Abstract
A polyphasic approach was applied to characterize 35 G. diazotrophicus isolates obtained from sugarcane varieties cultivated in Brazil. The isolates were analyzed by phenotypic (use of different carbon sources) and genotypic tests (ARDRA and RISA–RFLP techniques). Variability among the isolates was observed in relation to the carbon source use preference. Glucose and sucrose were used by all isolates in contrast to myo-inositol, galactose and ribose that were not metabolized. The results of the analysis showed the presence of two groups clustered at 68% of similarity. The genetic distance was higher when RISA-RFLP analysis was used. Analysis of 16S rDNA sequences from isolates showed that all of them belonged to the G. diazotrophicus species. Neither effect of the plant part nor sugarcane variety was observed during the cluster analysis. The observed metabolic and genetic variability will be helpful during the strain selection studies for sugarcane inoculation in association with sugarcane breeding programs.
Keywords: biological nitrogen fixation, genetic diversity, plant-bacteria interaction
INTRODUCTION
The endophytic bacterium G. diazotrophicus has been found colonizing different crop plants such as sugarcane, pineapple, coffee, sweet potato, Pennisetum pupureum, Eleusine coracana, Erianthus and insects such as mealy bugs (21). In addition to being able to fix nitrogen it has been shown to possess many other characteristics potentially valuable in the area of agriculture such as production of substances responsible for plant growth-promotion (10), antifungal and antibacterial properties (15,18).
One pre-condition to exploit the benefits derived from an efficient association, mainly related to the biotechnological potential for agriculture including biological nitrogen fixation, is to identify endophytic associations of microorganisms with different plants (8). It is already known that the genetic diversity within a species can be correlated with the spatial distribution, environmental effects and specificity of host plant interaction (12,26). These aspects could also be considered during the strain selection process concerning the inoculation efficiency within the process of host plant interaction and environmental conditions (6,16).
Previous studies (4,5) found that G. diazotrophicus isolated from sugarcane plants has a very low genetic diversity. In addition, the authors observed that the strains isolated from the Brazilian sugarcane varieties are more diverse than those isolated from the Mexican sugarcane plants. Nevertheless, the majority of the bacteria were isolated from few sugarcane varieties cultivated in both countries, which may have biased the results. In contrast, other studies using RAPD molecular analyses for G. diazotrophicus strains isolated from Indian sugarcane varieties showed a substantial genetic diversity (28). Since the first studies on the genetic diversity of G. diazotrophicus, the main factors that have been assigned as responsible for the limited genetic diversity are the limited distribution of host plants and its propagation by vegetative means as in the case of sugarcane using stem cuttings (setts). In this model of endophytic interaction, the bacteria is maintained from one generation to the next within the host plant environment that may protect the microbial natural population from competition for nutrient availability, survival in different soil types or other physico-chemical stress. The analysis of the phylogenetic micro diversity of the bacteria in association with the host plant may contribute to understand the “functional diversity of microbial communities and ultimately the ecosystem function as a whole” (26). These aspects may help to achieve a better understanding of the plant-bacteria relationship and perhaps to answer “why certain plant genotypes obtain much higher contribution from Biological Nitrogen Fixation than others”, question raised by Döbereiner et al. (7).
The aim of this study was to use the polyphasic analysis to characterize G. diazotrophicus isolates obtained from inner tissues of roots and leaves of different sugarcane varieties maintained in the Germoplasm Bank in Brazil.
MATERIALS AND METHODS
Isolates
A total of 35 isolates were tested (Table 1) and all of them were obtained as described by Barbosa et al. (3). Six reference strains (BR11334, BR11208, BR11284, BR11281, BR11280, BR11249) representing the different groups as described by Caballero-Mellado et al. (5) using multilocus enzyme electrophoresis analysis were also used for comparison.
Table 1.
