. monocytogenes strains isolated from food processing lines (15,16,37,38,47).
The lack of information regarding L. monocytogenes in fish processing plants in tropical countries is a cause of concern. In an effort to determine the distribution of L. monocytogenes in the only Brazilian gravlax salmon processing plant, we characterized the isolates from this industry by PFGE and identified the possible routes of contamination.
MATERIALS AND METHODS
Product manufacturing
Raw material was gutted Chilean ocean farm-raised salmon (Salmon salar), usually frozen fillets (90% of the time). When whole fishes were used they were transported to the plant under refrigeration for deheading and were manually filleted immediately before processing. In the processing plant the fillets were held at 4°C in their original container for defrosting and were submitted to visual inspection for bone removal. Fillets were washed with chlorinated water and transferred to stainless steel perforated trays to drain off the excess of water. The flesh side of the fillets was hand rubbed with a commercial mixture of NaCl, sodium nitrate and sodium nitrite and stored at 4°C for 24 hours in high density polypropylene (HDP) boxes. After the excess of salts being washed out with chlorinated water and drained, a mixture of sugar, NaCl, ground white pepper and dried dill was hand rubbed into the flesh side of the fillets and stored for 24 hours at 4°C in HDP boxes. Fillets were layered on stainless steel supports, sprayed with Jerez type wine and ripened for 48 hours at 4°C. Skin was manually removed and fillets were transferred to the slicing area. They were sliced with commercial slicing machine and manually separated in 100g portions before vacuum-packing. The packages were kept at 4°C or frozen (-20°C) according to the commercial demand. Table 1 shows the gravlax salmon processing flow chart. All but slicing and vacuum-packing operations took place in the same room with no physical barriers between the different areas. Gloves were used only to transfer the fillets to the slicing machine and portioning of the final product.
Table 1.
Flow chart of gravlax salmon processing, sample category, serogroup, number of isolates, PFGE profiles and clusters.
| Flow processing | Sample category Serogroup (n° of isolates) | Combined profiles of Apal and AscI | Clusters | |
|---|---|---|---|---|
Reception
|
raw salmon (S1) | 1(8) | P1C1, P2C1, P2C2, P2C12 | A |
| handlers (H1/H2) | 1(11), 4(1) | P7C14,P7C27,P11C18,P30C18 | A | |
| knife, table (U1/U2/U3) | 1(9) | P20C21,P30C18 | A/C | |
| drain, floor (E1/E2) | 1(10), 4(4) | P20C12,P21C18,P21C19,P21C23, P22C22,P30C18 | A/C | |
1st Curing
|
cured salmon (S2) | 1(1), 4(7) | P2C5,P3C3,P4C4,P4C7,P5C6 | A/B/C |
| handler (H3) | 1(3) | P30C13 | A | |
| table, box (U4/U5) | 1(5), 4(6) | P9C7,P21C12,P22C13 | A/B | |
| curing ingredients (I1) | *- | - | - | |
Salt removal
|
handler (H4) | - | - | - |
| boxes, table (U6/U7/U8) | - | - | - | |
| floor, drain (E3, E4) | 1(12) | P20C24,P21C28,P22C18,P30C28 | A/C | |
Washing
|
washed salmon (S3) | 1(5), 4(3) | P5C6,P7C9,P29C8 | A/B/C |
2nd Curing
|
cured salmon (S4) | 1(4), 4(2) | P1C2,P1C7,P8C2,P8C10 | A |
| handler (H5) | - | - | - | |
| table (U9) | - | - | - | |
| curing ingredients (I2/I3) | - | - | - | |
Jerez spraying
|
cured salmon (S5) | 4(5) | P9C7 | B |
| handler (H6) | 4(3) | P4C26 | B | |
| trolleys (U10) | - | - | - | |
Ripening
|
gravlax with skin (S6) | 1(7), 4(1) | P6C2,P7C14,P10C3,P11C19 | A/C/E |
| floor,door,wall (E5,E6,E7) | - | - | - | |
Skinning
|
gravlax without skin (S7) | 1(6), 4(2) | P5C6,P6C1,P6C8,P11C19,P12C7 | A/B |
| handler (H7) | - | - | - | |
| trolley,table,knife (U11/U12/U13) | 1(6) | P23C1 | A | |
Slicing
|
sliced gravlax (S8) | 1(5), 4(3) | P1C2,P5C11,P11C13,P13C17,P14C16 | A/B/C/F |
| handler (H8) | 1(7) | P11C1,P28C1,P27C1 | A | |
| conveyor belt, blade (U14/U15) | 1(5) | P24C19 | A | |
| floor, drain (E8, E9) | 1(12) | P22C13,P25C13,P26C13 | A | |
Portioning
|
handler (H9) | - | - | |
| tray, scale (U16/U17) | 1(6) | P17C13,P21C19 | A | |
Vacuum-packing
|
package gravlax (S9) | 1(6) | P1C1, P15C27 | A/B |
Cold storage
|
refrigerated gravlax (S10) | 1(3), 4(3) | P2C18,P4C6,P4C20,P16C18,P17C18 | A/B |
| Frozen storage | frozen gravlax (S11) | 1(2), 4(6) | P11C18,P18C15,P19C7 | A/B/D |
= no positive samples for L. monocytogenes, _ = serotype 4 strains points.
