Abstract
Fourteen strains of nitrogen-fixing bacteria were isolated from different agricultural plant species, including cassava, maize and sugarcane, using nitrogen-deprived selective isolation conditions. Ability to fix nitrogen was verified by the acetylene reduction assay. All potentially nitrogen-fixing strains tested showed positive hybridization signals with a nifH probe derived from Azospirillum brasilense. The strains were characterized by RAPD, ARDRA and 16S rDNA sequence analysis. RAPD analyses revealed 8 unique genotypes, the remaining 6 strains clustered into 3 RAPD groups, suggesting a clonal origin. ARDRA and 16S rDNA sequence analyses allowed the assignment of 13 strains to known groups of nitrogen-fixing bacteria, including organisms from the genera Azospirillum, Herbaspirillum, Pseudomonas and Enterobacteriaceae. Two strains were classified as Stenotrophomonas ssp. Molecular identification results from 16S rDNA analyses were also corroborated by morphological and biochemical data.
Keywords: endophytic bacteria, diazotrophs, selective isolation, molecular systematics
INTRODUCTION
Nitrogen-fixing bacteria are able to fix atmospheric nitrogen under different conditions: independently, in loose association with other organisms, or in strict symbiosis with them, such as in the Rhizobium-legume-plant symbiosis. The latter is the most efficient type of association between diazotrophic microorganisms and plants, and is of major importance for some agricultural practices, such as soybean crops in Brazil (Döbereiner, 1997).
Plant-interacting microorganisms can establish either mutualistic or pathogenic associations. Although the outcome is completely different, common molecular mechanisms that mediate communication between the interacting partners can be involved. Specifically, nitrogen-fixing bacterial symbionts of legume plants, collectively termed rhizobia, and phytopathogenic bacteria have adopted similar strategies and genetic traits to colonize, invade and establish a chronic infection in the plant host (Soto et al., 2006).
Besides leguminous plants, several economically important plant varieties are capable of developing associations with diazotrophic microorganisms (Döbereiner, 1997). Research in nitrogen-fixing bacteria associated with these plants increased mainly in the last two decades, after the work of Döbereiner on Azospirillum (Bashan, 1998; Baldani et al., 1997). Contribution of nitrogen to plant crops in nitrogen-deprived soils could be probably due to associations with endophytic diazotrophic bacteria, some of them yet unidentified (Döbereiner, 1997). In one study conducted in Brazil, it was demonstrated that some varieties of sugarcane can obtain a substantial amount of nitrogen via biological fixation (Urquiaga et al., 1992).
Bacterial colonization of internal tissues of healthy plants seems to be a very common phenomenon, including several species of non-diazotrophic bacteria and a wide range of plant species (Azevedo, 1998; Reinhold-Hurek & Hurek, 1998). Microaerophilic bacteria, such as Azospirillum spp., Burkholderia spp., and Herbaspirillum spp., colonize roots, shoots and leaves of maize, Pennisetum, rice and wheat (Döbereiner, 1997). Other facultative anaerobic species, including Citrobacter, Enterobacter, Erwinia, and Klebsiella, may establish associations with grasses and some strains are able to fix nitrogen (Eady, 1992). Nitrogen-fixing strains have also been reported for Acetobacter, Azotobacter, Campylobacter, and Pseudomonas (Döbereiner, 1989). However, studies are still in progress to determine whether bacteria occurring within plants can directly contribute with fixed nitrogen to their host.
Taxonomic identification of several unknown nitrogen-fixing organisms can be accomplished through sequencing of the nifH gene, which is also useful to analyze their genetic potential for the nitrogen fixation (Zehr et al., 1995). NifH genes can be employed as markers for the detection and study of the genetic diversity of diazotrophic organisms in microbial communities, like those in rice roots (Ueda et al., 1995) or forest soil (Widmer et al., 1999). Putative nitrogenase amino acid sequences revealed that more than half of the nifH products were derived from methylotrophic bacteria, such as Methylocella spp. The next most frequent sequence types were similar to those from Burkholderia (Izumi et al., 2006).
In this study, several bacterial strains were isolated from different plant sources and characterized by using phenotypic and genotypic methods in order to assess their taxonomic diversity and investigate their ability to fix atmospheric nitrogen and the occurrence of nifH-like genes.
MATERIAL AND METHODS
Isolation and phenotypic characterization of nitrogen-fixing bacteria
Bacteria were isolated from agricultural plants collected in rural areas close to the city of São Paulo, São Paulo State, Brazil. For the isolation, leaves and/or roots were externally decontaminated and macerated in sterile saline. Aliquots of macerated plant material were inoculated into selective semi-solid NFb medium (Hartmann and Baldani, 2006) and incubated at 30°C for 10 days. All primary cultures with positive growth were tested for nitrogen fixation by the acetylene reduction test (Turner & Gibson, 1980) after 1 or 24 hours incubation in the presence of acetylene. Diazotrophic strains were then isolated in pure culture by dilution plating, re-tested, and the nitrogen-fixing organisms selected for further study.