Characteristics of the bacterial isolates and reference strains used in this study.
| Isolates | Plant part | Sugarcane variety | Country of origin |
|---|---|---|---|
| BR11367 | roots | SP71–1406 | Brazil |
| BR11368 | Roots | SP70–443 | Brazil |
| BR11392 | Roots | SP70–1284 | Brazil |
| BR11373 | aerial part | SP70–443 | Brazil |
| BR11514 | Roots | PB-4672 | Brazil |
| BR11394 | Roots | PB-4612 | Brazil |
| BR11513 | Roots | CB-7437 | Brazil |
| BR11404 | aerial part | CB-7437 | Brazil |
| BR11509 | Roots | RB 739359 | Brazil |
| BR11369 | Roots | RB 739735 | Brazil |
| BR11383 | aerial part | Co 622 | India |
| BR11386 | Roots | Co 1007 | India |
| BR11950 | Roots | Co 740 | India |
| BR11397 | aerial part | Co 453 | India |
| BR11951 | Roots | Ragnar | India |
| BR11376 | aerial part | Ragnar | India |
| BR11379 | aerial part | Azul Casa Grande | India |
| BR11388 | Roots | CP 5376 | USA |
| BR11396 | aerial part | CP 5376 | USA |
| BR11316 | aerial part | CP 5323 | USA |
| BR11390 | Roots | CP 5330 | USA |
| BR11398 | aerial part | CP 5330 | USA |
| BR11952 | Roots | CP 5480 | USA |
| BR11953 | Roots | CP 5950 | USA |
| BR11391 | aerial part | CP 5950 | USA |
| BR11954 | Roots | CP 601 | USA |
| BR11387 | Roots | CP 5268 | USA |
| BR11401 | aerial part | CP 5268 | USA |
| BR11955 | Roots | H 493533 | USA |
| BR11377 | aerial part | B 4098 | Barbados |
| BR11384 | Roots | Pepe Cuca | Cuba |
| BR11393 | aerial part | Pepe Cuca | Cuba |
| BR11956 | aerial part | PR 1124 | Porto Rico |
| BR11378 | aerial part | Na 5679 | Argentina |
| BR11370 | Roots | Mex 5219 | Mexico |
| BR11280* | Stems | ND | Brazil |
| BR11281* | Stems | ND | Brazil |
| BR11284* | Roots | ND | Brazil |
| BR11208* | Stems | SP70–1143 | Brazil |
| BR11249* | Rhizoplane | Na5679 | Brazil |
| BR11334* | mealybug insect | ND | Mexico |
ND: Not determined
reference strains.
Carbon source metabolism
The main carbon source of the medium was omitted and the following carbon sources were tested: glucose, fructose, sucrose, arabinose, ribose, galactose, myo-innositol, sorbitol and adonitol (all provided by Sigma Chemical, St. Louis, MO, USA) as described by Barbosa et al. (3).
DNA extraction
The DNA of the strains was extracted either from liquid cultures or from colonies according to the methodology adapted from Audy et al. (1).
PCR amplification of the 16S rDNA and 16S-23S rDNA intergenic region
The amplification of 16S rDNA region followed the methodology described by Young et al. (31), using the pair of primers Y1 and Y3. The amplification of the 16S-23S rDNA intergenic spacer region was performed with primers pHr and p23Suni322anti (9,14).
Restriction of amplified DNA
Five microlitres of each PCR amplified products was digested with restriction endonucleases Hae III, Cfo I, Alu I, Msp I, Taq I and Hinf I (Invitrogen, SP, Brazil) for the 16S rDNA region (ARDRA-RFLP) and Hae III, Alu I, Msp I, Rsa I, and Pvu I (Invitrogen, SP, Brazil) for intergenic spacer region (RISA-RFLP). The banding pattern of digested products were separated by electrophoresis on a 3% agarose (Sigma Chemical, St. Louis, MO, USA) gel on 1 x TAE buffer (Trizma base – Promega Madison, Wisconsin, USA; Glacial Acetic Acid – Merck, Darmstadt, Germany; EDTA – Sigma Chemical, St. Louis, MO, USA), carried out at 60 V for 2 h 30 min, stained with ethidium bromide (Sigma Chemical, St. Louis, MO, USA), photographed using a Kodak Logic 100 (Kodak®, San Francisco, USA) equipment and size estimated by comparison against a 100 pb DNA ladder (Invitrogen, SP, Brazil).