Sampling procedure
A total of four hundred and fifteen samples were collected from November, 1999 to March, 2000. Table 1 shows the sampling points. 255 salmon samples (S) were collected in 9 different processing steps (S1-S9), and on each sampling day 5 samples of 100g each were taken from each sampling point. End product samples (S10 and S11) were taken after 2 days of cold storage and frozen storage, respectively. 44 non-food contact surfaces samples (E1-E9) (floor, drains and walls) and 63 food contact surfaces samples (U1-U17) (tables, knives, scale, boxes, perforated trays, trolleys and conveyor belts) were collected with moistened sterile cotton swabs. Samples from regular surfaces were collected by swabbing 5x25cm2 areas, while 25cm2 were swabbed from irregular surfaces or small surfaces. All swabs were collected in Letheen broth (Oxoid, Basingstoke, UK). Portions of 100g each of 1st curing ingredients (I1) (NaCl, sodium nitrite and sodium nitrate); 2nd curing ingredients (I2) (NaCl, sugar and ground white pepper) and dried dill (I3) were aseptically collected in plastic bags (15 samples). A total of 38 samples from food handlers (H1-H9) were collected according to Destro et al., (16). Samples were transported to the laboratory in insulated boxes and analyzed within 3 hours.
For the enumeration study samples from 3 different lots of gravlax salmon were collected on day of production and transported to the laboratory. These samples were stored at 4°C in a vertical refrigerator (Reubly, Brazil) for up to 90 days, according to the shelf life informed on each of the packages. Samples were microbiological and sensorial examined every 15 days from day zero and on each sampling day 4 gravlax packs were analyzed.
Bacteriological analysis
Bacteriological analysis was conducted as described by Farber and Daley (19). In brief: 25g or ml were homogenized in Listeria enrichment broth (UVM formulation) (Oxoid) and incubated at 30°C for 24 hours. 0.1ml were transferred to modified Fraser broth (Oxoid) and incubated at 35°C for 24-48 hours. Blackened Fraser tubes were streaked onto Palcam (Oxoid) and hemolytic ceftazidime lithium chloride (HCLA) agar plates (39) and incubated at 35°C for 24-48 hours. Three presumptive L. monocytogenes colonies per plate were submitted for identification employing Gram staining, motility at 25°C, catalase test, carbohydrates fermentation, -hemolysis and Api Listeria test (bioMérieux, Marcy L’Etoile, France). For L. monocytogenes enumeration study the same method and the most probable number technique (11) were used. Lactic acid bacteria (LAB) population was also determined for each of the samples (51).
Serology
Serogrouping was conducted on 179 L. monocytogenes isolates randomly chosen from the 429 obtained. These isolates represented the different sampling points and sampling dates (Table 1). Antiserum O types 1 and 4 (Difco, Detroit, USA) were used. Fifteen strains that could not be serogrouped with the former were tested against Denka Seiken antiserum (Tokyo, Japan) according to manufacturer’s instruction. Isolates from sampling points U3, U6, U7, U9, U11 and U12 lost viability and were not included in serology and PFGE analysis.
PFGE
PFGE was performed according to PulseNet protocol described by Graves and Swaminathan (24) using enzymes ApaI and AscI and the same 179 isolates as above. Lambda ladder PFG marker (New England Biolabs, Beverly, USA) was used as fragment size marker, L. monocytogenes H2446 as a reference strain and Salmonella Typhi as an outgroup. Similarities between combined restriction patterns, based on bands positions were derived from the Dice correlation coefficient. Profiles were clustered by unweighted pair group method using arithmetic averages (UPGMA) (NTSYSpc 2.0, Exeter Software, USA) with a maximum tolerance of 1.2% and a dendrogram was constructed. Normalization was done with Lambda ladder PFG marker that was included in every first, sixth and twelfth lane.
RESULTS
L. monocytogenes in the processing line: A total of 415 samples was tested and the overall incidence of Listeria sp and L. monocytogenes is shown in Table 2. L. monocytogenes was detected in 41% of salmon samples (S1) arriving into the plant. First curing (S2) and washing (S3) steps did not reduce the amount of positive samples. On the other hand, after the second curing (S4) and Jerez spraying (S5) processes L. monocytogenes was recovered from 24% and 32% of the samples, respectively. However, after been kept at 4°C for 48 hours (S6) this value increased to 73%. At the skin removal (S7) and slicing (S8) procedures, the recovery rate of positive salmon samples was still high (48% and 52%, respectively). L. monocytogenes was present in 36% of vacuum-packed samples (S9). Two days after processing, 35% of the frozen stored gravlax salmon (S11) and 15% of the cold stored (S10) samples were contaminated with the microorganism.
Table 2.