Strains were characterized by Gram staining, colony morphology and motility. Biochemical tests were performed by using API 20E and API 20NE strips (bioMerieux) and additional phenotypical tests were performed as described by Holt et al., (1994), Hartmann and Baldani (2006), and Schmid et al., (2006).
DNA extraction
Bacterial cultures were grown in LB broth at 30ºC for 18 h. Cells were pelleted for 20 min at 8000 ×g, resuspended in 2 mL of TE-lysozyme (50 mM Tris, pH 8.0; 50 mM EDTA; 1 mg/mL lysozyme), and incubated for 30 min at 37ºC. After lysozyme digestion, 4.5 mL of extraction buffer (100 mM Tris, pH 8.0; 100 mM EDTA; 1.5 M NaCl; 1% CTAB) and 22.5 μL of proteinase K (20 mg/mL) were added to the samples, which were then incubated for 1 h at 37ºC. After this step, 0.5 mL of 20% SDS and 100 μL of 5 M potassium acetate were added and samples were incubated at 65ºC for 20 min. The final procedures for DNA extraction and purification were done according to Bando et al. (1998).
Random Amplified Polymorphic DNA (RAPD) analysis
RAPD screening was performed using primers OPA 07, OPC 02, OPK 20, OPP 06, OPP 12 and OPL-14 (Operon Technologies, Alameda, CA). Reaction mixtures (25 μL) were prepared as follows: 30 ng of DNA, 1 X Taq buffer, 3.5 mM MgCl2, 200 μM each deoxynucleoside triphosphates, 0.4 μM primer, and 2 U Taq polymerase (Gibco BRL). Amplifications were performed in a MJ PTC-100 thermocycler using the following cycling program: initial denaturation at 94ºC for 5 min, followed by 40 rounds at 94ºC for 1 min, 35ºC for 1 min and 72ºC for 2 min., followed by a final extension at 72ºC for 7 min. Electrophoresis of RAPD products (10 μL) was carried on 1,4% agarose gels in 1 X TBE (0,1 M Tris, pH 8.3; 0,09 M boric acid; 0, 1 mM EDTA) at 110 V for 1h 30min. Gels were stained with ethidium bromide prior to image capture.
Amplified Ribosomal DNA Restriction Analysis (ARDRA)
Amplification of 16S rDNA was performed using 30 ng of DNA in 25 μL reactions containing 2 mM MgCl2, 20 μM each deoxynucleoside triphosphates, 0.3 μM each of primers 27f and 1525r (Stackebrandt and Goodfellow, 1991) and 2U Taq polymerase (Life Technologies) in 1 X Taq buffer. The reaction mixtures were incubated in a MJ PTC-100 thermocycler at 94ºC for 2 min and then cycled 30 times: 94ºC for 1 min, 55ºC for 1 min and 72ºC for 3 min. A final extension at 72ºC for 10 min was used.
A 5 μL aliquot of each PCR reaction was incubated with 3–5 units of one of the following restriction enzymes: Alu I, Hae III, Hha I, or Msp I at 37ºC or Taq I at 65ºC for 1h. The restriction products were then analysed by electrophoresis on 1, 5% agarose in 1X TBE at 110V for about 2h 30 min and visualized as described before.
In the analysis of ARDRA patterns, bands with the same gel mobility were considered equivalent, independent of their relative intensity. Only bands between 120 bp and 900 bp were considered for analysis. The results of the separate restriction profiles were combined (appended) into a single dataset and analysed by using the Jaccard similarity coefficient and UPGMA cluster analysis (NTSys-PC software package, Rohlf, 1992).
16S rDNA sequencing and phylogenetic analysis
Fragments of 16S rDNA obtained as described above were subjected to automated sequencing. PCR products were extracted once with chloroform and precipitated with 1 volume of isopropanol and 1/10 volume of LiCl 4 M. The DNA was centrifuged at 9500 ×g for 15 min, washed with 70% ethanol, dried and ressuspended in TE buffer (10 mM Tris, pH 8.0; 1 mM EDTA). Sequencing reactions were performed by using the “Thermo Sequenase fluorescent labelled primer cycle sequencing with 7-deaza-dGTP” kit (Amersham Pharmacia Biotech), according to the manufacturer’s instructions. The primers used in the sequencing reactions were p1659 (5’ CTGCTGCCTCCCGTAGGAGT 3’), 782r (5’ ACCAGGGTA TCTAATCCTGT 3’), 530f (5’ CAGCAGCCGCGGTAATAC 3’), and MG5f (5’ AAACTCAAAGGAATTGACGG 3’). Reaction products were analysed in an ALFexpress (Amersham Pharmacia Biotech) sequencer. Obtained sequences covered the totality of the amplified 16S rDNA fragments.
