Abstract
The most common bacterial mercury resistance mechanism is based on the reduction of Hg(II) to Hg0, which is dependent of the mercuric reductase enzyme (MerA) activity. The use of a 431 bp fragment of a conservative region of the mercuric reductase (merA) gene was applied as a molecular marker of this mechanism, allowing the identification of mercury resistant bacterial strains.
Keywords: merA gene, mercury resistant, bacteria, molecular marker
The contamination of aquatic environments by mercury (Hg) is of great concern because of its toxicity and ubiquity. Mercury levels have risen due to human activities, such as coal and petroleum burning , and industrial uses (e.g. lamps, catalysts, instruments, etc.), with a consequent release of mercury into the air, aquatic and terrestrial ecosystems. These activities can increase local mercury levels several thousand-fold above background (1,9,17). In aquatic environments, mercury can be naturally converted to its most toxic species, methylmercury a potent neurotoxin, which is bioaccumulated and biomagnified through the trophic chain (2,9).
Some bacteria developed a mercury resistance mechanism, based on a group of genes located in an operon (mer operon). It allows bacteria to detoxify Hg(II) into volatile metallic mercury through its enzymatic reduction (14,16,18,23,24). Bacterial mercury resistance has been found in a wide range of Gramnegative and Gram-positive bacteria. The operon varies in the number of genes present and is usually located on plasmids (3,5,8,19,25) and chromosomes (4,13,18,28); they are often components of transposons (5,11,15,20) and integrons (4,13,18) in a striking diversity of arrangements, often involving duplications and distributions of the enzymes, transporters or regulators among several replicons in one cell. Moreover, two major mer genes, the regulator merR and the merA, which codes the NADPH-dependent flavoenzyme mercuric reductase [E.C. 1.16.1.1] that is responsible for the reduction of Hg(II) to the volatile and low-toxicity elemental form, are each composed of discrete modules observed in paralogs with distinct but related roles in prokaryotic physiology. Most mercury resistance loci have some means of controlling the expression of the proteins involved in mercury resistance, the Hg(II) uptake and reduction system . This function is carried out by merR, a metal-specific activator/repressor of the mer operon that also encodes the merT, merP, and merA structural genes. These three components (transport, catalysis, and regulation) are the central functions of mercury resistance found in both Gram-negative and Grampositive eubacteria (3).
Recently, the microbial mercury resistance, dependent of MerA mechanism, has been used for the remediation of mercury contaminated wastewater (3,7,27). Therefore, the identification of Hg resistant strains is essential to the technological development of mercury bioremediation strategies.
The aim of our study was to determine a conservative region of the merA gene that could be used as a molecular marker of the mer operon, allowing the identification of mercury resistant bacterial strains.
Mercury resistance tests had been made using 100 bacterial strains, isolated from aquatic systems of the Amazon region, Brazil. Cells were grown in Luria-Bertani (LB) broth at 30°C for 24 hours. The growth in the slurries was assessed through optical density readings at 600 nm, and necessary dilutions were made to inoculate multi-wells plates according to the method described by Cursino et al. (6) with modifications. The cells were inoculated in agar nutritional medium containing mercury, with the help of a replicator. The mercury concentrations used varied from 0.5 μM Hg to 25 μM Hg, with 2.5 μM Hg intervals. The growth was evaluated after 24, 48 and 72 hours (12).
The alignment of the sequences, deposited in the GeneBank database until the 09/11/2005, that codes the mercuric reductase enzyme (MerA) in bacteria was analyzed using a matrix of similarity of the sequences. The BLASTn program was used with the initial searcher sequence of the partial sequence of the mercuric reductase enzyme, with length of 1141 base pairs (bp), for the Citrobacter freundii, access number AJ418049 (8). The alignment of the sequences, in which two regions with high similarity had been identified by visual approach in color key alignment score bigger or equal to 200, is shown in Fig. 1. Among the sequences deposited for the merA gene, 86% showed these consensus regions, suggesting that these regions are highly conserved within the merA gene. Therefore, they can be used for the development of a very specific molecular marker of the merA gene. The selected sequences were employed to design primers using the program available in Stanford Genomic Resources (21). The initial parameters were: Tm equal to 50 and 60°C, amount of CG 40%, and size of the primers between 18 and 25 nucleotides (nt). Primer F1merA hybridized with the Crick ribbon (3' – 5'), in position 251 the 268 nt of the gene merA, while primer F2merA hybridized with the Watson ribbon (5' – 3'), in position 676 up to 693 nt. The sequence of primers: Forward F1 merA – 5' TCGTGATGTTCGACCGCT 3' and Reverse F2 merA – 5’ TACTCCCGCCGTTTCCAAT 3’, 431 base pairs of merA fragment with an optimized PCR protocol.
