Abstract
The present study determined the molecular and resistance patterns of E. coli isolates from urinary tract of swine in Southern of Brazil. Molecular characterization of urinary vesicle samples was performed by PCR detection of virulence factors from ETEC, STEC and UPEC. From a total of 82 E. coli isolates, 34 (38.63%) harbored one or more virulence factors. The frequency of virulence factors genes detected by PCR were: pap (10.97%), hlyA (10.97%), iha (9.75%), lt (8.53%), sta (7.31%) sfa (6.09%), f4 (4.87%), f5 (4.87%), stb (4.87%), f6 (1.21%) and f41 (1.21%). Isolates were resistant to penicillin (95.12%), lincomycin (93.9%), erythromycin (92.68%), tetracycline (90.24%), amoxicillin (82.92%), ampicillin (74.39%), josamycin (79.26%), norfloxacin (58.53%), enrofloxacin (57.31%), gentamicin (39.02%), neomycin (37.8%), apramycin (30.48%), colistine (30.48%) and cefalexin (6.09%). A number of 32 (39.02%) E. coli isolates harbored plasmids.
Keywords: E. coli, plasmids, virulence factors, antimicrobial resistance, swine
The urinary tract infection (UTI) is defined by the presence of bacteria in the urinary tract (22). In humans, the disease is associated to many virulence factors present in the uropathogenic E. coli (UPEC), including haemolysin, aerobactin, adhesins, serum resistance, cytotoxic necrotizing factor (CNF), capsule production and uropathogenic-specific protein (14,15,21). E. coli adhesion to host cells is important to bacteria infection and persistence in urinary fluxes (6,10). The genes involved in biosynthesis of fimbria and adhesins present in UPEC are organized in operons denominated pap and sfa, coding for P and S fimbrial adhesins (2,3,24). IRGA homologue adhesin (IHA) is an outer membrane protein (OMP) found mainly in UPEC being also involved in adherence (13). Two toxin types are associated to UPEC: The alfa-haemolysin (HLY) and cytotoxic necrotizating factor 1 (CNF 1) are involved in host cell destruction necessary to bacteria persistence in urinary tract (4,8). Toxins from enterotoxigenic E. coli were described in swine urinary E. coli isolates as heat-labile toxin (LT) and vero toxin (VT) (3).
The antibiotic therapy may select resistant bacteria (5,22). The resistance to antimicrobial drugs may be carried by plasmids, as well as chromosomal mutations that occur spontaneously (9). Multi-drug resistance have been reported in human and swine E. coli isolated from urinary tract (4,11). The purpose of the present study was to determinate the pathotype, the plasmidial DNA content and the patterns of resistance to antimicrobial drugs in E. coli isolates from swine females with UTI.
Eighty two E. coli strains were isolated from sows from thirty breeding farms. Animals were considered with urinary infection according to microbiologic and urinary physic chemical patterns (22). The E. coli were isolated from urinary vesicle and urine samples collected from swine breeding farms in Southern of Brazil. One putative E. coli colony was identified by morphology and biochemical tests according to Quinn et al. (17). The Kirby-Bauer disc diffusion test (17) was used and the following drugs were tested: amoxicillin (10 μg), ampicillin (10 μg), tetracycline (30 μg), norfloxacin (10 μg), enrofloxacin (5 μg), cefalexin (30 μg), neomycin (30 μg) gentamicin (30 μg), penicillin (10 μg), lincomycin (2 μg), erythromycin (15 μg), apramycin (15 μg), josamycin (30 μg), and colistin (10 μg).
The E. coli isolates were characterized by multiplex PCR for fimbrial and toxin genotypification by using the amplification of the following regions: sta, stb, stx, cnf, hly, lt, f4, f5, f6, f41, f18, bfp, eae, sfa, pap, iha and usp. The primers and PCR conditions were previously described (1,2,4,7,12,15,16). Amplicons identities were confirmed by sequencing (Amersham Pharmacia Biotech). Plasmidial DNA extraction from E. coli isolates was performed by alkaline lysis, as previously described (4).