Sequences with high homology scores were retrieved from Genbank and RDP for phylogenetic analyses. Sequences were aligned using RDP alignment templates by using the GDE software package (Genetic Data Environment, v.2.2; gopher://megasun.bch.umontreal.ca:70/11/GDE). Distance matrices were calculated using DNADIST and the Jukes-Cantor model (Jukes & Cantor, 1969), as implemented in PHYLIP v. 3.5 (Felsenstein, 1989). A phylogenetic tree was constructed using the Neighbor-Joining method (Saitou & Nei, 1987), included in the PHYLIP package.
Detection of nifH genes by dot-blot hybridisation
Dot-blot hybridisation assays were carried out using standard protocols described elsewhere (Hybond N+, ECL System, Amersham Pharmacia Biotech). A fragment of 705 bp corresponding to the initial 5’-end of Azospirillum brasilense Sp7 nifH gene was amplified by PCR using specific primers PPf (5’ GCAAGTCCACCACCTCC 3’) and PPr (5’ TCGC GTGGACCTTGTTG 3’). PCR conditions comprised: 30 ng of DNA in 25 μL reactions containing 1, 5 mM MgCl2, 200 μM each deoxynucleoside triphosphates, 0.5 μM each primer, and 1,5 U Taq polymerase (Life Technologies) in 1 X Taq buffer. Amplification was performed with the following cycling program: initial denaturation at 94ºC for 3 min followed by 30 rounds of 94ºC for 30 s, 58ºC for 30 s and 72ºC for 45 s. A final extension of 72ºC for 7 min. was used.
The nifH gene fragments were labelled by using the horseradish peroxidase kit (ECL System, Amersham Pharmacia Biotech) and used as a probe in dot blot hybridisations. DNA preparations from strains COL, MANC, MAGDE3 and MI753 were spotted onto Hybond N+ membranes and these were hybridised and detected according to the manufacturers instructions (ECL System, Amersham Pharmacia Biotech). High stringency hybridisation conditions were used in the assays.
Hybridisation signals were detected by exposure to X Omat X-ray films.
RESULTS AND DISCUSSION
Isolation of nitrogen-fixing bacteria and detection of nifH-related gene sequences
Several bacteria were isolated from cassava (MAX1, MAX2, MAP1A, MANR, MANC and MAGDE3), guinea grass (Panicum maximum Jacq.) (COL), maize (MI753 and MIS), sugarcane (CAN9B, CANRA, CANRB, CANF3) and tomato (TOM), by using the selective media NFb described by Hartmann and Baldani (2006). These strains reduced acetylene on the chromatographic analyses performed, hence indicating their nitrogen fixation capability. However, because MANC, MAGDE3 and MI753 didn’t show any results after the 1-hour incubation period and a low ethylene production after the 24-hours period, a dot blot hybridization with a nifH probe was used to obtain further indications of their potential to fix nitrogen.
Dot blot hybridization (Fig. 1) revealed the presence of nifH-related sequences in DNA from strains COL, MANC, MAGDE3 and MI753 under the high stringency hybridization conditions used in the assays. The negative controls used, namely E. coli and human DNA, did not show any hybridization signal with the nifH probe, demonstrating the specificity of the used probe. Positive results in dot blot hybridization for strains MANC, MAGDE3 and MI753 corroborate their ability for nitrogen fixation, suggested by the acetylene reduction assay.
Figure 1.
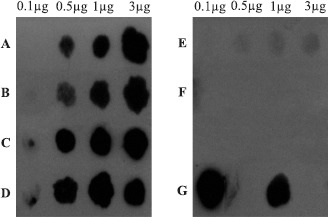
Dot blot hybridization using the nifH probe and genomic DNA (0.1–3.0 mg) from A) COL; B) MANC; C) MAGDE3; D) MI753; E) E. coli (negative control); F) human DNA (negative control). In G: 0.1 mg of a plasmid containing A. brasilense Sp7 nifH gene (left spot) and 1mg of genomic DNA from Azospirillum brasilense Sp7 (right spot).
RAPD analysis
RAPD analyses are useful for differentiating bacteria at the strain level (Tiedje, 1996). Fig. 2 shows the RAPD patterns generated with primer OPC 02. The results of all RAPD analyses are summarized in Table 1.
Figure 2.
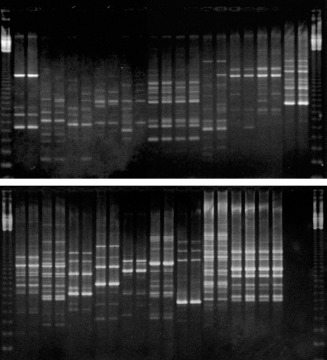
RAPD electrophoretic patterns obtained with primer OPC 02. In the upper lanes, from left to right: 1 kb ladder (Gibco BRL), Azospirillum lipoferum BR17, CANRA, Azospirillum brasilense Sp7, MAX1, MAX2, MANR, MAP1A, TOM, MANC, MAGDE3, Beijerinckia sp. strain, 1 kb ladder. In the lower lanes, from left to right: Rhizobium meliloti strain, Herbaspirillum rubrisubalbicans M4, Herbaspirillum seropedicae Z67, COL, MI753, MIS, Burkholderia brasilensis M130, CAN9B, CANRB, CANF3, blank sample, 1kb ladder. All samples are in duplicate.
