Abstract
An actinomycin-D producing strain was isolated from soil and characterized as Streptomyces sindenensis. The culture was subjected to UV irradiation and a mutant with 400% higher actinomycin-D production was isolated (400 mg/l-1 as compared to 80 mg/l-1 produced by the parent). Production medium was optimized and antibiotic yield with the mutant was enhanced to 850 mg/l-1 which is 963% higher as compared with the parent.
Keywords: Actinomycin-D, antibiotics, Streptomyces, mutation
The actinomycins are chromopeptide lactone antibiotics, of which more than 30 natives are known. Among the actinomycins, actinomycin-D (act-D) has been studied extensively and is used clinically for the treatment of Wilms’ tumor (3). Various strains of Streptomyces and Micromonospora are reported to produce different forms of actinomycins (9). Production and optimization of act-D by S. sindenensis is not reported in literature. Present study reports isolation of a high yielding mutant of S. sindenensis for the production of act-D. Bioprocess parameters for the mutant cultivation and optimum product formation were studied.
The producer microorganism, designated as C-5, was isolated from the soil sample, collected from steel plant effluents (Barabanki, U.P., India) and maintained on ISP-2 (International Streptomyces Project) agar slants containing glucose 4g, yeast extract 4g, malt extract 10g, CaCO3 2g, agar 20g and distilled water 1L, pH adjusted to 7–7.2 before sterilization. Morphological and cultural characteristics were studied using the ISP media recommended by Shirling and Gottlieb, 1966 (11). For Scanning Electron Microscopy (SEM) samples were prepared according to the methods described by Castillo et al. (2). The strain was characterized as S. sindenensis by 16S rRNA homology (data not shown) and has been deposited at MTCC (www.http://mtcc.imtech.res.in), Chandigarh, India (MTCC 8122). Nearly complete (1366 bp) 16S rRNA sequence of strain has been submitted in the NCBI Gen Bank database (accession number EF422787).
Antibiotic production was studied in shake flask with the production medium containing: soy bean meal 10 g, glycerol 15 ml, MgSO4 .7H2O 0.5 g, (NH4)2HPO4 0.5 g, K2HPO4 1.0 g, NaCl 3 g, CaCO3 2g, and distilled water 1L, pH adjusted to 7–7.2 before sterilization in autoclave at 15 lb PS i (1 Kg PS cm) for 15 minutes. Seed culture was prepared in 250 ml Erlenmeyer flask containing 50 ml of production medium by inoculating a loop full culture from the slant and incubating at 28°C on rotary shakers at 200 rpm for 48 h. Antibiotic production was observed in the same medium by inoculating 1L flask (200 ml medium) with 2.5% (v/v) of seed culture and growing under the same conditions for 168 h. Fermented broth was centrifuged and supernatant was extracted thrice with ethyl acetate, filtered and concentrated invaccuo. Antibiotic titer was estimated by the Reverse phase HPLC with ODS-3 column (outer dia.., 250 x 6.35 mm) with a particle size of 10 μ and Lambda-Max Spectrophotometer- LC 481 variable wavelength detector 254 nm, at a flow rate of 0.6 ml/min. with acetonirile:water (55:45) as eluting solvent. Act-D was eluted at 21.4 min retention time. Unless otherwise stated, all the chemicals and media components were purchased from Hi-media labs, Mumbai, India. Bacillus subtilis ATCC 6633 was used as the test strain for the bioassay of antibiotic production.
Spores of S. sindenensis were gently scrapped from the surface of ISP-2 agar plates, washed with sterile normal saline (0.85%) and filtered through glass wool. Spore suspension was diluted to have a count of 104 / ml as, determined by the viability observed on ISP-2 agar plate. Three ml of spore suspension was irradiated for 30 min. with a UV lamp (254 nm) placed about 30 cm above from the liquid surface and gently swirled (magnetically with a needle) in a Petri dish (covered by a piece of dialyzing membrane). Following irradiation, spores were kept in dark at 4°C overnight.