Occurrence of Listeria sp and L. monocytogenes in a gravlax salmon processing line.
| Sample type | Total of samples | Positive samples for for Listeria sp (%) | Positive samples L. monocytogenes (%) |
|---|---|---|---|
| Salmon (S) | 255 | 164 (64) | 105 (41) |
| Food contact surfaces (U) | 63 | 28(44) | 20 (32) |
| Non-food contact surfaces (E) | 44 | 24 (55) | 19 (43) |
| Food handlers (H) | 38 | 17(45) | 13 (34) |
| Ingredients (I) | 15 | 0 | 0 |
| Total | 415 | 233 (56) | 157 (38) |
Non-food contact surface samples from reception (E1, E2) and salt-removal areas (E3, E4) showed high frequency of L. monocytogenes (80% for each sampling point). In the slicing room area, samples from floor (E8) and drain (E9) presented the pathogen in 40% and 20% of the samples, respectively. No L. monocytogenes was detected in the samples collected in the ripening cold room (E5-E7) (Table 1).
Amongst the food contact surface, all samples from tables at reception (U2), salt removal (U7) and skin removal (U13) (Table 1) areas were positive for L. monocytogenes. Samples from table used for bone removal (U3) and the skin removal knives (U12) presented 50% and 67% of the microorganism, respectively. The pathogen was present in 40% of the samples from salting boxes (U5), ripening trolley (U11) after 48 hours in the cold room and weighting tray (U16). Lower frequency of positive samples was found in the salting table (U4) (33%) and raw fish filleting knives (U1) (25%). Boxes after salt removal (U6), table for applying second curing ingredients (U9) and the conveyor belt from the slicing machine (U14) were positive in 20% of the time. No L. monocytogenes was found in the washing boxes (U8), ripening trolley before entering the cold room (U10), slicing machine (U15) and scale (U17).
L. monocytogenes presence in food handlers’ samples varied from 0 to 80%. The highest incidence was found on handlers responsible for cleaning the raw fish (H1). Samples collected from employees responsible for first curing salts rubbing (H3), Jerez spraying (H6) and gravlax slicing (H8) harbored the pathogen in 40% of the time. Bone picking handler (H2) samples had 33% of L. monocytogenes presence. The pathogen was not found in other workers (H4, H5, H7 and H9) (Table 1).
Serology: Serology showed that most of L. monocytogenes strains belonged to serogroup 1 (73%), even though strains from serogroup 4 were also found. Both serotypes were spread along the processing line (Table 1).
PFGE: The two enzymes generated similar number of macrorestriction profiles being 28 and 30 for AscI and ApaI, respectively. Combination of both results originated 61 combined profiles and 6 distinct clusters, named A to F, could be identified. Table 1 presents the distribution of the 179 L. monocytogenes strains according to the combined PFGE profiles and the clusters. Cluster A grouped 38 profiles and 120 out of the 179 tested strains, while cluster B grouped 10 profiles and 35 strains. Clusters D, E and F were smaller and presented less diversity. It is interesting to note that clusters B, D, E and F grouped only serogroup 4 strains whereas strains from serogroups 1 and 4 were found in clusters A and C (Table 1).
Serogroup 4 strains belonging to clusters A and C were from non-food contact sources (E1 and E2) and from a food handler (H2). On the other hand the majority of the strains grouped in cluster B were from fish, and 8 out of the 14 strains isolated from final product (S10 and S11) belonged to this cluster (profiles P4C6, P4C20, P19C7) (Fig. 1 and Table 1).
Figure 1.

Dendrogram showing the relationship among PFGE-tested isolates (179) using ApaI and AscI. See Table 1 for identification of the strains belonging to each profile.
L. monocytogenes and LAB population during cold storage of gravlax salmon: Population of L. monocytogenes and LAB were determined every 15 days for up to 90 days. The experiment was repeated 3 times, and in each sampling day 4 packages were examined totalizing 84 samples of gravlax salmon. Microbiological analyses were conducted only when the sensorial characteristics (color, appearance, and smell) of the product were normal. Mean population of LAB was around 104CFU/g on day zero and increased to 107CFU/g by day 15th, staying at this value up to the 90th day of storage. On the other hand, L. monocytogenes population was low varying from <0.03 to 4.27 MPN/g on day zero and decreasing during storage (Fig. 2).
Figure 2.
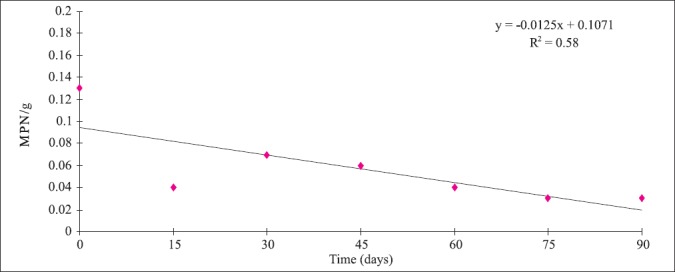
Mean population of L. monocytogenes in vacuum package gravlax salmon stored at 4°C for 90 days.
DISCUSSION
In the last few years several studies have been conducted on fish processing plants, mainly in smoking houses in Northern countries (15,22,26,36,52). However, studies in gravlax processing lines are still rare.
In this study the overall incidence of L. monocytogenes in the samples (38%) was higher than found in other studies (25,26,49), but similar to the results found by Hoffman, et al. (29). Also, the frequency of L. monocytogenes in raw salmon samples reported here (48%) was higher than those found by other researchers that vary from zero to 22% (6,15,26,29,36,49,52).