Table 1.
Results of genotypic analyses of the isolated nitrogen-fixing bacteria.
| Strain | Plant source and tissue | RAPD patterns | ARDRA group | Putative species assignment based on 16S rDNA data |
|---|---|---|---|---|
| COL | forage grass; leaf | unique | B | Herbaspirillum rubrisubalbicansb |
| CAN9B | sugarcane; leaf | unique | D | Enterobacteriaceaeb |
| CANRB | sugarcane; root | identical to CANF3 | D | Enterobacteriaceaeb |
| CANF3 | sugarcane; leaf | identical to CANRB | D | Enterobacteriaceaeb |
| MANC | cassava; leaf | identical to MAGDE3 | C | Stenotrophomonas sp.b |
| MAGDE3 | cassava; root | identical to MANC | C | Stenotrophomonas sp.b |
| MIS | maize; leaf | unique | N.a. | Pseudomonas stutzerib |
| CANRA | sugarcane; root | unique | N.a. | Azospirillum lipoferuma |
| TOM | tomato; leaf | unique | A | Azospirillum brasilensea |
| MAX1 | cassava; root | identical to MAX2 | A | Azospirillum brasilensea |
| MAX2 | cassava; root | identical to MAX1 | A | Azospirillum brasilensea |
| MANR | cassava; leaf | unique | A | Azospirillum brasilensea |
| MAP1A | cassava; root | unique | A | Azospirillum brasilensea |
| MI753 | maize; leaf | unique | N.a. | Acidovorax related |
Obtained by ARDRA analysis;
Obtained by 16S rDNA sequencing. N.a. = not assigned.
Using a combination of 6 different primers, it was possible to differentiate 8 out of the 14 bacterial strains under study and the clustering of the remaining 6 strains into 3 distinct groups. RAPD patterns of these 8 strains and of the 3 groups were unique and, due to their great variability, it was not feasible to consistently group them further. Nevertheless, this grouping was later allowed by ARDRA patterns.
The remaining 6 strains clustered into 3 distinct groups, each comprised by strains that yielded identical RAPD patterns with all primers tested: cluster 1) MANC, MAGDE3; 2) MAX1, MAX2; and 3) CANRB, CANF3. The occurrence of strains with identical RAPD patterns suggests that they share a common clonal origin. In one instance, strains with identical RAPD patterns were isolated from cassava plants collected at the same place (clusters 1 and 2). In another situation, identical strains (CANRB and CANF3) were isolated from the leaves and roots of the same sugarcane plant. In the latter case, it is reasonable to suppose that these strains are endophytic to the host plant.
ARDRA
ARDRA was proved to be useful for the classification of bacterial strains at different taxonomic levels, depending on selection of conserved or variable regions in the ribosomal genes for the analysis (Swings, 1996; Tiedje, 1996). ARDRA results (Fig. 3) revealed that some of the nitrogen-fixing strains tested were similar, or identical, to reference strains described in the literature. ARDRA groups (Fig. 4), defined by similarities in ARDRA patterns were comprised by: cluster A) Azospirillum brasilense Sp7, MAX1, MAX2, MAPIA, MANR, TOM; cluster B) COL, Herbaspirillum seropedicae Z67, and Herbaspirillum rubrisubalbicans M4; cluster C) MAGDE3 and MANC; and cluster D) CAN9B, CANRB, and CANF3.
Figure 3.
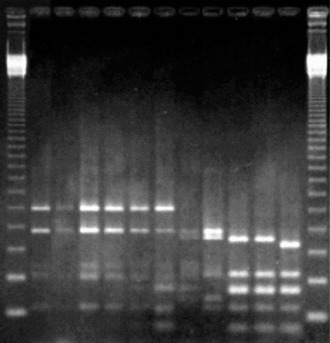
Agarose gel electrophoresis of amplified 16S rDNA digested with restriction endonuclease Alu I. From left to right: 123 bp ladder (Gibco BRL), Azospirillum brasilense Sp7, TOM, MAX1, MAX2, MAP1A, MANR, Azospirillum lipoferum BR17, CANRA, Herbaspirillum seropedicae Z67, COL, Herbaspirillum rubrisubalbicans M4, 123 bp ladder (Gibco BRL).
Figure 4.
