Abstract
Escherichia coli is one of the most important bacterial avian pathogens and a common inhabitant of the gastrointestinal tract of animals. Most pathogenic E. coli can not be differentiated biochemically or by classic microbiologic methods. Molecular typing methods, particularly PCR, facilitated epidemiological and ecological studies of bacteria. Here we describe the application of a random amplified polymorphic DNA- polymerase chain reaction (RAPD-PCR) for molecular genetic differentiation of E. coli isolates in Iran. In this study 58 E. coli isolates including 4 standard strains, 3 food originated isolates, 33 avian isolates, 8 isolates form diarrheic calves and 10 isolates from unweaned diarrheic lambs were analyzed by RAPD-PCR using primer 1247(5/-AAG AGC CCG T-3/). The RAPD analysis showed that these isolates could be grouped into 33 RAPD types and avian isolates were discriminated into 29 genotypes. It was shown that the primer could not differentiate E. coli isolated from lambs. Discriminatory index for entire isolates was 0.912 and for avian isolates was 0.990. We concluded that RAPD-PCR can be used as a method for molecular differentiation of E. coli isolates.
Keywords: Escherichia coli, RAPD-PCR, Molecular differentiation, Iran
INTRODUCTION
Escherichia coli is one of the most common and important avian bacterial pathogens and infections caused by E. coli are responsible for significant economic losses to the poultry industry. E. coli also causes intestinal and extra intestinal diseases in domestic and wild animals which lead to severe economic losses throughout the world (7). Colisepticemia, coligranuloma (Hjarre’s disease), air sac disease, coliform salpingitis, coliform cellulites, swollen-head syndrome, coliform peritonitis, coliform osteomyelitis/synovitis and coliform omphalitis/yolk sac infection are the different forms of E. coli infections in poultry (1).
Serological and bacteriological methods are not sensitive enough to differentiate all bacterial isolates. Molecular differentiation of different E. coli strains may aid in differing those that are specifically pathogenic for a certain host and give guidance for epidemiological studies of sources of infection and disease transmission. Those techniques of molecular and genetic differentiation involve multilocus enzyme electrophoresis (MLEE) (18), restriction fragment length polymorphism (RFLP) (12), pulsed field gel electrophoresis (15) and random amplification of polymorphic DNA (4,12). RAPD-PCR is quicker and more effective procedure than RFLP to differentiate different isolates of E. coli (12). It was shown that this method also is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains (16). Random amplified polymorphic DNA-Polymerase Chain Reaction (RAPD-PCR) or arbitrary primed-PCR (AP-PCR) (17,19) has been useful in typing microbial isolates such as Brucella spp. (2), Leptosphaeria maculans (5), Histoplasma capsulatum (10), Cryptococcus neoformans (3) and Salmonella enteritidis (11).
In this study we have tried to show genetic diversity of E. coli isolates from different hosts and regions in Iran and the susceptibility of poultry flocks being infected by different avian pathogenic E. coli (APEC) genotypes.
MATERIALS AND METHODS
Isolation and identification of E. coli isolates
31 avian pathogenic bacteria were isolated from normally sterile sites (pericardium, heart and liver) of chickens from different poultry flocks and regions and 2 fecal isolates were isolated from cloaca of clinically normal birds. Bacteria from dissected animals were isolated from necropsy specimens and cultured on 5% sheep blood agar and MacConkey agar plates. After 24 hours incubation period at 37°C, the typical colonies were tested biochemically for common characteristics of E. coli . In addition 8 isolates from diarrheic calves, 10 isolates form week-old lambs, 3 food-borne E.coli isolates and 4 standard isolates were used to compare genetically with avian isolates (Table 1). All these bacteria were cultured on Luria-Bertani agar and incubated overnight at 37°C for DNA extraction procedure.
Table 1.
