Abstract
The decolorization of molasses by 17 different fungi in 2 media was studied. Trichoderma viride showed the highest decolorization yield (53.5%) when cultivated at 30ºC for 7 days in Medium 1 which contained the molasses which was diluted to 40 g/L in distilled water. The other Trichoderma species and Penicillium sp. also gave similar results of 40-45%. Decolorization yield was increased by adding peptone and yeast extract to the production medium except Penicillium sp. Growth rate was not related to decolorization yet pH value was. When the pH decreased below 5.0 after the incubation, the decolorization yield increased. Although reducing sugar in culture broth decreased with decreasing color intensity, there was no connection between protein utilization and decolorizing activity.
Keywords: Molasses, decolorization, fungi
INTRODUCTION
Molasses is one of the most important by-products of sugar production processes. It is the most economical source of carbohydrate for ethanol and citric acid fermentation. Because molasses contains colored and polimerized substances, decolorization of molasses is essential from an environmental point of view (16). In various fermentations, molasses is used as carbon source, due to its rich sucrose and other saccharide content (11-13). Even though it is a suitable raw material for many processes, high concentration of organic matters remains in the fermentation as effluent. In addition to that, wastewater from factories using molasses contains a large amount of dark brown pigment, which is a kind of melanoidin (15). Melanoidin is one of the polymers, which has an unclear structure and is not easily decolorized by the usual biological or chemical treatments. Several studies have been made in order to remove color coming from melanoidin. In these studies, several fungi strains were screened. Biological decolorization of molasses melanoidin has been studied using fungi such as Aspergillus sp., Fusarium sp., Penicillium sp, Sclerotium sp., Pleurotus cystidious, Coriolus versicolor, Cladosporium sp. (15), Coriolus sp. (17), Aspergillus niger (13), Coriolopsis gallica and Paecilomyces variotii (3), the yeast Citeromyces (16) and bacteria such as Bacillus sp (10) and actinomycetes (8). Although much research has been made on decolorization of molasses by various bacteria and fungi, still many of them remain unstudied. The aim of this study is to find alternative fungal sources for decolorization of molasses. In this respect, decolorization efficiencies of different fungi were studied and compared.
MATERIAL AND METHODS
The microorganisms used in this study were obtained from the culture collection of Hacettepe University, Department of Biotechnology. They were maintained on PDA and slants were kept at 4ºC. Transfers were made at 1 month intervals.
The crude molasses was obtained from a sugar factory and stored at 4ºC and used in these experiments. It was dark brown in color. Two types of media were used for detecting the decolorization ability. The first medium (M1) contained the molasses which was diluted to 40 g/L in distilled water and the second (M2) contained 4 g/L peptone and 4 g/L yeast extract in addition to 40 g/L molasses. The pH was adjusted to 6.0 for both media and sterilized before inoculating. 1 ml spore suspension (107 ml-1) of respective strain was inoculated into 100 ml of fresh molasses medium in 250 ml flasks. Three sets of flasks were incubated at 30ºC in the incubator shaker at 150 rev min-1 for submerged cultivation for 7 days. After the cultivation, the flask contents were filtered through Whatman filter paper. In order to determine biomass in terms of dry weight, filter papers were dried until constant weight. The filtrate was checked for color decolorization, reducing sugar, total protein and pH.
The filtrate was centrifuged at 6000×g for 15 min and the color intensity of the solution was measured at 475 nm with a spectrophotometer (Jenway, 105 U.V., Vis spectrophotometer) against the original solution (15). The absorbance of 475 nm is used as a standard index to determine molasses decolorization (10). The results were calculated as the difference in decolorization between readings on days 0 7 and the decolorization yield was expressed as the degree of decrease in the absorbance at 475 nm against the initial absorbance at the same wavelength.
In order to detect the correlation between sugar and decolorization of molasses, sugar utilization studies were examined. Sugar was estimated as reducing sugar from the culture filtrate. The dinitrosalicylic acid (DNS) method was used to measure reducing sugar. Total reducing sugars were determined as glucose equivalents by a colorimetric method. The reagent was prepared by dissolving 1 g of DNS, 0.2 g of phenol, 0.05 g of Na2SO3, 1 g of NaOH and 20 g of sodium– potassium tartrate in 100 ml distilled water. The resultant liquid yielded a clear orange-yellow color, which was used to measure total reducing sugars. The intensity of the resulting color was detected using a spectrophotometer at 550 nm (6).
In order to detect the relationship between protein and decolorization of molasses, protein utilization studies were examined. Total proteins were assayed using the Lowry method (5).