Figure 1.

Demonstrative figure generated from the alignment of the sequences for bacterial merA gene, described by the pattern key alignment score.
Among the bacteria tested, nine strains showed the highest mercury resistance, they were able to grow in a medium containing 17.5 μM Hg (12). These strains were screened for the presence of the merA gene in the genomic and plasmidial DNA material. To test the specificity of the method, five bacterial strains sensitive to mercury were randomly selected and used as negative controls (Morganella morganii – CPqLMD 12, Serratia rubidea – CPqLMD 69, Proteus mirabilis – CPqLMD 70, Serratia marcescens - CPqLMD 63, Salmonella sp – CPqLMD 98). The standard strain Acinetobacter baumanni (provided by Dr. Nascimento from UFMG), in which the merA gene was shown previously, was used as positive control. The DNA extraction was made as described previously by Sambrook et al. (22) and submitted to PCR reaction. The best conditions of amplification reactions were performed in a total volume of 20 μL by using gradient thermal cycler (Mastercycler Gradient, Eppendorf, Hamburg, Germany). The PCR reaction mixture contained 1 U/μL Recombiant Taq polymerase (Invitrogen®), 1X Enzyme buffer, 200 μM of each dNTP (Invitrogen®), 1.25 mM MgCl2, 50 ng of DNA template and 0.5 μM of each primers. The optimized amplification conditions included a predesnaturation step at 94°C for 1 min; 30 cycles of 94°C for 1 min, 55°C for 1 min, and an extension step at 72°C for 1 min, and end extension at 72°C for 7 minutes. The PCR products were analyzed using gel electrophoresis (1.5% agarose gel, using 90 volts for 1 hour). The gel was dyed with ethidium bromide (0.5 mg/mL) and visualized in transilluminator, using a UV lamp at 365 nm.
Among the nine bacterial strains investigated for the presence of mer A gene, two strains showed it in genomic DNA: Enterobacter cloacae (CPqLMD 56) and Escherichia coli (CPqLMD 105); while it was detected in plasmid DNA of eight strains: Klebsiella pneumoniae (CPqLMD 47), Enterobacter cloacae (CPqLMD 56), Salmonella sp (CPqLMD 10), Provides rettgeri (CPqLMD 57), Escherichia coli (CPqLMD 105), Morganella morganii (CPqLMD 32), Provides rettgeri (CPqLMD 93), Morganella morganii (CPqLMD 40). Only one bacterial strain analyzed, Serratia marcescens (CPqLMD 79), did not show the target fragment either in the genomic or plasmid DNA (Fig. 2), suggesting an alternative Hg resistance mechanism as described elsewhere in the literature (3,10,11). The negative controls showed no amplification, while it was detected a fragment of 431 base pairs in the positive control (data not shown).
Figure 2.
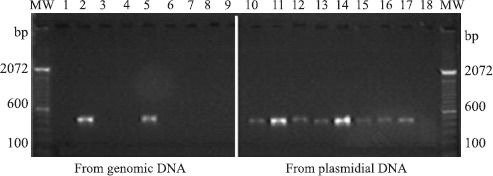
Amplification of443 pb merA fragment from mercury resistant strains using an optimized PCR protocol with primers F1 and F2 on an ethidium bromide-stained agarose gel. On the left hand (lines 1-9) samples from genomic DNA and on the right hand (lines 10-18) from plasmidial. Lines 2 and 11 – Enterobacter cloacae; Lines 5 and 14 – Escherichia coli; Lines 1 and 10 – Klebsiella pneumoniae; Lines 3 and 12 – Salmonella sp; Lines 4 and 13 – Providencia rettgeri; Lines 6 and 15 – Morganella morganii; Lines 7 and 16 – Providencia rettgeri; Lines 8 and 17 – Morganella morganii; Lines 9 and 18 – Serratia marcescens.
The presence of the merA gene, detected by PCR analysis in eight strains, was later confirmed by the measurement of the enzymatic activity of the MerA as described in Takeuchi et al. (11,24). While in the only strain, where merA gene was not detected by PCR, it was also not possible to measure mercuric reductase activity.