E. coliisolates, their locations and sources.
| Isolate | Location | Source | Isolate | Location | Source |
|---|---|---|---|---|---|
| 1 | Ardebil | Chicken | 30 | Qom | Chicken |
| 2 | Ardebil | Chicken | 31 | Qom | Chicken |
| 3 | Ardebil | Chicken | 32 | Tehran | Chicken |
| 4 | Ardebil | Chicken | 33 | Tehran | Chicken |
| 5 | Ardebil | Chicken | 34 | Tehran | Chicken |
| 6 | Ardebil | Chicken | 35 | Tehran | Chicken |
| 7 | Ardebil | Chicken | 36 | Tehran | Chicken |
| 8 | Ardebil | Chicken | 37 | Unknown | Calf |
| 9 | Garmsar | Chicken | 38 | Unknown | Calf |
| 10 | Ghazvin | Chicken | 39 | Unknown | Calf |
| 11 | Karaj | Chicken | 40 | Unknown | Calf |
| 12 | Karaj | Chicken | 41 | Unknown | Calf |
| 13 | Karaj | Chicken | 42 | Unknown | Calf |
| 14 | Karaj | Chicken | 43 | Unknown | Calf |
| 15 | Karaj | Chicken | 44 | Unknown | Calf |
| 16 | Karaj | Chicken | 45 | Qom | Lamb |
| 17 | Karaj | Chicken | 46 | Qom | Lamb |
| 18 | Karaj | Chicken | 47 | Qom | Lamb |
| 19 | Ardebil | Chicken | 48 | Qom | Lamb |
| 20 | Ardebil | Chicken | 49 | Qom | Lamb |
| 21 | Ardebil | Chicken | 50 | Qom | Lamb |
| 22 | Tehran | Food | 51 | Qom | Lamb |
| 23 | Tehran | Food | 52 | Qom | Lamb |
| 24 | Tehran | Food | 53 | Qom | Lamb |
| 25 | Qom | Chicken | 54 | Qom | Lamb |
| 26 | Qom | Chicken | 55 | ATCC1® 35218™ | Canine |
| 27 | Qom | Chicken | 56 | TM 295 | Human |
| 28 | Qom | Chicken | 57 | TM 296 | Human |
| 29 | Qom | Chicken | 58 | TM 297 | Human |
DNA extraction
Template DNA was prepared from bacteria grown overnight at 37°C on LB agar. Crude DNA extracts were obtained by suspending 4 colonies in 250 μl distilled water and boiling at 94–95°C for 15 minutes.
RAPD-PCR
Each polymerase chain reaction mixture consists of 3 μl of template DNA, 1 μl of 1.6 micromolar solution of primer oligonucleotide 1274, 5’-AAG AGC CCG-3’ (CinnaGen Inc., Tehran, Iran) (4,12), 2.5 μl 10X PCR buffer (CinnaGen Inc., Tehran, Iran), 2 mM MgCl2, 0.4 mM each nucleotides and 0.2 unit Smart T a q thermostable DNA polymerase (CinnaGen Inc., Tehran, Iran) in 25 μl PCR reaction mix. Amplification was performed in a TC-512 thermocycler (Techne, Cambridge, UK) and the program parameters for thermocycler were 2 cycles of 94 °C for 30 seconds, 42°C for 7 seconds and 72°C for 70 seconds; and 38 cycles of 94°C for 1 second, 42°C for 7 seconds and 72°C for 70 seconds; and 72°C for 5 minutes final extension.
The PCR products were analyzed by electrophoresis on 1.5% agarose gel at 80 V for 75 minutes. Gels were stained in a 0.5 μg/ ml ethidium bromide solution for 15 minutes and photographed by CCD Video Camera.
Analysis of RAPD data
Each isolate was scored for the presence or absence (1 or 0) of each band on agarose gel. The index of similarity (F) between samples was calculated using the formula of Nei and Li (13) F xy= 2nxy/ nx + ny . Where nxy is the number of RAPD bands shared by the two samples and nx and ny are the numbers of RAPD bands scored in each sample. The genetic distance (d) was calculated using the formula of Hillis and Mortiz (6): d= 1-F. The numerical index of discrimination (D) was calculated using the Simpson’s index of diversity (8).
RESULTS
The reproducibility of the RAPD technique was tested by repeated testing. The number of DNA bands for different isolates was between 0 and 11. Fifty eight E. coli isolates and 33 avian isolates could be differentiated to 34 and 29 RAPD – types, respectively. The discriminating index of 1247 primer was calculated 0.912 for all isolates and 0.990 for avian isolates. 700 bp, 900 bp and approximately 1 Kb DNA fragments were shared between 40 (96.9%), 35 (60.34%) and 33 (56.89%) isolates, respectively. Thirty one (93.9%) and 28 (84.8%) avian isolates were produced 700 bp and 900 bp DNA fragments in RAPD-PCR, respectively. Nine E. coli isolates from week-old lambs, 5 calves’ isolates, 2 food borne isolates and 1 avian isolate did not produce any DNA amplicon in repeated PCR reactions, using 1247 primer. Figs. 1 and 2 show banding patterns of some isolates after gel electrophoresis and ethidium bromide staining of PCR products.
Figure 1.