Polyphasic analysis based on carbon source utilization and genetic diversity (ARDRA and RISA – RFLP)
The clustering analysis of the isolates was based on the qualitative characteristic of presence (+) or absence (-) of pellicle veil formed in the semi-solid medium (carbon source) and bands in the gel (ARDRA and RISA – RFLP). According to these data, a binary matrix was constructed for each data independently prior to the consensus analysis using the polyphasic approach. The similarity among the isolates was estimated by UPGMA indices using the Jaccard coefficient (22) and represented graphically through a dendrogram generated with the NTSYS-pc (version 1.8, Exeter Software, Setauket, New York, USA).
16S rDNA sequencing and phylogenetic analysis
The PCR products were directly sequenced using a TE Dynamics Kit in the MegaBACE 1000 System (GE-Amersham Biosystem, Chalfont St. Giles, UK) with the primers designed by (27). The 16S rDNA nucleotide sequences were used to determine the phylogenetic position of the isolates among other G. diazotrophicus strains in the phylogenetic tree. Multiple sequence alignments were carried out with ClustalW (29) and the phylogenetic trees were constructed by the neighbour-joining method (25). To dendrogram construction the TreeCon (30) package, with the distance of Jukes and Cantor, was used (11). Others program parameters were maintained unchanged. Bootstrap analyses of 1.000 replicates were established.
Nucleotide sequence accession numbers
The nucleotide sequences of the 16S rDNA genes obtained were submitted to deposit at the Genbank. The identities of several nitrogen-fixing bacteria sequences obtained from this deposit that were used to construct the phylogenetic trees are represented by their accession numbers.
RESULTS AND DISCUSSION
All of the 35 isolates and the six reference strains of G . diazotrophicus were able to grow in the semi-solid nitrogenfree medium containing glucose or sucrose as carbon source (Fig. 1). Sorbitol was only used by 17 isolates originated either from the aerial part or the roots of the sugarcane varieties. On the other hand, arabinose was only used by three isolates, adonitol was used by the type strain BR11281 (PAL5) and the isolate BR11398 while fructose was used by two reference strains and one isolate. The carbon sources myo-inositol, galactose and ribose were not used by any isolates as well as by the reference strains tested. Similar result was also observed by Barbosa et al. (3) where a lower number of carbon sources were used by strains of G. diazotrophicus under nitrogen fixation condition.
Figure 1.
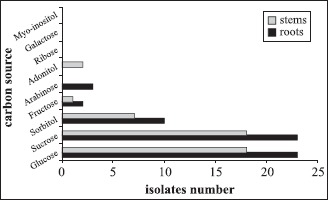
Utilization of carbon sources by different isolates and reference strains of G. diazotrophicus obtained from roots and aerial part of different sugarcane varieties maintained in the germoplasm bank.
The cluster analysis using the ARDRA technique showed the isolates randomly distributed forming two groups at 84% similarity (Fig. 2). Among the tested restriction enzymes, only HaeIII and HinfI were able to generate differential polymorphic bands of the 16S rDNA amplified region among the isolates tested .These two enzymes were able to distinguish a group of N2-fixing Burkholderia strains isolated from the rhizosphere and root interior of sugarcane, maize and teosinte plants grown in Brazil, Mexico and South Africa (20).
Figure 2.
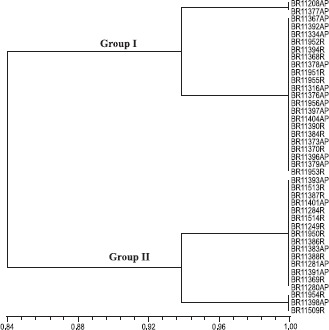
ARDRA-RFLP similarity cluster of 35 isolates, obtained from sugarcane varieties originated from different countries and maintained in the germoplasm bank, and 6 reference strains of G . diazotrophicus. Dendrogram was generated by the algorithm UPGMA and similarity matrix obtained from the use of Jaccard coefficient. AP: aerial part, R: roots.