Therefore, the results demonstrated the effectiveness of our approach, where a molecular marker for the merA gene was developed, and used to identify mercury resistant bacterial strains. This is an essential step in the development of bioreactors, which has a potential use in the bioremediation of mercury contamination.
ACKNOWLEDGEMENTS
The authors are grateful to Centro de Pesquisa Leônidas e Maria Deane (CPqLMD) – FIOCRUZ for providing the infrastructure for this project. This research was funded by PAPES-FIOCRUZ Program, grant 400173/2006-9, and to Conselho Nacional de Pesquisa (CNPq).
RESUMO
Região Conservada do Gene da Mercúrio Redutase (merA) como Marcador Molecular da Resistência Bacteriana ao Mercúrio
O mecanismo de resistência bacteriana ao mercúrio mais comum é baseada na redução do Hg(II) a Hg0, através da atividade da enzima mercúrio redutase (MerA). O uso do fragmento de 431 pb amplificado de uma região conservada do gene merA, que codifica a enzima MerA, foi utilizado como marcador molecular deste mecanismo, permitindo a identificação de bactérias resistentes ao mercúrio.
Palavras-chaves:gene merA; resistência ao mercúrio; bactéria; marcador molecular
REFERENCES
- 1.Adamis P.D.B., Panek A.D., Leite S.G.F., Eleutherio E.C.A. Factors involved with cadmium absorption by a wild-type strain of Saccharomyces cerevisiae. Braz. J. Microbiol. 2003;34:55–60. [Google Scholar]
- 2.Barbosa A.C., Souza J.R. Contaminação por mercúrio e o caso da Amazônia. Química e Sociedade. 2000;12 [Google Scholar]
- 3.Barkay T., Miller S.M., Summers A.O. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 2003;27:355–384. doi: 10.1016/S0168-6445(03)00046-9. [DOI] [PubMed] [Google Scholar]
- 4.Bass L., Liebert C.A., Lee M.D., Summers A.O., White D.G., Thayer S.G., Maurer J.J. Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 1999;43:2925–2929. doi: 10.1128/aac.43.12.2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown N.L., Misra T.K., Winnie J.N., Schmidt A., Seiff M., Silver S. The nucleotide sequence of the mercuric resistance operons of plasmid R100 and transposon Tn501: further evidence for mer genes which enhance the activity of the mercuric ion detoxification system. Mol. Gen. Genet. 1986;202:143–151. doi: 10.1007/BF00330531. [DOI] [PubMed] [Google Scholar]
- 6.Cursino L., Oberda S.M., Cecilio R.V., Moreira R.M., Chartone-Souza, Nascimento A.M.A. Mercury concentration in the sediment at different gold prospecting sites along the Carmo stream, Minas Gerais, Brazil, and frequency of resistant bacteria in the respective aquatic communities. Hydrobiologia (The Hague) 1999;394:5–12. [Google Scholar]
- 7.Felske A.D.M., Fehr W., Pauling B.V., Canstein H., Wagner-Döbler I. Functional profiling of mercuric reductase (merA) genes in biofilm communities of a technical scale biocatalyzer. BMC Microbiol. 2003;3:22. doi: 10.1186/1471-2180-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffin F.G., Foster T.J., Silver S., Misra T.K. Cloning and DNA sequence of the mercuric and organomercurial resistance determinats of plasmid pDU1358. Proc. Nal. Acad. Sci. 1987;84:3112–3116. doi: 10.1073/pnas.84.10.3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hacon S., Barrocas P.R.G., Vasconcellos A.C.S., Barcellos C., Wasserman J.C., Campos R.C., Ribeiro C., Azevedo-Carloni F.B. An overview of the mercury contamination research with emphasis on the Brazilian region. Cad. Saúde Pública. doi: 10.1590/s0102-311x2008000700003. (in press) [DOI] [PubMed] [Google Scholar]
- 10.Hobman J., Brown N. Bacterial mercury-resistance genes. Met. Ions Biol. Syst. 1997;34:527–568. [PubMed] [Google Scholar]
- 11.Inoue C., Sugawara K., Kusano T. The merR regulatory gene in Thiobacillus ferrooxidans is spaced apart from the mer structural genes. Mol. Microbiol. 1991;5(11):2707–2718. doi: 10.1111/j.1365-2958.1991.tb01979.x. [DOI] [PubMed] [Google Scholar]