Rifampicin (Rf) and streptomycin (S) were used as screening agents for identification of mutants among the survivors. After incubation in dark, spores were plated in triplicates on ISP-2 agar plates containing 25 μg ml-1of S and Rf each, incubated at 28°C and observed after 48 hr. Mutant colonies with different morphology and expressing, successively, resistance to S and Rf were isolated. Antibiotic production by mutants was studied with 10 μg ml-1 of S and Rf added in shake flasks. Parallel control flasks were also run for the parent culture. Confirmation for improved antibiotic production was done in shake flasks with no addition of S and Rf.
Effects of different carbon and nitrogen sources on antibiotic production were evaluated for medium optimization (Table 1). Glycerol was replaced with different sugars (1%). Amino acids were supplemented to complete production medium (0.1%). Mutant was cultivated in optimized medium using NBS BioFlow 110 bench top stirred bioreactor.
Table 1.
Influence of C & N sources on act-D production by parent and mutant.
| C & N sources | dry cell weight g l-1 | act-D, mg l-1 | ||
|---|---|---|---|---|
| parent | M-46 | parent | M-46 | |
| replacement of glycerol with (1%) | ||||
| Fructose | 7.2 | 6.2 | 190 | 660 |
| Lactose | 6.8 | 6.0 | 156 | 510 |
| Maltose | 7.0 | 6.0 | 130 | 590 |
| Mannose | 7.5 | 7.0 | 70 | 369 |
| Xylose | 7.5 | 6.2 | 110 | 480 |
| production medium | 7.0 | 6.1 | 80 | 400 |
| supplimentation of production medium with (0.1%) | ||||
| L-asparagine monohydrate | 6.8 | 6.0 | 66 | 440 |
| DL-aspartic acid | 7.5 | 6.2 | 148 | 220 |
| L-glutmate | 7.0 | 5.5 | 141 | 284 |
| L-histidine | 6.8 | 6.0 | 110 | 335 |
| Hydroxy L-proline | 6.9 | 6.5 | 90 | 330 |
| DL-isoleucine | 6.8 | 7.2 | 88 | 286 |
| DL-serine | 6.8 | 6.0 | 64 | 266 |
| DL-threonine | 7.2 | 6.5 | 126 | 620 |
| L-tryptophan | 7.5 | 6.2 | 74 | 438 |
| Optimized medium | 6.8 | 6.2 | 215 | 710 |
| Optimized medium (bioreactor) | 7.0 | 6.8 | 270 | 850 |
The rate of survival of UV irradiated cell progenies was 1%. Fifty colonies, showing resistance for higher concentration of Rf and S (25 μg ml-1 each) and higher zone of inhibition against B. subtilis were selected for further studies. One such mutant designated as M-46, produced 5 folds higher act-D (400 mg/l-1) as compared to the parent strain (80 mg/l-1).
The parent and M-46 were studied for their cultural characteristics with respect to the utilization of nitrate, urea and sodium citrate. M-46 utilized Sodium citrate while the parent did not. M-46 better utilized other substrates (starch, asparagine, yeast extract and tyrosine) as evidenced by the colony size and pigment production. Growth of parent and M-46 was also observed on medium ISP 2 - 7 media. The parent strain did not produce melanin on ISP-6 agar where as M-46 produced brown color indicating the production of melanin. On ISP-2 agar plates, M-46 showed vigorous sporulation and higher pigment production. As shown in SEM images (Fig. 1) M-46 spores were stouter (0.7 × 0.5 μm) as compared to spores produced by the parent strain (0.7 × 0.3 μm).
Figure 1.