These differences can be caused by salmon farming and catching techniques, geographic location of the farm and climate, presence of L. monocytogenes in culture water and use of GMP (Good Manufacturing Practices) and HACCP (Hazard Analysis and Critical Control Points) in the primary production sites (12). Miettinen and Wirtanen (34) showed that the number of samples contaminated with Listeria spp was typically higher after rainy periods decreasing during dry ones.
The high incidence of L. monocytogenes in the final product (35% for frozen stored and 15% for cold stored gravlax) found in this study is a cause of concern since this is a ready-to-eat product. In a study conducted in Denmark, the incidence of L. monocytogenes in gravlax salmon varied from 25 to 33% for the refrigerated product (32). Lower incidence was observed in Iceland by Hartemink and Georgsson (28) who found 8% of L. monocytogenes positive gravlax samples.
An increase in L. monocytogenes frequency in the samples collected after ripening (S6) was observed and could be explained either by cross-contamination in the cold room during the 48 hours storage or by resuscitation of stressed bacterium cells during the cold storage. The first hypothesis can not be sustained since no L. monocytogenes was found in environmental samples from this cold room (E5-E7) (Table 1). This is an interesting result since cold rooms are usually considered important points for product cross-contamination.
The frequency of L. monocytogenes in environmental samples differs from plant to plant and is dependent, besides other factors, on time of samples collection (after cleaning procedures or during production) and status of facility maintenance. The samples in this study were collected during production which may explain the high incidence of L. monocytogenes on non-food contact surfaces and food contact surfaces, respectively (Table 2). Similar results were observed by Gay and Cerf (23) but these values are higher than other reports for seafood processing environments (4,5,16,36). On the other hand Rorvik et al. (44) found L. monocytogenes in 63% of the drains of 40 salmon smokehouses.
This fluctuation is not uncommon Hoffman et al. (29) and Thimothe et al. (49) found a broad variation amongst L. monocytogenes positive environmental samples. Depending on the plant the occurrence varied from 1.2% to 43.8% for the former, and from zero to 29.8% for the latter.
The overall percentage of L. monocytogenes positive samples for handlers found in this study is higher than to those reported for other seafood plants (Table 2) (16,49). Whether this can represent difference in hygienic status of the operators or the use or not of gloves remains unclear.
Since hands may play an important role in foodborne pathogen dissemination/ cross contamination, the findings of this study could be cause of concern. However, one can not overestimate the importance of the food handlers in disseminating L. monocytogenes in the evaluated plant once the incidence of the pathogen in other samples is also high. In this plant workers have multitasks and depending on the necessity they have to work with both raw material and final product in the same day. This could explain the contamination of food handlers by L. monocytogenes of the same PFGE profile present in fish and environment.
The prevalence of serogroup 1 strains amongst the L. monocytogenes isolates is according to other studies (15,43). It is important to emphasize that strains belonging to serogroup 4, were found in the final product.
It can be seen in table 1 that cluster A strains were widespread in this plant being isolated from raw material arriving into the plant to final product, as well as from all the other sample types. Clusters D, E and F on the other hand, were restricted to the product only. Predominance of specific L. monocytogenes genetic types in seafood processing environment was also verified by others (26,42,49,52).
The Cluster A strains are introduced into the plant via raw fish. These strains were first found on the fish flesh immediately after opening the transport container. This group has a good adaptation power, resisting the environmental stress, and from raw material it was able to establish in the different niches of the plant. Besides that, the constant re-introduction of L. monocytogenes strains from this cluster into the plant, via raw material, may also have contributed to its maintenance. According to Thimothe et al. (49) it is very difficult to differentiate between a truly persistent subtype and a persistent re-introduction of specific subtype.
There is some disagreement between the most important sources of L. monocytogenes contamination of processing lines. While many authors have emphasized the importance of raw material as a source of this contamination (4,30,33,34), others highlighted the importance of equipment and environment as contamination sources (22,49). Even though, these authors mentioned that raw fish can not be excluded as the source of equipment and environmental contamination.
The strains used in this study were submitted to RAPD typing together with 3 isolates obtained 4 years later from a different fish product processed in the same plant. It was seen that two of the new strains presented the same RAPD profile as several cluster A strains. The third strain showed the same RAPD profile of strains from cluster B. This results indicates that cluster A strains are resident on this plant (2).
Strains grouped in cluster B were first found in food contact surfaces in the dry salting area, and from this step on they could be found on the fish up to final product (Table 1), all of them belonged to serogroup 4. A similar pattern of contamination could be seen for strains from cluster C that were first detected in food contact and non-food contact surfaces and handlers’ samples and later in the product (Table 1).
Slicing machine may have contributed to the introduction of cluster F strains to the product, since they were found only after slicing; however, L. monocytogenes was not detected in the samples collected at this point (U15, Table 1). Strains of cluster E were found on salmon at two steps before skin removal (salting and ripening – S2 and S6) (Table 1) not being detected after skin removal suggesting that these strains were adapted to the salmon skin. Other studies have reported that contamination decreases after skin removal (4,17,31,43).