In the case of the RISA, the formation of two groups was also observed, but at a lower similarity level (56%) when compared with the ARDRA analysis (Fig. 3). The RISA-RFLP analyses allowed the detection of a higher variation among the isolates when compared to the 16S rDNA, confirming other studies carried out with nitrogen-fixing bacteria (2,19,25).
Figure 3.
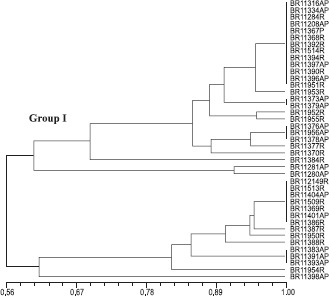
RISA-RFLP similarity cluster among 35 isolates obtained from sugarcane varieties originated from different countries and 6 reference strains of G. diazotrophicus. AP: aerial part, R: roots.
The polyphasic approach based on the consensus analysis of the phenotypic and genotypic (ARDRA and RISA) characteristics showed that the isolates formed two great groups with 68% of similarity (Fig. 4). The formation of two groups, with 70 % similarity, was also observed by Loganathan et al. (13) when analyzing nine new strains of G. diazotrophicus isolated from Eleusine coracana and compared to the type strain PAL5 of G. diazotrophicus using the RAPD technique. Perin et al. (17) analyzed the diversity of 123 isolates of G . diazotrophicus originated from sugarcane using the Enzyme-Linked Immunosorbent Assay (ELISA) technique and also observed the formation of two groups with distance of 35% between them.
Figure 4.
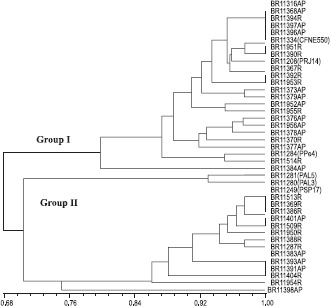
Consensus dendrogram obtained by combining the data of utilization of carbon sources, ARDRA and RISA-RFLP among 35 isolates from sugarcane and 6 reference strains of G. diazotrophicus. Dendrogram was generated by the algorithm UPGMA and similarity matrix obtained from the use of Jaccard coefficient. AP: aerial part, R: roots.
The cluster analysis based on polyphasic approach showed a random distribution of the isolates in relation to the part of the plant, sugarcane variety and country origin and therefore suggests a certain independence of genetic evolution of this bacterium in relation to its origin. Similar results were also observed by Rosado et al. (23) for isolates of Paenibacillus azotofixans (nowadays P. durus).
Sequencing of 16S rDNA unit of eight isolates, representatives of different branches within the ARDRA and RISA-RFLP clusters, showed that all of them belongs to the G. diazotrophicus species according to the phylogenetic analysis (Fig. 5). These isolates clustered together with the reference strains obtained from sugarcane, pineapple and coffee plants but separated from the species G. azotocaptans and G. johannae. Recently, a new nitrogen-fixing and cellulotic species of the genus Gluconacetobacter, named G. kombuchae, was isolated from a Kombucha tea plants grown in India and clustered quite distant from the already known diazotrophic Gluconacetobacter species (24).
Figure 5.
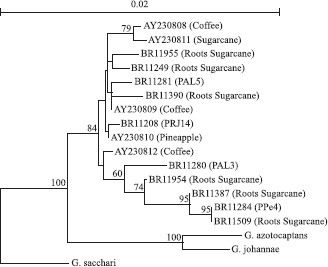
Phylogenetic analysis of the 16S rDNA sequences of different nitrogen-fixing isolates obtained from several sugarcane varieties maintained in the germoplasm bank. The tree was constructed by Neighbour-joining. The bootstrap values (1.000 replicas), program Treecon are indicated in the grouping base. The scale bar indicates the distance in substitutions by nucleotide. Reference sequences used in this analysis were obtained from the Genebank: BR11281 (PAL5 – ATCC49037); AF127413: Gluconacetobacter sacchari; AF111841: G. johannae; AF192761: G. azotocaptans; AY230814: PAL3, sugarcane strain; AY230810: UAP-AC10, pineapple strains; AY230809: UAP-AC7, pineapple strains; AY230811: CFNE550, sugarcane strains; AY230808: UAP-cf51, coffee strains.