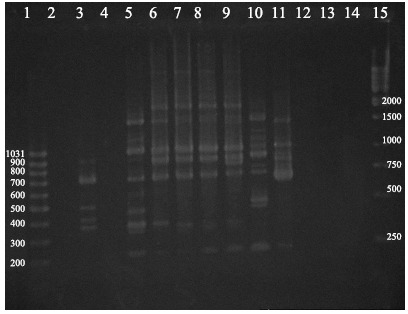
RAPD patterns of E. coli isolates using primer 1247. Lanes 2, 4, 12 and 14 are empty. Lanes 3 and 5–11 are samples 55 and 4–10, respectively. Lanes 1 and 15 are molecular ladders and lane 13 is negative blank control.
Figure 2.

RAPD patterns of E. coli isolate using primer 1247. Lanes 2–12 are samples 55, 56, 48, 49, 18, 19, 20, 21, 37, 38 and 39, respectively. Lane 13 is negative blank control and lanes 1 and 14 are molecular ladders.
DISCUSSION
Several methods had been used for typing and differentiating E. coli isolates. The RAPD-PCR is more sensitive and more cost effective than other molecular methods (9,12,14,16). Here we describe the application of a random amplified polymorphic DNA- polymerase chain reaction (RAPD-PCR) for molecular genetic differentiation of E. coli isolates in Iran. Oligonucleotide primer 1247 was discriminating for all E. coli isolates especially avian isolates.
The avian E. coli isolates were highly discriminated based on our research which this result complies with experiences of Maurer et al. (12) and Chansiripornchai et al. (4) in United States and Thailand, respectively. Samples 1 to 8 (Table 1) were isolated from ill birds in different poultry houses of the same poultry farm. The least genetic distance was between samples 6 and 7 which were 0.07 and the most was between samples 1 and 6 with 0.5 of genetic distance. Samples 9 and 10 were isolated from cloaca of healthy birds from different poultry farms in Garmsar and Ghazvin regions, respectively. We can not show genetic similarity between them or genetic difference between these isolates and avian pathogenic isolates which were cultured from colisepticemic cases. We also isolated samples 11 to 18 from colisepticemia avian cases in different poultry farms in Karaj. As shown before for samples 1 to 8 we could not confirm any genetic relatedness between these samples but the amplicon patterns of samples 19 and 20 which were isolated from a poultry farm in Ardebil, was the same. The banding patterns of samples 19, 20 and 35, which later one was isolated from a poultry house in Tehran, were alike. Samples 16 and 25 were genetically similar but were isolated from dead birds in different regions. No single bacterial type or clone appears to be associated with avian collibacilosis in Iran. Finally, based on the same E. coli RAPD-types from different regions, we concluded that the avian E. coli were not geographically restricted. On the other hand different E. coli genetic types were isolated from same regions, flocks and poultry houses. Similar findings have been reported for avian isolates (4,12).
ACKNOWLEDGMENTS
This research was supported by University of Tehran Research fund. The authors thank Mr. Ghaffari, Mr. Ashrafi, and Mr. Khormali for their technical assistances.
RESUMO
Diferenciação molecular de Escherichia coli aviária por RAPD-PCR
Escherichia coli é um dos patógenos aviários mais importantes e um habitante comum do trato gastrointestinal de animais. A maioria das cepas patogênicas não pode ser diferenciada por métodos bioquímicos ou outros métodos microbiológicos clássicos. Métodos de tipagem molecular, particularmente PCR, têm facilitado os estudos epidemiológicos e ecológicos a respeito desse microrganismo. Nesse estudo, descrevemos a aplicação do RAPD-PCR para a diferenciação
molecular de isolados de E.coli do Irã. No estudo, 58 isolados, incluindo 4 isolados padrão, 3 isolados de alimentos, 33 isolados de aves, 8 isolados de bezerros diarréicos e 10 isolados de carneiros diarréicos foram analisados por RAPD-PCR com o primer 1247 (5’-AAG AGC CCG T-3’). A análise mostrou que esses isolados podiam ser agrupados em 33 tipos RAPD, sendo os isolados de aves agrupados em 29 genótipos diferentes. Verificou-se que o primer utilizado não diferenciou os isolados de carneiros. O índice discriminatório para todos os isolados foi 0,912 e para os isolados de aves foi 0,990. Concluiu-se que o RAPD-PCR pode ser usado como método para diferenciação molecular de isolados de E. coli .