Eliminating L. monocytogenes from the food processing environment is a very hard task that has been challenging food processors. In this plant this would be even more difficult since it had some structural problems by the time the samples were collected. Examples of such problems are porous floor, lack of physical barriers between raw and semi-processed products, free circulation of employees and multi-task employees. At the same time, the inner part of the drains was covered with stainless steel indicating that facilities were starting to be improved.
The population of L. monocytogenes found in this study was low (<0.03 MPN/g to 4.3 MPN/g). Only two studies were found reporting L. monocytogenes population on seafood and they were also low: 0.23 MPN/g in cooked lobster (21) and <100 MPN/g in crab meat (40).
It was also observed in this study that the pathogen population did not increase during the storage at 4°C. This behavior is not constant in naturally contaminated products as verified by Cortesi et al. (13) and by Jorgensen et al. (32). They noted that in some samples the population can increase and in others it can decrease. Dalgaard and Jorgensen (14) observed that L. monocytogenes growth in naturally contaminated cold-smoked fish is slower than in artificially ones. They emphasize that predictive model developed based on spiked samples may result in overestimated growth values.
LAB population may have had some influence on L. monocytogenes multiplication on gravlax salmon. Rorvik et al. (45) and Nilsson et al. (35) noted that L. monocytogenes multiplication can be suppressed by LAB in cold smoked salmon. It’s well known that these bacteria can produce bacteriocins that can kill or inhibit microorganism development (1,48).
CONCLUSION
The importance of raw material as the first source of L. monocytogenes to this plant was determined. It was also identified that some strains found in the plant have high capacity to persist in the environment. There are several measures that could be taken to diminish the contamination of the environment and of the product. Amongst them one can suggest the implementation of GMP and HACCP. Also, post-packaging treatments could be applied to the product to decrease the frequency of L. monocytogenes positive samples.
Even though the results of this study did not show the growth of L. monocytogenes in gravlax salmon, this possibility can not be excluded. Other studies with naturally contaminated samples should be conducted in order to evaluate the real risk of this product to the population.
ACKNOWLEDGMENTS
This research was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 98/12368-0 and 01/ 05990-1) and by Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the fellowship to the first author. We thank Deborah A. Lewis for the review of the manuscript, Cristiano Andrigheto and Priscila de A. Trindade for help during the PFGE study and Elsa M. Mamizuka for the PFGE facilities. The authors are indebted to Oxoid Brazil for the Denka Seiken antiserum.
RESUMO
Epidemiologia de Listeria monocytogenes em uma linha de processamento de salmão gravlax
Listeria monocytogenes é um patógeno de grande preocupação para as indústrias alimentícias, principalmente aquelas produtoras de alimentos prontos para consumo (RTE). Este microrganismo pode sobreviver às etapas de cura e defumação a frio, além de tolerar temperaturas de refrigeração. A presença de L. monocytogenes em pescados RTE com vida de prateleira longa representa um risco para a população susceptível, sendo o salmão gravlax deste tipo de produto. No presente estudo avaliou-se a incidência e disseminação de L. monocytogenes em 415 amostras de salmão gravlax obtidas de diferentes etapas de processamento de uma indústria localizada no Estado de São Paulo. A presença de L. monocytogenes foi confirmada em amostras de salmão (41%), superfícies de contato (32%) e não contato (43%) e manipuladores (34%), porém não se isolou o microrganismo em nenhum ingrediente. Do total de cepas isoladas, 179 destas foram escolhidas aleatoriamente e submetidas a sorologia e tipagem por PFGE. A maioria dos isolados pertenceu ao sorogrupo 1 (73%), sendo identificados 61 pulsotipos quando se combinou os resultados de sorologia e PFGE e 6 clusters foram distribuídos em um dendrograma. O cluster A agrupou a maioria das cepas (120). Pode-se sugerir que as cepas foram introduzidas na linha de processamento por meio da matéria prima e contaminando o produto final. Estes resultados indicam que a eliminação de L. monocytogenes deste estabelecimento requer um grande esforço, ainda que o microrganismo não se multiplicou no produto final estocado a 4°C por 90 dias.