RESULTS AND DISCUSSION
Fungal strains were used to test their ability to decolorize molasses. Results presented in Fig. 1 show that Trichoderma sp. can be good alternatives as they have 40-55% decolorization efficiency. Although there are studies conducted for decolorization of different dyes by Trichoderma species, in literature no similar study on the molasses decolorization with Trichoderma species was found (1,14). In our study, Penicillium species with a molasses decolorization of 44% were detected as the second most successful fungus (Fig. 1). Contrary to our study, Sirianuntapiboon et al. (15) could not achieve the decolorization of molasses by Penicillium species. On the other hand, in another study there is a close result (40%) to that of ours with Penicillium species (4). In this study, Penicillium was followed by Rhizopus with a molasses decolorization of 38%. When compared to other fungi relatively with lower molasses decolorizations, 25% and 20% were achieved for A. alternata and B. bassiana, respectively (Fig. 1). Literature review revealed that there are no studies with these particular fungi on the molasses decolorization, except pulp in study with Rhizopus (9).
Figure 1.
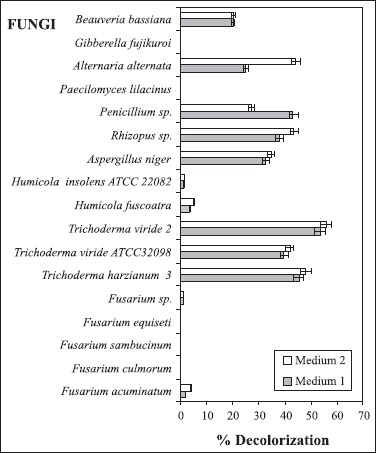
Decolorization activity of fungi in M1 and M2. Each data given are the mean value of three independent experiments. Standard deviations are shown on the graphs.
The molasses decolorizations of Gibberella fujikuroi, Paecilomyces lilacinus, Fusarium species and Humicola species in our study were not successful (Fig. 1). There is no study published on molasses decolorization with Humicola species, G.fujikuroi and P. lilacinus. However, successful decolorization studies with different Paecilomyces species were found (3,17). In parallel to our study, the molasses decolorization with Fusarium species has not been achieved (15). Pigments, which appear in the media, could also impede us to detect the molasses decolorization in the Fusarium species. In addition, A. niger, which is used for molasses decolorization in literature, showed 33% decolorization in our study and it was taken as a reference for the comparison with the other fungi within the scope of our study (Fig. 1) (7,15).
Growths of fungi were also compared during the molasses decolorization. Although the highest growth was detected in F. sambucinum, its decolorization could not be achieved. Furthermore, it was found that the growth of Trichoderma species, which show successful molasses decolorization, were not high (0.18-0.40 g/100 ml) when compared with other fungi. In particular, T. harzianum 3 showed a decolorization of 46% despite its growth of 0.18 g/100 ml (Table 1). These findings show that excess in growth does not affect the decolorization to a great extent. It indicates that enzymes, which resulted in the growth media are effective in the molasses decolorization. Although Rhizopus species and A. alternata with the molasses decolorizations of 38% and 25% respectively showed higher growths (0.74 g/100 ml and 0.76 g/100 ml) when compared with others. Decolorization of microorganisms was achieved either by adsorption of the pigment on to mycelia or by its enzymatic degradation (10). In this case, most of the decolorization was achieved by enzymes in Trichoderma species.
Table 1.
pH, total protein, reduced sugar and dry weight of fungi in M1.