Palavras-chave:Escherichia coli , RAPD-PCR, diferenciação molecular, Irã
REFERENCES
- 1.Barnes H.J., Vaillancourt J., Gross W.B. Diseases of Poultry. 11. Iowa: Iowa State Press; 2003. Colibacillosis; pp. 631–652. [Google Scholar]
- 2.Behroozikhah A.M., Keyvanfar H., Feizabadi M.M., Tabatabayi A.H., Alamian S. Differentiation of Iranian strains of Brucella spp .by random amplification of polymorphic DNA. Arch. Razi Inst. 2005;59:13–23. [Google Scholar]
- 3.Brandt M.E., Hutwanger L.C., Kuykendall R.J., Pinner R.W. The Cryptococcal disease active surveillance group. Comparison of multilocus enzyme electrophoresis and random amplified polymorphic DNA analysis for molecular subtyping of Cryptococcus neoformans. J. Clin. Microbiol. 1995;33(7):1890–1895. doi: 10.1128/jcm.33.7.1890-1895.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chansiripornchai N., Ramasoota P., Sasipreeyajan J., Svenson S.B. Differentiation of avian pathogenic Escherichia coli (APEC) strains by random amplified polymorphic DNA (RAPD) analysis. Vet. Microbiol. 2001;80:75–83. doi: 10.1016/s0378-1135(00)00380-1. [DOI] [PubMed] [Google Scholar]
- 5.Goodwin P.H., Annis S.L. Rapid identification of genetic variation and pathotype of Leptosphaeria maculans by random amplified polymorphic DNA assay. Appl. Environ. Microbiol. 1991;57:2482–2486. doi: 10.1128/aem.57.9.2482-2486.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hillis D.M., Mortiz C. Molecular Systematics. Saunderland, MA: Sinauer Associates; 1990. [Google Scholar]
- 7.Hirsh D.C. Veterinary Microbiology. (Hirsh DC, Maclachlan NJ and Walker RL eds.) London: Blackwell publishing; 2004. Enterobacteriaceae: Escherichia; pp. 61–68. [Google Scholar]
- 8.Hunter P.R., Gaston M.A. Numerical index of the discriminatory abilty of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 1988;26(11):2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. 1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson J.R., Murray A.C., Gajewski A., Sullivan M., Snippes P., Kuskowski M.A., Smith K.E. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 2003;47(7):2161–2168. doi: 10.1128/AAC.47.7.2161-2168.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kersulyte D., Woods J.P., Keath E.J., Goldman W.E., Berg D.E. Diversity among clinical isolates of Histoplasma capsulatum detected by polymerase chain reaction with arbitrary primers. J. Bacteriol. 1992;174(22):7075–7079. doi: 10.1128/jb.174.22.7075-7079.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin A.W., Usera M.A., Barrett T.J., Goldsby R.A. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella enteritidis. J. Clin. Microbiol. 1996;34(4):870–876. doi: 10.1128/jcm.34.4.870-876.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maurer J.J., Lee M.D., Lobsinger C., Brown T., Maier M., Thayer S.G. Molecular typing of avian Escherichia coli isolates by Random Amplification of Polymorphic DNA. Avian Dis. 1998;42:431–451. [PubMed] [Google Scholar]
- 13.Nei M., Li W.H. Mathematical model for studying genetic variation of restriction endonucleases. Proc. Natl. Acad. Sci. 1979;76(10):5269–5273. doi: 10.1073/pnas.76.10.5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osek J. Virulence factors and genetic relatedness of Escherichia colistrains isolated from pigs with post-weaning diarrhea. Vet. Microbiol. 2000;71:211–222. doi: 10.1016/s0378-1135(99)00168-6. 2000. [DOI] [PubMed] [Google Scholar]
- 15.Singer R.S., Jeffrey J.S., Carpenter T.E., Cooke C.L., Atwill E.R., Johnson W.O., Hirsh D.C. Persistence of cellulitis-associated Escherichia coliDNA fingerprints in successive broiler chicken flocks. Vet. Microbiol. 2000;75:59–71. doi: 10.1016/s0378-1135(00)00205-4. 2000. [DOI] [PubMed] [Google Scholar]
- 16.Wang G., Whittam T.S., Berg C.M., Berg D.E. RAPD (arbitrary primer) PCR is more sensitive than multilocus enzyme electrophoresis for distinguishing related bacterial strains. Nucleic Acids Res. 1993;21(25):5930–5933. doi: 10.1093/nar/21.25.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Welsh J., McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Whittam T.S., Wilson R.A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect. Immun. 1988;56(9):2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams J.G.K., Kubelik A.R., Livak K.J., Rafalski J.A., Tingey S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–6536. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]