In conclusion, these results showed that data from the phenotypic and genotypic techniques show here were efficient to reveal intra-specific variability among the G. diazotrophicus isolates at different levels and can be applied in a selection of strains for sugarcane inoculation studies. The absence of effect from origin of isolation, as sugarcane variety or part of the plant, on the genetic diversity suggest that this nitrogen-fixing species has maintained its genetic structure along the sugarcane breeding programs carried out by different countries in the world.
ACKNOWLEDGMENTS
The authors would like to thank Universidade Federal Rural do Rio de Janeiro, Estação Experimental de Campos dos Goytacazes for the assessment of the sugarcane Germoplasm Bank and also PESAGRO-RJ. This work was partially funded by Empresa Brasileira de Pesquisa Agropecuária – Embrapa grant n° 02.02.5.13, PRONEX II grant n° E-26/171.208/2003; PADCT III grant n° CIBio 77.97.1138.00, and FAPERJ – “Bolsa Cientista do Nosso Estado”.
RESUMO
Caracterização polifásica de isolados de Gluconacetobacter diazotrophicus obtidos de diferentes variedades de cana-de-açúcar.
Foi realizado a caracterização polifásica de 35 isolados obtidos de variedades de cana-de-açúcar cultivadas no Brasil, através de testes fenotípicos (uso de fontes diferentes de carbono) e genotípicos (técnicas de ARDRA e RISA-RFLP). Houve variação entre os isolados com relação à utilização de fontes de carbono. Glicose e sacarose foram usadas por todos isolados, diferentemente de mio-inositol, galactose e ribose que não foram metabolizados. Os resultados da análise polifásica dos dados confirmam a formação de dois grupos, que apresentaram 68% de similaridade. Observou-se maior distância genética entre os isolados quando a técnica de RISA-RFLP foi aplicada. O sequênciamento da região 16S do rDNA mostrou que todos os isolados pertencem à espécie G. diazotrophicus. Não foi observado efeito da parte da planta ou variedade de cana-de-açúcar no agrupamento dos isolados. Em conjunto, esses resultados poderão auxiliar no estudo de seleção de estirpes para inoculação em cana-de-açúcar, orientando programas de melhoramento vegetal.
Palavras chaves: fixação biológica de nitrogênio, diversidade genética, interação planta-bactéria
REFERENCES
- 1.Audy P., Braat C.A., Saidon G., Huang H.C., Latoche A. A rapid and sensitive PCR-based assay for concurrent detection of bacteria causing common and halo blights in bean seed. Phytopathol. 1996;86:361–366. [Google Scholar]
- 2.Azevedo M.S., Teixeira K.R.S., Kirchhof G., Hartmann A., Baldani J.I. Influence of soil and host plant crop on the genetic diversity of Azospirillum amazonense isolates. Pedobiol. 2005;49:565–576. [Google Scholar]
- 3.Barbosa E.A., Perin L., Reis V.M. Uso de fontes de carbono por G. diazotrophicus isolados de plantas de cana-de-açúcar. Pesq. Agropec Bras. 2006;41:827–833. [Google Scholar]