Palavras-chave: Listeria monocytogenes; salmão gravlax; PFGE
REFERENCES
- 1.Ahn C., Stiles M.E. Antibacterial activity of lactic acid bacteria isolated from vacuum-packaged meats. J. Appl. Bact. 1990;69(3):302–310. doi: 10.1111/j.1365-2672.1990.tb01520.x. [DOI] [PubMed] [Google Scholar]
- 2.Alves V.F., De Martinis E.C., Destro M.T., Vogel B.F., Gram L. Antilisterial activity of a Carnobacterium piscicola isolated from Brazilian smoked fish (surubim [Pseudoplatystoma sp]) and its activity against a persistent strain of Listeria monocytogenes isolated from surubim. J. Food Prot. 2005;68(10):2068–2077. doi: 10.4315/0362-028x-68.10.2068. [DOI] [PubMed] [Google Scholar]
- 3.Antony M.M., Jeyasekaran G., Shakila R.J., Shamugan S.A. Microbiological quality of raw shrimps processed in seafood processing plants of Tuticorin, Tamilnadu, India. Asian Fish. Sci. 2002;15:305–314. [Google Scholar]
- 4.Autio T., Hielm S., Miettinen M., Sjoberg A.M., Aarnisalo K., Bjorkroth J., Mattila-Sandholm T., Korkeala H. Sources of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl. Environ. Microbiol. 1999;65(1):150–155. doi: 10.1128/aem.65.1.150-155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Embarek P.K., Hansen L.T., Enger O., Huss H.H. Occurrence of Listeria spp. in farmed salmon and during subsequent slaughter; comparison of Listertest Lift and the USDA method. Food Microbiol. 1997;14:39–46. [Google Scholar]
- 6.Ben Embarek P.K. Presence, detection and growth of Listeria monocytogenes in seafoods: a review. Int. J. Food Microbiol. 1994;23(1):17–34. doi: 10.1016/0168-1605(94)90219-4. [DOI] [PubMed] [Google Scholar]
- 7.Bille J., Rocourt J. WHO International Multicenter Listeria monocytogenes Subtyping Study-rationale and set-up of the study. Int. J. Food Microbiol. 1996;32(3):251–262. doi: 10.1016/s0168-1605(96)01140-3. [DOI] [PubMed] [Google Scholar]
- 8.Boerlin P., Boerlin-Petzold F., Bannerman E., Bille J., Jemmi T. Typing Listeria monocytogenes isolates from fish products and human listeriosis cases. Appl. Environ. Microbiol. 1997;63(4):1338–1343. doi: 10.1128/aem.63.4.1338-1343.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brett M.S., Short P., McLauchlin J. A small outbreak of listeriosis associated with smoked mussels. Int. J. Food Microbiol. 1998;43(3):223–229. doi: 10.1016/s0168-1605(98)00116-0. [DOI] [PubMed] [Google Scholar]
- 10.Brosch R., Brett M., Catimel B., Luchansky J.B., Ojeniyi B., Rocourt J. Genomic fingerprinting of 80 strains from the WHO multicenter international typing study of Listeria monocytogenes via pulsed-field gel electrophoresis (PFGE) Int. J. Food Microbiol. 1996;32(3):343–355. doi: 10.1016/s0168-1605(96)01147-6. [DOI] [PubMed] [Google Scholar]
- 11.Bunning V.K., Crawford R.G., Tierney J.T., Peeler J.T. Thermotolerance of heat-shocked Listeria monocytogenes in milk exposed to high-temperature, short-time pasteurization. Appl. Environ. Microbiol. 1992;58(6):2096–2098. doi: 10.1128/aem.58.6.2096-2098.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colburn K.G., Kaysner C.A., Abeyta C., Wekell M.M. Listeria species in a California coast estuarine environment. Appl. Environ. Microbiol. 1990;56(7):2007–2011. doi: 10.1128/aem.56.7.2007-2011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cortesi M.L., Sarli T., Santoro A., Murru N., Pepe T. Distribution and behavior of Listeria monocytogenes in three lots of naturally-contaminated vacuum-packed smoked salmon stored at 2 and 10 degrees C. Int. J. Food Microbiol. 1997;37(2-3):209–214. doi: 10.1016/s0168-1605(97)00064-0. [DOI] [PubMed] [Google Scholar]
- 14.Dalgaard P., Jorgensen L.V. Predicted and observed growth of Listeria monocytogenes in seafood challenge tests and in naturally contaminated cold-smoked salmon. Int. J. Food Microbiol. 1998;40(12):105–115. doi: 10.1016/s0168-1605(98)00019-1. [DOI] [PubMed] [Google Scholar]
- 15.Dauphin G., Ragimbeau C., Malle P. Use of PFGE typing for tracing contamination with Listeria monocytogenes in three cold-smoked salmon processing plants. Int. J. Food Microbiol. 2001;64(1-2):51–61. doi: 10.1016/s0168-1605(00)00442-6. [DOI] [PubMed] [Google Scholar]
- 16.Destro M.T., Leitão M.F., Farber J.M. Use of molecular typing methods to trace the dissemination of Listeria monocytogenes in a shrimp processing plant. Appl. Environ. Microbiol. 1996;62(2):705–711. doi: 10.1128/aem.62.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eklund M.W., Peterson M.E., Poysky F.T., Paranjpye R.N., Perlroy G.A. Control of bacterial pathogens during processing of cold-smoked and dried salmon strips. J. Food Prot. 2004;67(2):347–351. doi: 10.4315/0362-028x-67.2.347. [DOI] [PubMed] [Google Scholar]