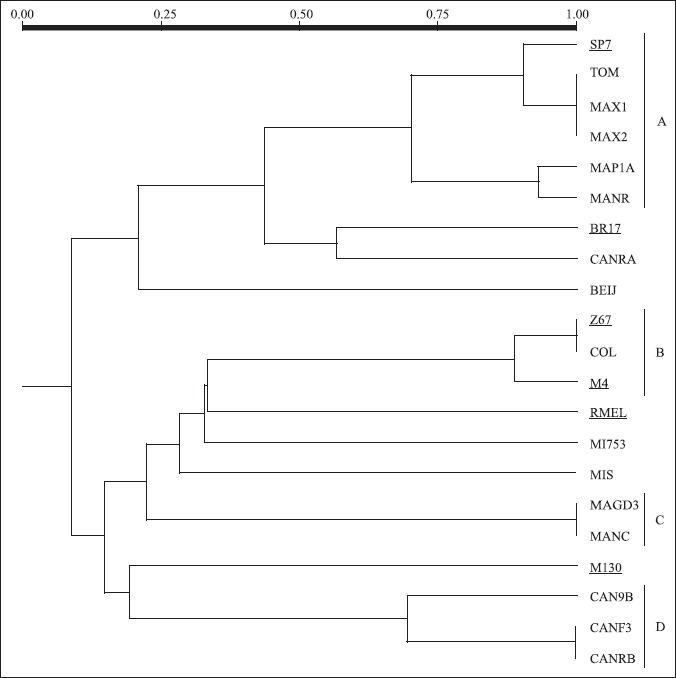
Dendrogram based on UPGMA cluster analysis of Jaccard coefficients obtained for the combined ARDRA restriction profiles.
ARDRA patterns of strains A. brasilense Sp7 and A. lipoferum BR17 are consistent with those obtained by Grifoni et al. (1995) and Han and New (1998), using the enzyme Alu I. In agreement with those authors, Alu I digestion yields patterns which are useful for the rapid and reliable identification of different species of Azospirillum. Alu I patterns of MAX1, MAX2, MANR, MAP1A, and TOM were similar to that of Azospirillum brasilense, whereas the pattern of strain CANRA was similar to that of Azospirillum lipoferum. These genotypic results were further corroborated by morphologic and physiologic properties of these isolates, consistent with the description of the two species (Hartmann and Baldani, 2006).
ARDRA patterns of strain COL are similar to those of strains assigned to the genus Herbaspirillum, suggesting its identification as either Herbaspirillum seropedicae or Herbaspirillum rubrisubalbicans M4. Assignment to H. rubrisubalbicans was later confirmed by 16S rDNA sequence analysis, though this strain was able to use N-acetylglucosamine as the sole carbon source, a characteristic of H. seropedicae (Baldani et al., 1997b).
Phylogenetic analysis of 16S rDNA sequences
Strains COL, CAN9B, CANF3, MAGDE3, MIS, and MI753 were subjected to 16S rDNA phylogenetic analysis. All sequences obtained in the current study were deposited in GenBank under acession numbers: AF214642, AF214639, AF214640, AF214643, AF214645, AF214644, respectively. Sequencing data were compared to bacterial sequences deposited at the RDP (Bonnie et al., 1999) and GenBank (Altshul et al., 1997) databases and sets of retrieved sequences were analysed as described previously.
Strain MAGDE3, which formed an ARDRA group with strain MANC with all enzymes tested, was grouped with several strains of Stenotrophomonas maltophilia and one strain of S. africana (Fig. 5). The similarity of RAPD patterns between these two strains (Table 1) suggests that both belong to Stenotrophomonas, which was further confirmed by phenotypic data obtained by using the API 20NE kit.
Figure 5.
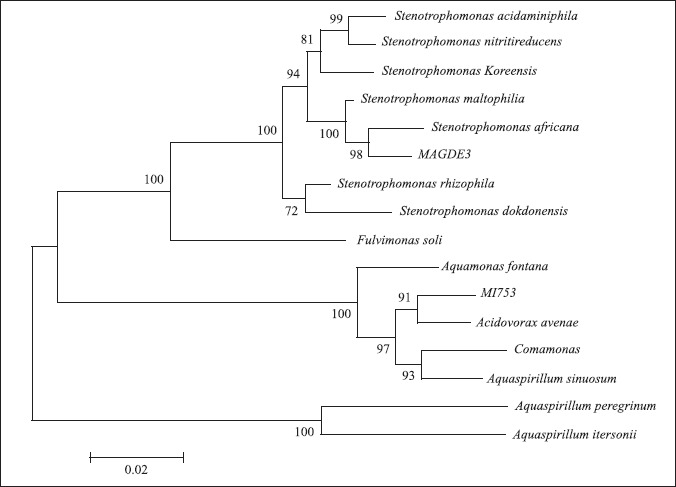
Phylogenetic tree obtained for strains MAGDE3 and MI753 using the Neighbor-Joining method and p-distance, based on their 16S rDNA sequence. Number indicate the results of the bootstrap analysis with 1000 replicates.