| Fungi | pH | T.P. | R.S. | D.W. |
|---|---|---|---|---|
| Fusarium acuminatum | 8.2 ± 0.1 | 1.39 ± 0.04 | 3.28 ± 0.15 | 0.36 ± 0.02 |
| Fusarium culmorum | 9.2 ± 0.3 | 1.15 ± 0.1 | 3.24 ± 0.06 | 0.80 ± 0.1 |
| Fusarium sambucinum | 6.7 ± 0.11 | 1.15 ± 0.09 | 3.24 ± 0.11 | 0.98 ± 0.08 |
| Fusarium equiseti | 6.6 ± 0.11 | 1.15 ± 0.12 | 3.24 ± 0.1 | 0.46 ± 0.05 |
| Fusarium sp. | 8.9 ± 0.37 | 1.35 ± 0.06 | 3.24 ± 0.09 | 0.58 ± 0.06 |
| Trichoderma harzianum 3 | 4.5 ± 0.19 | 1.99 ± 0.13 | 3.24 ± 0.14 | 0.18 ± 0.05 |
| Trichoderma viride ATCC32098 | 4.6 ± 0.12 | 1.86 ± 0.11 | 3.24 ± 0.12 | 0.40 ± 0.05 |
| Trichoderma viride 2 | 4.5 ± 0.1 | 1.38 ± 0.08 | 3.20 ± 0.13 | 0.36 ± 0.08 |
| Humicola fuscoatra | 6.3 ± 0.2 | 1.25 ± 0.12 | 2.40 ± 0.08 | 0.46 ± 0.02 |
| Humicola insolens ATCC 22082 | 5.5 ± 0.23 | 0.81 ± 0.05 | 3.24 ± 0.15 | 0.46 ± 0.05 |
| Aspergillus niger | 2.9 ± 0.13 | 1.17 ± 0.09 | 3.28 ± 0.1 | 0.62 ± 0.09 |
| Rhizopus sp. | 2.3 ± 0.15 | 0.49 ± 0.08 | 3.24 ± 0.13 | 0.74 ± 0.11 |
| Penicillium sp. | 4.9 ± 0.29 | 1.42 ± 0.11 | 3.28 ± 0.17 | 0.30 ± 0.02 |
| Paecilomyces lilacinus | 6.4 ± 0.32 | 1.20 ± 0.14 | 3.20 ± 0.18 | 0.44 ± 0.04 |
| Alternaria alternata | 2.7 ± 0.24 | 0.74 ± 0.1 | 3.24 ± 0.18 | 0.76 ± 0.09 |
| Gibberella fujikuroi | 7.1 ± 0.4 | 1.52 ± 0.2 | 3.16 ± 0.2 | 0.32 ± 0.03 |
| Beauveria bassiana | 4.2 ± 0.25 | 1.33 ± 0.24 | 2.92 ± 0.11 | 0.70 ± 0.1 |
T.P: Total Protein (mg/ml); R.S.: Reduced sugar (mg/ml); D.W: Dry weight (g/100 ml).
Each data given are the mean value of three independent experiments. Standard deviations are shown on the graphs.
After the incubation, pH was detected. The results show that pH generally increased in Fusarium species, while no decolorization was observed. The pH values in Trichoderma species, which show higher decolorization, were often measured as acidic (pH 4.5). It was observed that the molasses decolorization was achieved in fungi, where the acidic condition is created with the pH value of lower than 4.9 after productions (Table 1). In this respect, it was thought that acidic conditions after incubation might be related with decolorization. Therefore, it can be said that enzymes formed by fungi, especially those showing an enzymatic molasses decolorization, were optimum in acidic conditions and thereby lead to more successful decolorization. According to Sirianuntapiboon et al. (16), the pH value decreased to values below 4 after incubation in their decolorization study. It can be concluded that the decrease in pH has a critical role in decolorization (2).
Adikane et al. concluded that the utilization of organic nitrogen (protein) and carbon source (reducing sugar) has a critical role in decolorization (2). However, when the remaining reducing sugar after the production of fungi in our study was considered, there was no difference between reducing sugar amounts remained in the media of decolorized and non-decolorized fungi (Table 1). Similar to our results, Sirianuntapiboon et al. (16) showed that the reducing sugar in culture broth rapidly decreased with decreasing color intensity.
When the remaining protein after production was considered, the difference of the protein utilization in fungi was higher than the difference of reducing sugar utilization in this media. A. alternata, Rhizopus sp. and H. insolens ATCC 22082 showed the highest protein utilizations. Since higher decolorization efficiencies were observed in A. alternata and Rhizopus, and lower decolorization efficiency was observed in H. insolens ATCC 22082, no relation with protein utilization could be found. In addition, such correlations have not been found in other studies.
In another part of our study, we tried to achieve more efficient molasses decolorization by adding peptone and yeast extract as additional nutrients to the production medium. It was seen that the efficiency of decolorization increased in all fungi except Penicillium sp, in small amounts, when additional nutrition sources were added (Fig. 1). This shows that the decolorization efficiency can be increased with the optimization of the media. Lower decolorization efficiency in Penicillium sp can be explained with the pH effect in decolorization activity. Using fungi, which showed high decolorization efficiency, was suitable. On the other hand, the ones with low decolorization should not be preferred as it increases production costs. Results were confirmed with the fact that the pH values, protein, reducing sugar are similar to those mentioned above (Table 2).