- 18.Ericsson H., Eklow A., Danielsson-Tham M.L., Loncarevic S., Mentzing L.O., Persson I., Unnerstad H., Tham W. An outbreak of listeriosis suspected to have been caused by rainbow trout. J. Clin. Microbiol. 1997;35(11):2904–2907. doi: 10.1128/jcm.35.11.2904-2907.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farber J.M., Daley E. Presence and growth of Listeria monocytogenes in naturally-contaminated meats. Int. J. Food Microbiol. 1994;22(1):33–42. doi: 10.1016/0168-1605(94)90005-1. [DOI] [PubMed] [Google Scholar]
- 20.Farber J.M., Daley E.M., Mackie M.T., Limerick B. A small outbreak of listeriosis potentially linked to the consumption of imitation crab meat. Lett. Appl. Microbiol. 2000;31(2):100–104. doi: 10.1046/j.1365-2672.2000.00775.x. [DOI] [PubMed] [Google Scholar]
- 21.Farber J.M. Symposium on microbiology update: old friends and new enemies Listeria monocytogenes. J. AO AC. 1991;74(4):701–704. [PubMed] [Google Scholar]
- 22.Fonnesbech Vogel B., Huss H.H., Ojeniyi B., Ahrens P., Gram L. Elucidation of Listeria monocytogenes contamination routes in cold-smoked salmon processing plants detected by DNA-based typing methods. Appl. Environ. Microbiol. 2001;67(6):2586–2595. doi: 10.1128/AEM.67.6.2586-2595.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gay M., Cerf O. Significance of temperature and preincubation temperature on survival of Listeria monocytogenes at pH 4.8. Lett. Appl. Microbiol. 1997;25(4):257–260. doi: 10.1046/j.1472-765x.1997.00216.x. [DOI] [PubMed] [Google Scholar]
- 24.Graves L.M., Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed- field gel electrophoresis. Int. J. Food Microbiol. 2001;65(1-2):55–62. doi: 10.1016/s0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 25.Gudmundsdottir S., Gudbjornsdottir B., Einarsson H., Kristinsson K.G., Kristjansson M. Contamination of cooked peeled shrimp (Pandalus borealis) by Listeria monocytogenes during processing at two processing plants. J. Food Prot. 2006;69(6):1304–1311. doi: 10.4315/0362-028x-69.6.1304. [DOI] [PubMed] [Google Scholar]
- 26.Gudmundsdottir S., Gudbjornsdottir B., Lauzon H.L., Einarsson H., Kristinsson K.G., Kristjansson M. Tracing Listeria monocytogenes isolates from cold-smoked salmon and its processing environment in Iceland using pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2005;101(1):41–51. doi: 10.1016/j.ijfoodmicro.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 27.Hara Y., Izumisawa M., Ishii K. Contamination of Listeria monocytogenes in ready-to-eat seafoods in Japan. Jpn. J. Food Microbiol. 2003;20:63–67. [Google Scholar]
- 28.Hartemink R., Georgsson F. Incidence of Listeria species in seafood and seafood salads. Int. J. Food Microbiol. 1991;12(2-3):189–195. doi: 10.1016/0168-1605(91)90069-2. [DOI] [PubMed] [Google Scholar]
- 29.Hoffman A.D., Gall K.L., Norton D.M., Wiedmann M. Listeria monocytogenes contamination patterns for the smoked fish processing environment and for raw fish. J. Food Prot. 2003;66(1):52–60. doi: 10.4315/0362-028x-66.1.52. [DOI] [PubMed] [Google Scholar]
- 30.Huss H.H., Jorgensen L.V., Vogel B.F. Control options for Listeria monocytogenes in seafoods. Int. J. Food Microbiol. 2000;62(3):267–274. doi: 10.1016/s0168-1605(00)00347-0. [DOI] [PubMed] [Google Scholar]
- 31.Jemmi T., Keusch A. Behavior of Listeria monocytogenes during processing and storage of experimentally contaminated hot- smoked trout. Int. J. Food Microbiol. 1992;15(3-4):339–346. doi: 10.1016/0168-1605(92)90067-d. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen L.V., Huss H.H. Prevalence and growth of Listeria monocytogenes in naturally contaminated seafood. Int. J. Food Microbiol. 1998;42(1-2):127–131. doi: 10.1016/s0168-1605(98)00071-3. [DOI] [PubMed] [Google Scholar]
- 33.Miettinen H., Aarnisalo K., Salo S., Sjoberg A.M. Evaluation of surface contamination and the presence of Listeria monocytogenes in fish processing factories. J. Food Prot. 2001;64(5):635–639. doi: 10.4315/0362-028x-64.5.635. [DOI] [PubMed] [Google Scholar]
- 34.Miettinen H., Wirtanen G. Ecology of Listeria spp in a fish farm and molecular typing of Listeria monocytogenes from fish farming and processing companies. Int. J. Food Microbiol. 2006;112(2):138–146. doi: 10.1016/j.ijfoodmicro.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 35.Nilsson L., Huss H.H., Gram L. Inhibition of Listeria monocytogenes on cold-smoked salmon by nisin and carbon dioxide atmosphere. Int. J. Food Microbiol. 1997;38(2-3):217–227. doi: 10.1016/s0168-1605(97)00111-6. [DOI] [PubMed] [Google Scholar]