S. maltophilia is very common in a range of environments, and is usually referred as an agent of nosocomial infections, representing an opportunistic pathogen to humans. It is also found as a growth promoting bacteria in the rhizosphere of several different plant species (Hauben et al., 1999). Stenotrophomonas maltophilia isolates display intrinsic resistance to many commonly prescribed antimicrobials, particularly β-lactams and aminoglycosides. They can also evolve broad spectrum resistance to a cross section of other drugs that have been used to treat infections (Denton and Kerr, 1998).
However, we could not find any report in the literature related to endophytic Stenotrophomonas strains, as described in the current study and supported by RAPD data.
The ability of Stenotrophomonas MAGDE3 and MANC to fix atmospheric nitrogen was suggested through acetylene reduction test and corroborated by detection of homologous nifH gene sequences in these organisms (Fig. 1). In accordance with Liba et al., (2006), nitrogen fixation ability was a new character not previously reported for Stenotrophomonas, and only one publication refers to a Stenotrophomonas-like strain able to fix atmospheric nitrogen (Elo and Haahtela, 1999).
The analysis of 16S rDNA sequences from CAN9B and CANF3 positioned these strains among organisms from the Enterobacteriaceae family, but a more precise identification was not possible due to the lack of resolution of the 16S rDNA for the differentiation of species in this group of organisms. Biochemical data generated by using API 20E kit suggested the assignment of these two organisms to Enterobacter agglomerans, but the agreement was only acceptable. More analyses are necessary for the taxonomic assignment of CAN9B, CANF3, and CANRB. However, it is well known that many members of the Enterobacteriaceae are capable of fixing nitrogen. Furthermore, occurrence of Pantoea herbicola (former Enterobacter agglomerans, Beij et al., 1988) was reported in roots, shoots and dry leaves of sugarcane (Gracioli et al., 1986). This species is an endophyte of beet, associated to the roots (Jacobs et al., 1985). Other related genera, such as Klebsiella, also have endophytic representatives (Azevedo, 1998). In this study, the isolation of Enterobacteriaceae-related organisms is in agreement with these findings.
Analysis of the 16S rDNA sequence of strain MIS identified the strain as Pseudomonas stutzeri, species for which nitrogen fixation (Andrade et al., 1997; Puente and Bashan, 1994) and endophytism were already observed (Puente and Bashan, 1994). Biochemical results obtained using the API 20NE kit agreed with the molecular identification.
In the phylogenetic tree (Fig. 5) strain MI753 groups with Acidovorax avenae, but with low similarity, and phenotypic results are inconclusive. So additional characterization of this strain is necessary to confirm the identification. Although it was not showed that Acidovorax avenae possess nitrogen-fixing capabilities, these were demonstrated for some Aquaspirillum species, which also groups with this strain. Because the taxonomy of strains that seem to be related to MI753 is not well established yet, it is possible that this strain can be reassigned to another species or genera if further studied.
In this paper, we described the isolation and molecular identification of several nitrogen-fixing strains in association with different plant species. Isolation of strains belonging to well characterized diazotrophic taxa, such as Azospirillum spp. and Herbaspirillum spp., demonstrated the adequacy of the methods employed. In addition, RAPD characterization of some isolates yielded data to support the occurrence of clonal strains in different organs of the plant and the putative endophytic nature of these isolates. Taxonomic identification of several strains could be successfully achieved by ARDRA and by phylogenetic analysis of 16S rDNA sequence data.
Isolation of some unusual strains is suggestive that the biodiversity of plant-associated microorganisms is yet poorly explored. This was demonstrated by the isolation of a putative nitrogen-fixing Stenotrophomonas and Enterobacteriaceae-related strains that couldn’t be identified to known species.
ACKNOWLEDGEMENTS
ELR received a fellowship from FAPESP (96/12279–2). We thank Dr. Marie-Anne van Sluys, Department of Botany, University of São Paulo, for valuable help and suggestions on DNA sequencing.
RESUMO
Caracterização molecular de bactérias fixadoras de nitrogênio isoladas de plantas brasileiras no estado de São Paulo
Quatorze linhagens de bactérias fixadoras de nitrogênio foram isoladas de diferentes espécies de plantas, incluindo cassava, milho e cana-de-açúcar, usando condições seletivas desprovidas de nitrogênio. A capacidade de fixar nitrogênio foi verificada por ensaio de redução de acetileno. Todas as linhagens fixadoras de nitrogênio testadas apresentaram hibridização positiva com sonda de gene nifH derivada de Azospirillum brasilense. As linhagens foram caracterizadas por RAPD, ARDRA e sequenciamento do gene 16S rDNA. As análises de RAPD revelaram 8 genótipos, as 6 linhagens restantes foram agrupadas em 3 grupos de RAPD, sugerindo uma origem clonal. ARDRA e seqüências de 16S rDNA foram alocadas em 13 grupos conhecidos de bactérias fixadoras de nitrogênio, incluindo organismos dos gêneros Azospirillum, Herbaspirillum, Pseudomonas e Enterobacteriaceae. Duas linhagens foram classificadas como Stenotrophomonas ssp. Os resultados da identificação molecular baseados em sequencias de 16S rDNA corroboram com dados obtidos em testes morfológicos e bioquímicos.