From E. coli isolates, 34 (38.63%) harbored one or more virulence factor revealed by PCR amplification. UPEC were found in 14.63% (12/82), ETEC in 15.85% (13/82) and in 10.97% (9/82) were amplified virulence factors of both, ETEC and UPEC. Brito et al. (4) described the occurrence of ETEC and UPEC in swine with UTI. According to Russo and Jonhson (19) the current E. coli pathotype classification are performed by a combination of virulence traits, and not by genetically source. ETEC were reported in swine urinary strains suggesting the ascending intestinal origin of UTI (4,23). The pathogenesis of urinary tract infections depends of the E. coli skills to adhere, persist and multiply in the host (5). The genes involved in bacteria adherence detected in our study were pap (10.97%), iha (9.75%), sfa (6.09%), f4 (4.87%), f5 (4.87%), f6 (1.21%) and f41 (1.21%). The pap, iha and sfa elements are reported as important to adhesion of UPEC (13,24). Pap gene was found in 54.8% of Brazilian UPEC isolates studied by Brito et al. (4), although in our study, the frequency of this gene was lower (10.97%). According to Brito (4) afa, Bfp and sfa adhesins were not found in UPEC isolates from Brazil. In our study we found sfa in 6.09% E. coli isolates. The f4 and f5 fimbriae genes were amplified alone or in association with virulence factors typical of UPEC.
The presence of haemolysis in blood agar was observed in four isolates, although the hlyA gene was detected by PCR in nine isolates. This difference may be associated to silent expression or mutation in hly genes in E. coli (20,21). Brito et al. (4) reported the presence of 25.8% of haemolytic UPEC isolates. In our study lt, sta and stb toxins genes commonly found in ETEC were detected, respectively, in 8.53%, 7.31% and 4.87% of E. coli isolates. Lt was previously described in swine UPEC by Brito et al. (4).
The presence of usp gene in dog and cat E. coli isolates permitted the proposition of the role these animals as an alternative reservoir for human UTI (14,15). In contrast, we amplified usp in one E. coli isolated and this may suggest genotypic differences among swine and human urinary E. coli isolates.
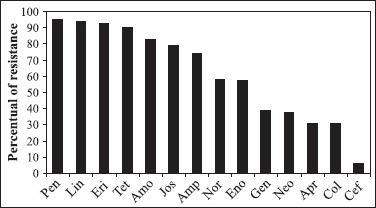
The antimicrobial resistances of UPEC isolates are presented in Fig. 1. Brito et al. (4) report a higher resistance of swine UPEC to tetracycline and ampicillin. In swine UPEC 95.12% (78/82) were resistant to four or more antimicrobial groups. The more frequent patterns of resistance were to beta lactam, lincomicin, tetraciclyne, quinolone, macrolide, aminoglycoside and polymixin groups (data not shown). Plasmids and other genetic elements, like integrons, may be encountered in UPEC and are associated to coding virulence factors and MDR (4,5,18). In our study plasmids were found in 39.02% (32/82) of E. coli isolate.
Figure 1. Antimicrobial drugs resistance patterns of urinary E. coli isolates from swine in Southern of Brazil. Penicillin (Pen), Lincomycin (Lin), Erythromycin (Eri), Tetracycline (Tet), Amoxicillin (Amo), Josamycin (Jos), Ampicillin (Amp), Norfloxacin (Nor), Enrofloxacin (Eno), Gentamicin (Gen), Neomycin (Neo), Apramycin (Apr), Colistin (Col) and Cefalexin (Cef).
Some E. coli isolates maintain virulence factors of both ETEC and UPEC simultaneously, suggesting the elevated genetic relationship between urinary and intestinal strains. Multi-drug resistance was widely found in swine urinary isolates.