- 36.Norton D.M., McCamey M.A., Gall K.L., Scarlett J.M., Boor K.J., Wiedmann M. Molecular studies on the ecology of Listeria monocytogenes in the smoked fish processing industry. Appl. Environ. Microbiol. 2001;67(1):198–205. doi: 10.1128/AEM.67.1.198-205.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peccio A., Autio T., Korkeala H., Rosmini R., Trevisani M. Listeria monocytogenes occurrence and characterization in meat-producing plants. Lett. Appl. Microbiol. 2003;37(3):234–238. doi: 10.1046/j.1472-765x.2003.01384.x. [DOI] [PubMed] [Google Scholar]
- 38.Pourshaban M., Gianfranceschi M., Gattuso A., Menconi F., Aureli P. Identification of Listeria monocytogenes contamination sources in two fresh sauce production plants by pulsedfield gel electrophoresis. Food Microbiol. 2000;17:393–400. [Google Scholar]
- 39.Poysky F.T., Paranjpye R.N., Lashbrook L.C., Peterson M.E., Perlroy G.A., Eklund M.W. Selective and differential medium for isolation of Listeria monocytogenes from foods. J. Food Prot. 1993;56:326–329. doi: 10.4315/0362-028X-56.4.326. [DOI] [PubMed] [Google Scholar]
- 40.Rawles D., Flick G., Pierson M., Diallo A., Wittman R., Croonenberghs R. Listeria monocytogenes occurrence and growth at refrigeration temperatures in fresh blue crab (Callinectes sapidus) meat. J. Food Prot. 1995;58:1219–1221. doi: 10.4315/0362-028X-58.11.1219. [DOI] [PubMed] [Google Scholar]
- 41.Rocourt J., Jacquet C., Reilly A. Epidemiology of human listeriosis and seafoods. Int. J. Food Microbiol. 2000;62(3):197–209. doi: 10.1016/s0168-1605(00)00336-6. [DOI] [PubMed] [Google Scholar]
- 42.Rorvik L.M., Aase B., Alvestad T., Caugant D.A. Molecular epidemiological survey of Listeria monocytogenes in seafoods and seafood-processing plants. Appl. Environ. Microbiol. 2000;66(11):4779–4784. doi: 10.1128/aem.66.11.4779-4784.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rorvik L.M., Caugant D.A., Yndestad M. Contamination pattern of Listeria monocytogenes and other Listeria spp. in a salmon slaughterhouse and smoked salmon processing plant. Int. J. Food Microbiol. 1995;25(1):19–27. doi: 10.1016/0168-1605(94)00080-p. [DOI] [PubMed] [Google Scholar]
- 44.Rorvik L.M., Skjerve E., Knudsen B.R., Yndestad M. Risk factors for contamination of smoked salmon with Listeria monocytogenes during processing. Int. J. Food Microbiol. 1997;37(2-3):215–219. doi: 10.1016/s0168-1605(97)00057-3. [DOI] [PubMed] [Google Scholar]
- 45.Rorvik L.M., Yndestad M., Skjerve E. Growth of Listeria monocytogenes in vacuum-packed, smoked salmon, during storage at 4 degrees C. Int. J. Food Microbiol. 1991;14(2):111–117. doi: 10.1016/0168-1605(91)90097-9. [DOI] [PubMed] [Google Scholar]
- 46.Rorvik L.M. Listeria monocytogenes in the smoked salmon industry. Int. J. Food Microbiol. 2000;62(3):183–190. doi: 10.1016/s0168-1605(00)00334-2. [DOI] [PubMed] [Google Scholar]
- 47.Senczek D., Stephan R., Untermann F. Pulsed-field gel electrophoresis (PFGE) typing of Listeria strains isolated from a meat processing plant over a 2-year period. Int. J. Food Microbiol. 2000;62(1-2):155–159. doi: 10.1016/s0168-1605(00)00395-0. [DOI] [PubMed] [Google Scholar]
- 48.Tagg J.R., Dajani A.S., Wannamaker L.W. Bacteriocins of gram-positive bacteria. Bacteriol. Rev. 1976;40(3):722–756. doi: 10.1128/br.40.3.722-756.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thimothe J., Nightingale K.K., Gall K., Scott V.N., Wiedmann M. Tracking of Listeria monocytogenes in smoked fish processing plants. J. Food Prot. 2004;67(2):328–341. doi: 10.4315/0362-028x-67.2.328. [DOI] [PubMed] [Google Scholar]
- 50.Vaz-Velho M., Duarte G., McLauchlin J., Gibbs P. Characterization of Listeria monocytogenes isolated from production lines of fresh and cold-smoked fish. J. Appl. Microbiol. 2001;91(3):556–562. doi: 10.1046/j.1365-2672.2001.01425.x. [DOI] [PubMed] [Google Scholar]
- 51.Vedamuthu E.R., Raccach M., Glatz B.A., Seitz E.W., Reddy M.S. Acid-producing microorganisms. In: Vanderzant C., Splittstoesser D.F, editors. Compendium of methods for the microbiological examination of foods. Washington: APHA; 1992. pp. 225–238. [Google Scholar]
- 52.Vogel B.F., Jorgensen L.V., Ojeniyi B., Huss H.H., Gram L. Diversity of Listeria monocytogenes isolates from cold-smoked salmon produced in different smokehouses as assessed by Random Amplified Polymorphic DNA analyses. Int. J. Food Microbiol. 2001;65(1-2):83–92. doi: 10.1016/s0168-1605(00)00503-1. [DOI] [PubMed] [Google Scholar]