Palavras-chave: bactéria endofítica, diazotróficas, isolamento seletivo, sistemática molecular.
REFERENCES
- 1.Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade G., Esteban E., Velasco L., Lorite M.J., Bedmar E.J. Isolation and identification of N2-fixing microorganisms from the rhizosphere of Capparis spinosa (L.) Plant Soil. 1997;197:19–23. [Google Scholar]
- 3.Azevedo J.L. Microorganismos endofíticos. Ecologia Microbiana. In: Melo I.S, Azevedo J.L, editors. Jaguariúna: Embrapa-CNPMA; 1998. pp. 117–137. [Google Scholar]
- 4.Baldani J.I., Caruso L., Baldani V.L.D., Goi S.R., Döbereiner J. Recent Advances in BNF with non-legume plants. Soil Biol. Biochem. 1997;29:911–922. [Google Scholar]
- 5.Baldani V.L.D., Reis V.M., Baldani J.I., Kimura O., Döbereiner J. Seropédica: Embrapa-CNPAB; 1997. Bactérias fitopatogênicas fixadoras de N2 em associação com plantas; p. 25. 25. [Google Scholar]
- 6.Bashan Y. Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol. Adv. 1998;16:729–770. [Google Scholar]
- 7.Beij A., Mergaert J., Gavini F., Izard D., Kersters K., Leclerc H., De Ley J. Subjective synonymy of Erwinia herbicola, Erwinia milletiae and Enterobacter agglomerans and redefinition of the taxon by genotypic and phenotypic data. Int. J. Syst. Bacteriol. 1988;38:77–88. [Google Scholar]
- 8.Bando S.Y., Valle G.R.F., Martinez M.B., Trabulsi L.R., Moreira-Filho C.A. Characterization of enteroinvasive Escherichia coli and Shigella strains by RAPD analysis. FEMS Microbiol. Lett. 1998;165:159–165. doi: 10.1111/j.1574-6968.1998.tb13141.x. [DOI] [PubMed] [Google Scholar]
- 9.Denton M., Kerr K.G. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 1998;11:57–80. doi: 10.1128/cmr.11.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Döbereiner J. Biological nitrogen fixation in the tropics: social and economic contributions. Soil Biol. Biochem. 1997;29:771–774. [Google Scholar]
- 11.Dõbereiner J. São Paulo: IEA/USP; 1989. Avanços recentes na pesquisa em fixação biológica de nitrogênio no Brasil; p. 23. [Google Scholar]
- 12.Eady R.R. Vol. 1. New York: 1992. The dinitrogen-fixing bacteria. In: The Prokaryotes, Springer-Verlag; pp. 534–553. [Google Scholar]
- 13.Elo S., Haahtela K. Nitrogen-fixing bacteria isolated from forest soils in Finland. In: Current plant science and biotechnology in agriculture. In: Pedrosa F.O, Hungria M., Yates M.G, Newton W.E, editors. Vol. 38. Dordrecht: Kluwer Academic Publishers; 1999. p. 190. [Google Scholar]
- 14.Felsenstein J. Phylip-phylogeny inference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 15.Gracioli L.A., Freitas J.R., Ruschel A.P. Bactérias fixadoras de nitrogênio nas raízes, colmos e folhas de cana-de-açúcar (Saccharum sp.) Rev. Microbiol. 1986;14:191–196. [Google Scholar]
- 16.Grifoni A., Bazzicalupo M., Di Serio C., Fancelli S., Fani R. Identification of Azospirillum strains by restriction fragment length polymorphism of the 16S rDNA and of the histidine operon. FEMS Microbiol. Lett. 1995;127:85–91. doi: 10.1111/j.1574-6968.1995.tb07454.x. [DOI] [PubMed] [Google Scholar]
- 17.Hartmann A., Baldani J.I. Vol. 5. New York: 2006. The Genus Azospirillum. In: The Prokaryotes, Springer; pp. 115–140. [Google Scholar]
- 18.Holt J.G., Krieg N.R., Sneath P.H.A., Staley J.T., Williams S.T. Baltimore: Williams & Wilkins; 1994. Bergey’s manual of determinative bacteriology; p. 787. [Google Scholar]
- 19.HAN S.O., NEW P.B. Variation in nitrogen fixing ability among natural isolates of Azospirillum. Microb. Ecol. 1998;36:193–201. doi: 10.1007/s002489900106. [DOI] [PubMed] [Google Scholar]
- 20.Hauben L., Vauterin L., Moore E.R.B., Hoste B., Swings J. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 1999;49:1749–1760. doi: 10.1099/00207713-49-4-1749. [DOI] [PubMed] [Google Scholar]