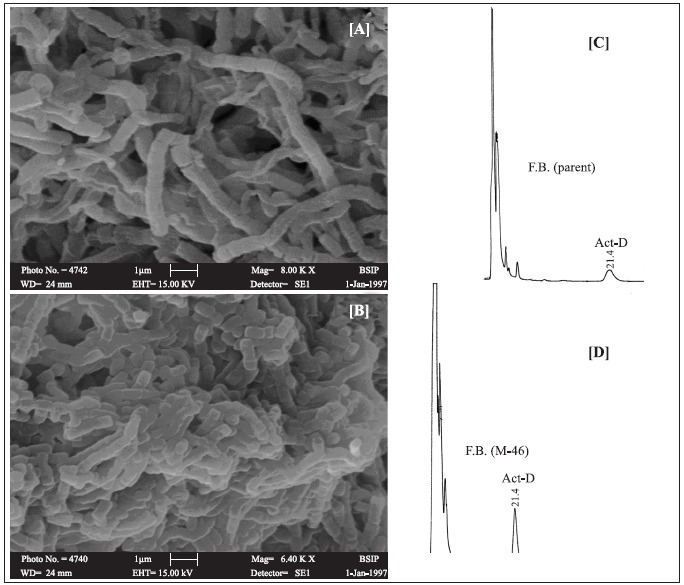
A & B, showing SEM images of parent and mutant strain and C & D showing the HPLC profile of fermented broth (F.B.) of parent and mutant strains respectively.
Influence of different carbon and nitrogen sources on act-D production by parent strain and M-46 is given in Table 1. Most of the medium components that favoured act-D production by the parent strain also favoured mutant. It is evidenced from Fig. 1 (C & D) that act-D production by the mutant was enhanced. Negative effect exerted by glutamate and aspartic acid (29 and 45% decreased production) on antibiotic production by the mutant was the only contradiction observed. Replacement of glycerol with fructose gave an enhancement of 65 and 138% in the antibiotic titer of M-46 and parent strain respectively (Table 1). Incorporation of DL-threonine in the production medium also favoured act-D production by the mutant as well as parent strain (55 and 57% respectively). Finally a production medium containing: soy bean meal 10 g, fructose 15 g, DL-threonine 1 g, MgSO4 .7H2O 0.5 g, (NH4)2HPO4 0.5 g, K2HPO4 1.0 g, NaCl 3 g, CaCO3 2g, and distilled water 1L, pH adjusted to 7–7.2, was used for act-D production by M-46 giving an enhancement of 77% (710 mg/l-1) as compared to the yield obtained with M-46 in normal production medium. When M-46 was cultivated in stirred bioreactor with aeration and agitation rates of 1.5 vvm and 600 rpm respectively, productivity was enhanced by 963% (850 mg/l-1) as compared to the parent strain, cultivated in shake flasks with non-optimized medium (80 mg/l-1).
In our study, we have observed an enhancement in act-D synthesis by the introduction of Rifampicin and Streptomycin resistance into S. sindenensis. Introduction of certain mutations into rpo gene that confer resistance to Rifampicin are reported to activate the antibiotic production by Streptomyces spp. (5). DNA-dependent RNA polymerase (RNAP), which is composed of an essential catalytic core enzyme (α2 β β ω and one of the sigma (σ) factors, is the central enzyme for the expression of genomic information in all organisms. Rifampicin (Rif) inhibits transcription initiation by blocking the subunit of bacterial RNA Polymerase (7). Resistance to streptomycin is brought about by mutation in the rpsL gene which encodes for the S12 protein of the 30S subunit of the ribosome (10).
In our studies, fructose was found to induce a substantial increase in act-D production by the mutant and parent both. Inbar and Lipidot (6) showed that carbon atoms of an intracellular glutamate pool of S. parvullus were not derived biosynthetically from the culture medium glutamate source but rather from fructose catabolism. Foster and Katz (4) found that in case of S. parvullus use of L-glutamate and L-aspartate as a C-source exerts catabolic repression on synthesis of tryptophan oxygenase, an enzyme needed for the synthesis of actinomycin. Perhaps in case of M-46 this catabolic repression has got expressed at a higher level as L-glutamate and DL-aspartic acid supplementations were found to inhibit the production of act-D significantly (29 and 45% respectively). Some nitrogen sources may get incorporated in antibiotic molecules as precursors or their amino groups transfer to specific intermediate products (1). Katz and Goss (8) have reported up to 83% enhancement in act-D production by S. chrysomallus with the addition of DL-valine in the production medium. In our studies, positive effects of DL-threonine, L-valine and proline could be due to their direct incorporation in the peptide chains attached to the chromophore (actinocin) of act-D molecule.