RESUMO
Fatores de virulência e resistência aos antimicrobianos de Escherichia coli isoladas do trato urinário de suínos do Sul do Brasil
O presente estudo teve por objetivo determinar os padrões moleculares e de resistência aos antimicrobianos de isolados de E. coli provenientes do trato urinário de suínos no Sul do Brasil. Os fatores estudados dividiram os patotipos ETEC, STEC e UPEC. Trinta e quatro (38,63%) isolados avaliados apresentavam um ou mais dos fatores de virulência pesquisados. A freqüência dos genes de virulência detectados foram: pap (10,97%), hlyA (10,97%), iha (9,75%), lt (8,53%), sta (7,31%) sfa (6,09%), f4 (4,87%), f5 (4,87%), stb (4,87%), f6 (1,21%) e f41 (1,21%). Os isolados foram resistentes à penicilina (95,12%), lincomicina (93,9%), eritromicina (92,68%), tetraciclina (90,24%), amoxacilina (82,92%), ampicilina (74,39%), josamicina (79,26%), norfloxacina (58,53%), enrofloxacina (57,31%), gentamicina (39,02%), neomicina (37,8%), apramicina (30,48%), colistina (30,48%) e cefalexina (6,09%). Trinta e dois (39,02%) isolados de E. coli continham plasmídeos.
Palavras chave: E. coli, plasmídeos, fatores de virulência, re-sistência antimicrobiana, suínos.
REFERENCES
- 1.Blanco J.E., Blanco M., Blanco J., Mora A., Balaguer L., Mourino M., Juarez A., Jansen W.H. O serogroups, biotypes, and eae genes in Escherichia coli strains isolated from diarrheic and healthy rabbits. J. Clin. Microbiol. 1996;34:3101–3107. doi: 10.1128/jcm.34.12.3101-3107.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanco M., Blanco J.E., Alonso M.P., Mora A., Balsalobre C., Munoa F., Juárez A., Blanco J. Detection of pap, sfa and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res. Microbiol. 1997;148:745–755. doi: 10.1016/s0923-2508(97)82450-3. [DOI] [PubMed] [Google Scholar]
- 3.Bouguenec C., Archambaud M., Labigne A. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 1992;30:1189–1193. doi: 10.1128/jcm.30.5.1189-1193.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brito B.G., Leite D.S., Linhares R.E., Vidotto M.C. Virulence-associated factors of uropathogenic Escherichia coli strains isolated from pigs. Vet. Microbiol. 1999;65:123–132. doi: 10.1016/s0378-1135(98)00277-6. [DOI] [PubMed] [Google Scholar]
- 5.Brito B., Vidotto M., Berbel M., Tagliari K. Fatores de virulência presentes em amostras de E. coli uropatogênicas – UPEC para suínos. Ciência Rural. 2004;34:645–652. [Google Scholar]
- 6.Chen S.L., Hung C.S., Xu J., Reigstad C.S., Magrini V., Sabo A. Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc. Natl. Acad. Sci. USA. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Costa M., Silva M., Spricigo D., Witt N., Marchioro S., Kolling L. Caracterização epidemiológica, molecular e perfil de resistência aos antimicrobianos de amostras de Escherichia coli isoladas de criatórios suínos do sul do Brasil. Pesq. Vet. Bras. 2006;26:5–8. [Google Scholar]
- 8.Davis J.M., Rasmussen S.B., O’Brien A.D. Cytotoxic necrotizing factor type 1 production by uropathogenic Escherichia coli modulates polymorphonuclear leukocyte function. Infect. Immun. 2005;73:5301–5310. doi: 10.1128/IAI.73.9.5301-5310.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doss S.A. Chromosomally-mediated antibiotic resistance and virulence. J. Med. Microbiol. 1994;40:305–306. doi: 10.1099/00222615-40-5-305. [DOI] [PubMed] [Google Scholar]
- 10.Gyles C., Fairbrother J., Gyles C. In: Escherichia coli Pathogenesis of bacterial infections in animals. Thoen O., Prescott J., editors. Ames: Iowa State University Press; 2004. pp. 193–214. [Google Scholar]
- 11.Gulsun S., Oguzoglu N., Inan A., Ceran N. The virulence factors and antibiotic sensitivities of Escherichia coli isolated from recurrent urinary tract infections. Saudi Med. J. 2005;26:1755–1758. [PubMed] [Google Scholar]