- 21.Izumi H., Anderson I.C., Alexander I.J., Killham K., Moore E.R.B. Diversity and expression of nitrogenase genes (nifH) from ectomycorrhizas of Corsican pine (Pinus nigra) Environ. Microbiol. 2006;8(12):2224–2230. doi: 10.1111/j.1462-2920.2006.01104.x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobs M.J., Bugbee W.M., Gabrielson D.A. Enumeration, location and characterization of endophytic bacteria within sugar beet. Can. J. Botany. 1985;63:1262–1265. [Google Scholar]
- 22.Jukes T.H., Cantor R.R. Munro H.N. New York: Academic Press; 1969. Evolution of protein molecules. In: Mammalian protein metabolism; pp. 21–132. [Google Scholar]
- 23.Liba C.M., Ferrara F.I.S., Manfio G.P., Fantinatti-Garboggini F., Albuquerque R.C., Pavan C., Ramos P.L., Moreira-Filho C.A., Barbosa H.R. Nitrogen-fixing chemo-organotrophic bacteria isolated from cyanobacteria-deprived lichens and their ability to solubilize phosphate and to release amino acids and phytohormones. J. Appl. Microbiol. 2006;101(5):1076–86. doi: 10.1111/j.1365-2672.2006.03010.x. [DOI] [PubMed] [Google Scholar]
- 24.Maidak B.L., Cole J.R., Lilburn T.G., Parker Jr C.T., Saxman P.R., Stredwick J.M., Garrity G.M., Li B., Olsen G.J., Pramanik S., Schmidt T.M., Tiedje J.M. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 2000;28:173–174. doi: 10.1093/nar/28.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puente M.E., Bashan Y. The desert epiphyte Tillandsia recurvata harbours the nitrogen-fixing bacterium Pseudomonas stutzeri. Can. J. Botany. 1994;72:406–408. [Google Scholar]
- 26.Reinhold-Hurek B., Hurek T. Life in grasses: diazotrophic endophytes. Trends Microbiol. 1998;4:139–44. doi: 10.1016/s0966-842x(98)01229-3. [DOI] [PubMed] [Google Scholar]
- 27.Rohlf F.J. New York: Exeter Software; 1992. NTSYS-pc numerical taxonomy and multivariate analysis system. [Google Scholar]
- 28.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 29.Schmid M., Baldani J.I., Hartmann A. Vol. 5. New York: 2006. The Genus Herbaspirillum. n: The Prokaryotes, Springer; pp. 141–150. [Google Scholar]
- 30.Soto M.J., Sanjuan J., Olivares J. Rhizobia and plant-pathogenic bacteria commom infections weapons. Microbiology. 2006;152(11):3167–74. doi: 10.1099/mic.0.29112-0. [DOI] [PubMed] [Google Scholar]
- 31.Stackebrandt E., Goodfellow M. Chichester: John Wiley & Sons; 1991. Nucleic acid techniques in bacterial systematics; p. 329. [Google Scholar]
- 32.Swings J. Exploration of prokaryotic diversity employing taxonomy. In Microbial diversity and ecosystem function. In: Allsopp D, Colwell R.R, Hawksworth D.L, editors. Egham: CAB International; 1996. p. 482. [Google Scholar]
- 33.Tiedje J.M. Approaches to the comprehensive evaluation of prokaryote diversity of a habitat. In: Microbial diversity and ecosystem function. In: Allsopp D, Colwell R.R, Hawksworth D.L, editors. Egham: CAB International; 1996. p. 482. [Google Scholar]
- 34.Turner G.L., Gibson A.H. London: John Wiley & Sons; 1980. Measurement of nitrogen fixation by indirect means. In: Methods for evaluating biological nitrogen fixation; pp. 111–139. [Google Scholar]
- 35.Ueda T., Suga Y., Yahiro N., Matsuguchi T. Remarkable N2-fixing bacterial diversity detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 1995;177(5):1414–1417. doi: 10.1128/jb.177.5.1414-1417.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Urquiaga S., Cruz K.H.S., Boddey R.M. Contribution of nitrogen fixation to sugarcane: Nitrogen-15 and nitrogen balance estimates. Soil Sci. Soc. Am. J. 1992;56:105–114. [Google Scholar]
- 37.Widmer F., Shaffers B.T., Porteous L.A., Seidler R.J. Analysis of a nifH gene pool complexity in soil and litter at a Douglas Fir forest site in the Oregon cascade mountain range. Appl. Environ. Microbiol. 1999;65(2):374–380. doi: 10.1128/aem.65.2.374-380.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zehr J., Mellon M., Braun S., Litaker W., Steppe T., Paerl H. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 1995;61:2527–2532. doi: 10.1128/aem.61.7.2527-2532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]