Table 2.
pH, total protein, reduced sugar and dry weight of fungi in M2.
| Fungi | pH | T.P. | R.S. | D.W. |
|---|---|---|---|---|
| Fusarium acuminatum | 8.6 ± 0.12 | 3.36 ± 0.1 | 1.40 ± 0.2 | 0.50 ± 0.09 |
| Fusarium culmorum | 9.3 ± 0.2 | 3.08 ± 0.14 | 1.64 ± 0.1 | 0.73 ± 0.11 |
| Fusarium sambucinum | 8.6 ± 0.14 | 3.14 ± 0.1 | 1.60 ± 0.14 | 0.85 ± 0.12 |
| Fusarium equiseti | 7.80 ± 0.3 | 2.61 ± 0.08 | 1.59 ± 0.1 | 0.59 ± 0.08 |
| Fusarium sp. | 9.0 ± 0.4 | 3.40 ± 0.08 | 1.63 ± 0.22 | 0.88 ± 0.14 |
| Trichoderma harzianum | 34.9 ± 0.08 | 1.23 ± 0.04 | 1.20 ± 0.2 | 0.34 ± 0.12 |
| Trichoderma viride ATCC32098 | 4.8 ± 0.14 | 2.57 ± 0.11 | 1.25 ± 0.1 | 0.42 ± 0.2 |
| Trichoderma viride | 24.6 ± 0.12 | 1.54 ± 0.04 | 1.52 ± 0.3 | 0.28 ± 0.08 |
| Humicola fuscoatra | 6.5 ± 0.22 | 3.37 ± 0.15 | 1.36 ± 0.14 | 0.54 ± 0.15 |
| Humicola insolens ATCC 22082 | 5.7 ± 0.2 | 1.47 ± 0.01 | 1.40 ± 0.22 | 0.86 ± 0.12 |
| Aspergillus niger | 3.3 ± 0.05 | 1.27 ± 0.08 | 1.38 ± 0.2 | 1.06 ± 0.2 |
| Rhizopus sp. | 4.1 ± 0.22 | 2.19 ± 0.08 | 1.28 ± 0.17 | 0.82 ± 0.08 |
| Penicillium sp. | 7.5 ± 0.3 | 1.52 ± 0.05 | 1.32 ± 0.22 | 0.62 ± 0.09 |
| Paecilomyces lilacinus | 6.8 ± 0.2 | 2.67 ± 0.04 | 1.44 ± 0.32 | 0.86 ± 0.12 |
| Alternaria alternata | 3.5 ± 0.3 | 3.05 ± 0.1 | 1.32 ± 0.2 | 1.28 ± 0.14 |
| Gibberella fujikuroi | 8.2 ± 0.4 | 2.47 ± 0.1 | 1.26 ± 0.08 | 0.38 ± 0.04 |
| Beauveria bassiana | 4.3 ± 0.1 | 1.29 ± 0.1 | 1.20 ± 0.05 | 0.71 ± 0.05 |
T.P.: Total Protein (mg/ml); R.S.: Reduced sugar (mg/ml); D.W.: Dry weight (g/100 ml);
Each data given are the mean value of three independent experiments. Standard deviations are shown on the graphs.
In our study, it can be concluded that Trichoderma, Rhizopus and Penicillium species can be utilized for the molasses decolorization. Although not as high as to other fungi, molasses decolorization activities of A. alternata and B. bassiana (25% and 20% respectively), can be evaluated as an important outcome since they have not been studied before. It was concluded that more efficient decolorization activities could be achieved through optimizations. However, it was observed that the decrease in pH after incubation was more effective in the decolorization for the studied species of fungi.
RESUMO
Triagem de fungos para descoloramento de melaço
Este estudo avaliou o descoloramento do melaço por 17 fungos em dois meios. Trichoderma viride apresentou o melhor rendimento de descoloramento (53,5%) quando cultivado a 30ºC por 7 dias no meio 1, composto de melaço diluído a 40 g/L em água destilada. As outras espécies de Trichoderma e Penicillium sp apresentaram rendimento da ordem de 40-45%. O rendimento de descoloramento aumentou com a adição de peptona e extrato de levedura ao meio de produção, com exceção de Penicillium sp. A taxa de crescimento não teve relação com o descoloramento, mas o pH sim. Quando o pH diminuiu para abaixo de 5,0 depois da incubação, o rendimento de descoloramento foi maior. Embora os açúcares redutores no meio de cultura tenham diminuído com a diminuição da intensidade de cor, não houve relação entre utilização de proteína e atividade descolorizante.