- 4.Caballero-Melado J., Martinez-Romero E. Limited genetic diversity in the endophytic sugarcane bacteria Acetobacter diazotrophicus. Appl. Environ. Microbiol. 1994;60:1532–1537. doi: 10.1128/aem.60.5.1532-1537.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caballero-Mellado J., Fuentez-Ramirez L.E., Reis V.M., Martinez-Romero E. Genetic structure of Acetobacter diazotrophicus population and identification of a new genetic distance group. Appl. Environ. Microbiol. 1995;61:3308–3013. doi: 10.1128/aem.61.8.3008-3013.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Cello F., Bevivino A., Chiarini L., Fani R., Paffetti D., Tabacchioni S., Dalmastri C. Biodiversity of a Burkhoderia cepacea population isolated from maize rhizosphere at different plant growth stages. Appl. Environ. Microbiol. 1997;63:4485–4493. doi: 10.1128/aem.63.11.4485-4493.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Döbereiner J., Baldani V.L.D., Reis V.M. Endophytic occurrence of diazotrophic bacteria in non-leguminous crops. In: Fendrik I., Gallo M. del, Vanderleyden J., Zamaroczy M., editors. Azospirillum and Related Microorganisms: Genetics, Physiology, Ecology. Berlim: Springer-Verlag; 1995. pp. 3–13. [Google Scholar]
- 8.Hartmann A., Eckert B., Stoffels M., Olivares F.L., Weber O.B., Reis V., Baldani V.L.D., Baldani J.I., Schloter M., Kirchhof G. Molecular approaches for the investigation of the diversity and localization of diazotrophic bacteria. In: Pedrosa F.O., Hungria M., Yates M.G., Newton W.E., editors. Nitrogen Fixation: From Molecules to Crop Productivity. Dordrecht: Kluwer Academic; 2000. pp. 441–442. [Google Scholar]
- 9.Honeycutt R.J., Sobral B.W.S., Mcclelland M. tRNA intergenic spacers reveal polymorphisms diagnostic for Xantomonas albilineans. Microbiol. 1995;141:3229–3239. doi: 10.1099/13500872-141-12-3229. [DOI] [PubMed] [Google Scholar]
- 10.James E.K., Olivares F.L., Oliveira A.L.M., Reis Jr. F.B., da Silva L.G., Reis V.M. Further observation on the interaction between sugar cane and G. diazotrophicus under laboratory and greenhouse condition. J. Exp. Bot. 1998;52:747–760. doi: 10.1093/jexbot/52.357.747. [DOI] [PubMed] [Google Scholar]
- 11.Jukes T.H., Cantor C.R. Evolution of protein molecules. In: Munro H.N., editor. Mammalian Protein Metabolism. Vol. 3. New York: Academic; 1969. pp. 21–132. [Google Scholar]
- 12.Kennedy A.C. Bacterial diversity in agroecosystems. Agric. Ecosyst. Environ. 1999;74:65–76. [Google Scholar]
- 13.Loganathan P., Sunita R., Parida A.K., Nair S. Isolation and characterization of two genetically distant groups of G. diazotrophicus from a new host plant Eleusine coracana. J. Appl. Microbiol. 1999;87:167–172. [Google Scholar]
- 14.Massol-Deya A.A., Odelson D.A., Hickey R.F., Tiedje J.M. Bacterial community fingerprinting of amplified 16 and 16–23S ribosomal DNA gene sequences and restriction endonuclease analysis (ARDRA) In: Akkermans AD.L., Van Elsas J.D., De Bruijn F.J., editors. Molecular Microbial Ecology Manual. Dordrecht: Kluwer Academic; 1995. pp. 1–8. [Google Scholar]
- 15.Muthukumarasamy R., Revathi G., Seshadri S., Lakshminarasimhan C. G. diazotrophicus (syn. Acetobacter diazotrophicus), a promising diazotrophic endophyte in tropics. Cur. Sci. 2002;83:137–145. [Google Scholar]
- 16.Oliveira I.A., Vasconcellos M.J., Seldin L., Paiva E., Vargas M.A., Sá N.M.H. Random amplified polymorphic DNA analysis of effective Rhizobium sp. associated with beans cultivated in Brazilian Cerrado soils. Brazil. J. Microbiol. 2000;31:39–44. [Google Scholar]