- 12.Jesus M.S. Manaus, Amazonas, Brasil: CDD 589.95, 98p. (M. Sc. Dissertation. Programa Multi-Institucional em Biotecnologia, UFAM); 2007. Identificação e Caracterização de Bactérias Resistentes ao Mercúrio em Sistemas Aquáticos da Região Amazônica: uma nova proposta de biorremediação. [Google Scholar]
- 13.Kholodii G.Y., Yurieva O.V., Lomovskaya O.L., Gorlenko Z., Mindlin S.Z., Nikiforov V.G. Tn5053, a mercury resistance transposon with integron’s ends. J. Mol. Biol. 1993;230(4):1103–1107. doi: 10.1006/jmbi.1993.1228. [DOI] [PubMed] [Google Scholar]
- 14.Komura I., Izaki K. Mechanism of mercuric chloride resistance in microorganisms. I. Vaporization of a mercury compound from mercuric chloride by multiple drug resistance strain of Escherichia coli. J. Biochem. 1971;70:885–893. doi: 10.1093/oxfordjournals.jbchem.a129718. [DOI] [PubMed] [Google Scholar]
- 15.Mindlin S., Kholodii G., Gorlenko Z., Minakhina S., Minakhin L., Kalyaeva E., Kopteva A., Petrova M., Yurieva O., Nikiforov V. Mercury resistance transposons of Gram-negative environmental bacteria and their classification. Res. Microbiol. 2001;152:811–822. doi: 10.1016/s0923-2508(01)01265-7. [DOI] [PubMed] [Google Scholar]
- 16.Misra T.K. Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid. 1992;25:4–16. doi: 10.1016/0147-619x(92)90002-r. [DOI] [PubMed] [Google Scholar]
- 17.Nascimento M.A., Chartone-Souza E. Operon mer: Bacterial resistance to mercury and potential for bioremediation of contaminated environments. Genet. Mol. Res. 2003;2:92–101. [PubMed] [Google Scholar]
- 18.Osborn A.M., Bruce K.D., Strike , Ritchie D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997;19:239–262. doi: 10.1111/j.1574-6976.1997.tb00300.x. [DOI] [PubMed] [Google Scholar]
- 19.Otth L., Solís G., Wilson M., FernÁndez H. Susceptibility of Arcobacter butzleri to heavy metals. Braz. J. Microbiol. 2005;36:286–288. [Google Scholar]
- 20.Partridge S.R., Brown H.J., Stokes H.W., Hall R.M. Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 2001;45:1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saccharomyces Genomic Database. Gene/Seq. Resources (Design Primers). Standford Genomic Resources of Satanford University. Available at: http://genome-www.stanford.edu/. Accessed 9 November 2005.
- 22.Sambrook J., Fritsch E.F., Maniatis T. 2. Cold Spring Harbor, NY: Laboratory Press; 1989. Molecular Cloning – a laboratory manual. [Google Scholar]
- 23.Silver S. Bacterial resistances to toxic metal ions. Gene. 1996;179:9–19. doi: 10.1016/s0378-1119(96)00323-x. [DOI] [PubMed] [Google Scholar]
- 24.Summers A.O. Organization, expression and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 1986;4:607–634. doi: 10.1146/annurev.mi.40.100186.003135. [DOI] [PubMed] [Google Scholar]
- 25.Summers A.O., Silver S. Mercury resistance in a plasmid- bearing strain of Escherichia coli. J. Bacteriol. 1972;112:1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi F., Iwahori K., Kamimura K., Sugio T. Isolation and some properties of Thiobacillus ferrooxidans strains with differing levels of mercury resistance from natural environments. J. Biosci. Bioeng. 1999;88(4):387–392. doi: 10.1016/s1389-1723(99)80215-1. [DOI] [PubMed] [Google Scholar]
- 27.Wagner-Döbler I., von Canstein H., Li Y., Timmis K.N., Deckwer W.D. Removal of mercury from chemical wastewater by microorganisms in technical scale. Environ. Sci. Technol. 2000;34:4628–4634. [Google Scholar]
- 28.Wang Y., Moore M., Levinson H.S., Silver S., Walsh C., Mahler I. Nucleotide sequence of a chromosomal mercury resistance determinant from a Bacillus sp. with broad-spectrum mercury resistance. J. Bacteriol. 1989;171:83–92. doi: 10.1128/jb.171.1.83-92.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]