Palavras-chave: melaço, descoloramento, fungos
REFERENCES
- 1.Abd El-Rahim W.M., Moawad H., Khalafallah M. Microflora involved in textile dye waste removal. J. Basic Microbiol. 2003;43:167–174. doi: 10.1002/jobm.200390019. [DOI] [PubMed] [Google Scholar]
- 2.Adikane H.V., Dange M.N., Selvakumari K. Optimization of anaerobically digested distillery molasses spent wash decolorization using soil as inoculum in the absence of additional carbon and nitrogen source. Bioresour. Technol. 2006;97:2131–2135. doi: 10.1016/j.biortech.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Calvo A.M., Galletti G.C., Gonzalez A.E. Paper wastewater analyses by pyrolysis gas-chromatography mass-spectrometry during biological decolorization with the fungi Coriolopsis gallica and Paecilomyces variotii. J. Anal. Appl. Pyrol. 1995;33:39–50. [Google Scholar]
- 4.Jiménez A.M., Borja R., Martín A. Aerobic-anaerobic biodegradation of beet molasses alcoholic fermentation wastewater. Process Biochem. 2003;38:1275–84. [Google Scholar]
- 5.Lowry O.H., Rosenbrough N.J., Farr A.L., Randal R.J. Protein measurement with the folin reagent. J. Biol. Chem. 1951;19:265–275. [PubMed] [Google Scholar]
- 6.Miller G.L. Use of dinitrosalicilic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 7.Miranda M.P., Benito G.G., San Cristobal N., Nieto C.H. Color elimination from molasses wastewater by Aspergillus niger. Bioresour. Technol. 1996;57:229–235. [Google Scholar]
- 8.Murata M., Terasawa N., Homma S. Screening of microorganisms to decolorize a model melanoidin and the chemical properties of a microbially treated melanoidin. Biosci. Biotech. Biochem. 1991;56:1182–1187. [Google Scholar]
- 9.Nagarathnamma R., Bajpai P. Decolorization and detoxification of extraction -stage effluent from chlorine bleaching of kraft pub by Rhizopus oryzae. Appl. Environ. Microbiol. 1999;65:1078–1082. doi: 10.1128/aem.65.3.1078-1082.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakajima-Kambe T., Shimomura M., Nomura N., Chanpornpong T., Nakahara T. Decolorization of molasses wastewater by Bacillus sp. under thermophilic and anaerobic conditions. J. Biosci. Bioeng. 1999;87:119–121. doi: 10.1016/s1389-1723(99)80021-8. [DOI] [PubMed] [Google Scholar]
- 11.Pinotti T.I, Carvalho P.M.B., Garcia K.M.G., Silva T.R., Hagler A.N., Leite S.G.F. Media components and amino acid supplements influencing the production of fruity aroma by Geotrichum candidum. Braz. J. Microbiol. 2006;37:494–498. [Google Scholar]
- 12.Rossi I.H., Monteiro A.C., Machado J.O., Andrioli J.L., Barbosa J.C. Shiitake (Lentinula edodes) production on a sterilized bagasse substrate enriched with rice bran and sugarcane molasses. Braz. J. Microbiol. 2003;34:66–71. [Google Scholar]
- 13.Rossi I.H., Monteiro A.C., Machado J.O., Barbosa J.C. Supplementation of sugarcane bagasse with rice bran and sugarcane molasses for shiitake (Lentinula edodes) spawn production. Braz. J. Microbiol. 2003;34:61–65. [Google Scholar]
- 14.Sadhasivam S., Saritha E., Savitha S. Comparison of the efficacy of live and autoclaved mycelium of Trichoderma harzianum on the removal of trypan blue. Bull. Environ. Contamin. Toxicology. 2005;75:1046–1053. doi: 10.1007/s00128-005-0855-0. [DOI] [PubMed] [Google Scholar]
- 15.Sirianuntapiboon S., Somchai P., Ohmomo S., Atthasumpunna P. Screening of filamentous fungi having the ability to decolorize molasses pigment. Agric. Biol. Chem. 1988;52:387–392. [Google Scholar]
- 16.Sirianuntapiboon S., Zohsalam P., Ohmomo S. Decolorization of molasses wastewater by Citeromyces sp. WR-43-6. Process Biochem. 2004;39:917–924. [Google Scholar]
- 17.Terasawa N., Murata M., Homma S. Decolorization of brown pigments in foods by immobilized mycelia of Coriolus versicolor IFO 30340 and Paecilomyces canadensis NC-1. J. Food Sci. 2000;65:870–875. [Google Scholar]