- 12.Johnson J.R., Russo T.A., Tarr P.I., Carlino U., Bilge S.S., Vary J.C., Stell A.L. Molecular epidemiological and phylogenetic associations of two novel putative virulence genes, iha and iroN (E. coli), among Escherichia coli isolates from patients with urosepsis. Infect. Immun. 2000;68:3040–3047. doi: 10.1128/iai.68.5.3040-3047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson J.R., Jelacic S., Schoening L.M., Clabots C., Shaikh N., Mobley H.L., Tarr P.I. The IrgA homologue adhesin Iha is an Escherichia coli virulence factor in murine urinary tract infection. Infect. Immun. 2005;73:965–971. doi: 10.1128/IAI.73.2.965-971.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurazono H., Yamamoto S., Nakano M., Nair G.B., Terai A., Chaicumpa W. Characterization of a putative virulence island in the chromosome of uropathogenic Escherichia coli possessing a gene encoding a uropathogenic-specific protein. Microb. Pathog. 2000;28:183–189. doi: 10.1006/mpat.1999.0331. [DOI] [PubMed] [Google Scholar]
- 15.Kurazono H., Nakano M., Yamamoto S., Ogawa O., Yuri K., Nakata K., Kimura M., Makino S., Nair G.B. Distribution of the usp gene in uropathogenic Escherichia coli isolated from companion animals and correlation with serotypes and size-variations of the pathogenicity island. Microbiol. Immunol. 2003;47:797–802. doi: 10.1111/j.1348-0421.2003.tb03437.x. [DOI] [PubMed] [Google Scholar]
- 16.Nakazato G., Gyles C., Ziebell K., Keller R., Trabulsi L.R., Gomes T.A., Irino K., Da Silveira W.D., Pestana De Castro A.F. Attaching and effacing Escherichia coli isolated from dogs in Brazil: characteristics and serotypic relationship to human enteropathogenic E. coli (EPEC) Vet. Microbiol. 2004;101:269–277. doi: 10.1016/j.vetmic.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Quinn P., Carter M., Markey B., Carter G. Clinical veterinary microbiology. London: Wolfe; 1994. [Google Scholar]
- 18.Rijavec M., Starcic E.M., Ambrozic A.J., Reissbrodt R., Fruth A., Krizan-Hergouth V., Zgur-Bertok D. High prevalence of multidrug resistance and random distribution of mobile genetic elements among uropathogenic Escherichia coli (UPEC) of the four major phylogenetic groups. Curr. Microbiol. 2006;53:158–162. doi: 10.1007/s00284-005-0501-4. [DOI] [PubMed] [Google Scholar]
- 19.Russo T.A., Johnson J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 20.Schmidt H., Karch H. Enterohemolytic phenotypes and genotypes of shiga toxin-producing Escherichia coli O111 strains from patients with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 1996;34:2364–2367. doi: 10.1128/jcm.34.10.2364-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Silveira W., Benetti F., Lancelotti M., Ferreira A., Solferini V., Brocchi M. Biological and genetic characteristics of uropathogenic Escherichia coli strains. Rev. Inst. Med. Trop. São Paulo. 2001;43:303–310. doi: 10.1590/s0036-46652001000600001. [DOI] [PubMed] [Google Scholar]
- 22.Sobestiansky J., Barcellos D., Moraes N., Carvalho L., Oliveira S. Clínica e patologia suína. Goiânia: Art 3 impressos especiais; 1999. [Google Scholar]
- 23.Starcic M., Johnson J.R., Stell A.L., Van der G.J., Hendriks H.G., van der Goot, van Vorstenbosch C., van Dijk L., Gaastra W. Haemolytic Escherichia coli isolated from dogs with diarrhea have characteristics of both uropathogenic and necrotoxigenic strains. Vet. Microbiol. 2002;85:361–377. doi: 10.1016/s0378-1135(02)00003-2. [DOI] [PubMed] [Google Scholar]
- 24.Yuri K., Nakata K., Katae H., Tsukamoto T., Hasegawa A. Serotypes and virulence factors of Escherichia coli strains isolated from dogs and cats. J. Vet. Med. Sci. 1999;61:37–40. doi: 10.1292/jvms.61.37. [DOI] [PubMed] [Google Scholar]