ACKNOWLEDGEMENT
This study was financially supported by Council of Scientific and Industrial Research, India.
RESUMO
Produção de actinomicina-D por um mutante de uma nova cepa de Streptomyces sindenensis
Uma cepa produtora de actinomicina-D foi isolada de solo e caracterizada como Streptomyces sindenensis. A cultura foi submetida à radiação UV, e um mutante capaz de produzir 400% mais actinomicina-D foi isolado (400mg/L comparado a 80mg/L produzido pela cepa parental). O meio de produção do antibiótico foi otimizado e o rendimento aumentou para 850 mg/L, ou seja, 963% mais alto que a cepa parental.
Palavras-chave: actinomicina-D, antibióticos, Streptomyces, mutação
REFERENCES
- 1.Aharonowitz Y. Nitrogen metabolite regulation of antibiotic biosynthesis. Ann. Rev. Microbiol. 1980;34:209–234. doi: 10.1146/annurev.mi.34.100180.001233. [DOI] [PubMed] [Google Scholar]
- 2.Castillo U.F., Strobel G.A., Ford E.J., Hess W.M., Porter H., Jensen J.B., Albert H., Robison R., Condron M.A., Teplow D.B., Stevens D., Yaver D. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiology. 2002;148:2675–2685. doi: 10.1099/00221287-148-9-2675. [DOI] [PubMed] [Google Scholar]
- 3.Farber S., D’Angio G., Evans A., Mitus A. Clinical studies of actinomycin D with special reference to Wilms’ tumor in children. 1960. J. Urol. 2002;168:2560–2562. doi: 10.1016/S0022-5347(05)64213-9. [DOI] [PubMed] [Google Scholar]
- 4.Foster J.W., Katz E. Control of actinomycin D biosynthesis n Streptomyces parvullus: Regulation of tryptophan oxygenase activity. J. Bacteriol. 1981;148:670–677. doi: 10.1128/jb.148.2.670-677.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu H., Ochi K. Novel approach for improving the productivity of antibiotic-producing strains by inducing combined resistant mutations. Appl. Environtl. Microbiol. 2001;6:1885–1892. doi: 10.1128/AEM.67.4.1885-1892.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Inbar I., Lapidot A. Metabolic regulation in Streptomyces parvulus during actinomycin D synthesis, studied with 13C- and 15N-labeled precursors by 13C and 15N nuclear magnetic resonance spectroscopy and by gas chromatography-mass spectrometry. J. Bacteriol. 1988;170:4055–4064. doi: 10.1128/jb.170.9.4055-4064.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Inaoka T., Kosaku T., Yada H., Yoshida M., Ochi K. RNA polymerase mutation activates the production of a dormant antibiotic 3,3’-neotrehalosadiamine via an autoinduction mechanism in Bacillus subtilis. J. Biol. Chem. 2004;279:3885–3892. doi: 10.1074/jbc.M309925200. [DOI] [PubMed] [Google Scholar]
- 8.Katz E., Goss W.A. Controlled biosynthesis of actinomycin with sarcosine. J. Biochem. 1958;73:458–465. doi: 10.1042/bj0730458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurosawa K., Bui V.P., Van E.J.L., Willis L.B., Lessard P.A., Ghiviriga I., Sambandan T.G., Rha C.K., Sinskey A.J. Characterization of Streptomyces MITKK-103, a newly isolated actinomycin X2-producer. Appl. Microbiol. Biotechnol. 2006;72:145–154. doi: 10.1007/s00253-005-0240-2. [DOI] [PubMed] [Google Scholar]
- 10.Shima J., Hesketh A., Okamato S., Kawamoto S., Ochi K. Induction of actinorhodin production by rpsL mutations that confer streptomycin resistance in S. lividans and S. coelicolor A3(2) J. Bacteriol. 1996;178:7276–7284. doi: 10.1128/jb.178.24.7276-7284.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shirling E.B., Gottlieb D. Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 1966;16:313–340. [Google Scholar]