- 17.Perin L., Baldani J.I., Reis V.M. Diversidade de Gluconacetobacter diazotrophicus isolada de plantas de cana-de-açúcar cultivadas no Brasil. Pesq. Agropec. Bras. 2004;39:763–770. [Google Scholar]
- 18.Piñon D., Casas M., Blanch M., Fontaniella B., Blanco Y., Vicenti C., Solas M.T., Legaz M.E. G. diazotrophicus, a sugar cane endosymbionte produces a bacteriocin against X. albilineans, a sugar cane pathogen. Res. Microbiol. 2002;153:345–351. doi: 10.1016/s0923-2508(02)01336-0. [DOI] [PubMed] [Google Scholar]
- 19.Reis Junior F.B., Da Silva M.F., Teixeira K.R.S., Urquiaga S., Reis V.M. Identificação de isolados de Azospirillum amazonense associados à Brachiaria spp., em diferentes épocas e condições de cultivo e produção de fitormônio pela bactéria. Rev. Bras. Ciênc. Solo. 2004;28:103–113. [Google Scholar]
- 20.Reis V.M., Estrada-de los Santos P., Tenorio-Salgado S., Vogel J., Stoffels M., Guyon S., Mavingui P., Baldani V.L.D., Schmid M., Baldani J.I., Balandreau J., Hartmann A., Caballero-Mellado J. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 2004;54:2155–2162. doi: 10.1099/ijs.0.02879-0. [DOI] [PubMed] [Google Scholar]
- 21.Reis V.M., Baldani J.I., Baldani V.L.D., Döbereiner J. Biological nitrogen fixation in gramineae and palm trees. Crit. Rev. Plant Sci. 2000;19:227–247. [Google Scholar]
- 22.Rohlf F.J. New York: State University of New York; 1994. NTSYS-pc Numerical taxonomy and multivariate analysis system; p. 318. [Google Scholar]
- 23.Rosado A.S., de Azevedo F.S., da Cruz D.W., Van Elsas J.D., Seldin L. Phenotypic and genetic diversity of Paenibacillus azotofixans strains from the rhizoplane or rhizosphere soil of different grasses. J. Appl. Microbiol. 1998;84:216–226. [Google Scholar]
- 24.Saravanan V.S., Madhaiyan M., Osborne J., Thangaraju M., Sa T.M. Ecological Occurrence of Gluconacetobacter diazotrophicus and Nitrogen-fixing Acetobacteraceae Members: Their Possible Role in Plant Growth Promotion. Microb. Ecol. 2008;55:130–140. doi: 10.1007/s00248-007-9258-6. [DOI] [PubMed] [Google Scholar]
- 25.Saitou N., Nei M. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Schloter M., Lebuhn M., Heulin T., Hartmann A. Ecology and evolution of bacterial microdiversity. FEMS Microbiol. Rev. 2000;24:647–660. doi: 10.1111/j.1574-6976.2000.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 27.Soares-Ramos J.R.L., Ramos H.J.O., Cruz L.M., Chubatsu L.S. Comparative molecular analyses of Herbaspirillum strains by RAPD, RFLP, and 16S rDNA sequencing. Gen. Mol. Biol. 2003;26:537–543. [Google Scholar]
- 28.Suman A., Shasany A.K., Singh M., Shahi H.N., Gaur A., Khanuja S.P.S. Molecular assessment of diversity among endophytic diazotrophs isolated from subtropical Indian sugarcane. World J. Microbiol. Biotechnol. 2001;17:39–45. [Google Scholar]
- 29.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van de Peer Y., De Wachter R. TREECON: A software package for the construction and drawing of evolutionary trees. Comput. Applic. Biosci. 1993;9:177–182. doi: 10.1093/bioinformatics/9.2.177. [DOI] [PubMed] [Google Scholar]
- 31.Young J.P.W., Downer H.L., Eardly B.D. Phylogeny of phototrophic Rhizobium strain BTAil by polymerase chain reaction-based sequencing of a 16S rRNA segment. J. Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]


