Abstract
Listeria monocytogenes is a bacterium capable to adhere to the surfaces of equipment and utensils and subsequently form biofilms. It can to persist in the food processing environmental for extended periods of time being able to contaminate the final product. The aim of this study was to trace the contamination route of L. monocytogenes on a fresh mixed sausage processing line, from raw material to the final product. The isolates obtained were characterized by serotyping and molecular typing by pulsed-field gel electrophoresis (PFGE) using the restriction enzymes ApaI and AscI. L. monocytogenes was detected in 25% of the samples. The samples of raw material were not contaminated, however, the microorganism was detected in 21% of the environmental samples (food contact and non-food contact), 20.8% of the equipments, 20% of the food worker’s hands, 40% of the mass ready to packaging and in all the final products samples, demonstrating that the contamination of final product occurred during the processing and the importance of cross contamination. PFGE yielded 22 pulsotypes wich formed 7 clusters, and serotyping yielded 3 serotypes and 1 serogroup, however, the presence of serotypes 4b and 1/2b in the final product is of great concern for public health. The tracing of contamination showed that some strains are adapted and persisted in the processing environment in this industry.
Keywords: Listeria monocytogenes, PFGE, serotyping, persistent strains
INTRODUCTION
Listeria monocytogenes represents an important hazard to human health because it is capable of causing listeriosis, an atypical foodborne disease with a high fatality rate, ranging from 25 to 30% in susceptible populations (15). Contamination of products by this microorganism occurs more commonly during their processing and it is often the result of persistence of strains in the food processing plant (9, 20, 30). Beef, poultry, seafood and dairy processing plants can be contaminated by L. monocytogenes (3, 5, 7) and studies have shown that this bacterium can survive in the plant for extended periods of time (17, 18, 20, 28).
Thirteen serotypes have been reported for L. monocytogenes. Three of them (1/2a, 1/2b and 4b) are associated with the majority of sporadic cases of human listeriosis and the serotype 4b is linked to almost all recent outbreaks of listeriosis. Serotyping may be used to investigate foodborne disease outbreaks and to identify sources of contamination and routes of spread in the food processing environment, but this technique is less discriminatory than molecular typing methodologies (22). Nevertheless, it provides relevant information for quick discrimination of isolates obtained during suspicious outbreaks, and it improves the efficiency of other more sensitive typing methods (14). Pulsed-field gel electrophoresis (PFGE) have been successful used in characterization of L. monocytogenes, mainly in epidemiological investigation and the tracing of contamination of food processing plants. This method has a very high discriminatory power and is reproducible (12).
Pork meat and processed pork products have been the sources of outbreaks of listeriosis (30), however, there has been sporadic information on the occurrence in processed pork meat products in Brazil. Fresh mixed sausage (a sausage made of raw pork meat, beef meat and pork fat) is a very popular Brazilian product, thus, the evaluation about the prevalence and contamination route of Listeria monocytogenes in these products is a cause of concern. The aim of this work was to trace the contamination route of L. monocytogenes on a fresh mixed sausage processing line, from raw material to the final product, of a slaughterhouse in the city of Pelotas-RS, Brazil, using serology and PFGE.
MATERIALS AND METHODS
Sampling
All material were collected from a fresh mixed sausage processing line of a slaughterhouse under state inspection, in Pelotas, RS, Brazil, and the sampling followed the method recommended by APHA (1). Sixty-eight samples [10 from raw material, 19 from processing environment and surfaces (food contact surfaces and non-food contact surfaces), 5 from food worker’s hands, 24 from equipment, 5 from mass ready for packaging and 5 from the final product] from 15 sampling points [raw material (pork, beef and pork fat), equipments (disintegrator, mincer, mixer, packaging equipment, tieing equipment), environment and surfaces (working tables, boxes, troleys, drains), handlers, mass ready for packing and final product] were analyzed in a period of 5 months. Other raw materials involved in sausage mixing, such spices and preservers, don’t have been tested in this study. All the final products analyzed were made from the same batches of raw materials tested. All samples were collected prior to processing, except the samples of final product and of mass ready for packaging.
Isolation of L. monocytogenes
Isolation was carried out according to the methodology described by Farber et al. (10), using Listeria enrichment broth (UVM formulation) incubated for 24h at 30°C as primary enrichment. Fraser broth was used as secondary enrichment and was incubated for 48h at 35°C. Aliquots were streaked onto Oxford and Palcam agar plates that were incubated at 35°C for 48 hours. Three to five characteristic colonies from each plate were submitted to biochemical identification. The tests used were catalase production; motility at 25°C (motility test medium, Difco, USA); ß-hemolysis production (Tryptone soy agar + 7% horse blood) and carbohydrate fermentation (dextrose, ramnose, xylose and manitol, Vetec, BR). All media were from Oxoid (UK) except where noted.
Pulsed Field Gel Electrophoresis
The CDC (12) protocol was used for PFGE analysis. Briefly, a suspension of pure culture was prepared and optical density (A610nm) determined (Ultrospec 2000, Pharmacia, USA). Density was adjusted to 1.25 – 1.35, lysozyme solution (10mg mL-1, Pharmacia) was added and the mixture was incubated at 37°C for 10min. 300µL of SSP solution [1.2% Agarose Pulsed Field (USB, USA); 1% sodium dodecyl sulphate (Sigma, USA), 0.2mg mL-1 proteinase K (Pharmacia)] were added and the mixture transferred to PFGE molds (BioRad, USA). The blocks were incubated in lysis solution (50mM Tris-HCl, 50mM EDTA pH 8,0, 1% sarcosine (Sigma, USA) and 0.15mg mL-1 proteinase K) under agitation (175rpm; incubator shaker, Innova 4000, New Brunswick Scientific) for 2h at 54°C. Blocks were digested overnight with ApaI (160U block-1) and AscI (25U block-1) (New England Biolabs, USA), incubated at 25°C and 37°C, respectively. Digested products were subjected to PFGE in a CHEF-DR III (BioRad, USA) using TBE at 14°C. The electrophoretic parameters used were as follows: initial switch time, 40 seg; run time, 22h; angle 120°; gradient 6.0v cm-1; temperature, 14°C, ramping factor, linear. The gels were stained with ethidium bromide solution and pattern images were acquired in Photo Documentation System (Vilber Lourmat, Germany). In addition to the L. monocytogenes isolates obtained from the processing plant, two standard strains isolated from food, Salmonella Enteritidis and L. monocytogenes 1529, were used as an outgroup and were typed by PFGE.
The electrophoretic profiles generated with each enzyme were compared and grouped. The size of bands of each profile was calculated using the EDAS 120 system (Eastman Kodak, USA), and the correlation among the profiles was evaluated with NTSYSpc 2.0 (Numerical Taxonomy and Multivariate Analysis System) program, using the Dice coefficient of similarity (23). The same program was used for the construction of the dendrogram based on NJOIN (Neighbor-joining method) group analysis (25).
RESULTS AND DISCUSSION
Global contamination by Listeria monocytogenes: a total of 68 samples were collected from a processing line of fresh mixed sausage in Pelotas, RS, and the overall prevalence of L. monocytogenes in the samples was 25%. The raw material was not contaminated (0/10). Nevertheless, the environment and the equipment (9/43), the handlers (1/5), the mass ready to packing (1/5), and the final products (5/5) were more frequently contaminated. In a previous study, our group has shown that L. monocytogenes was prevalent in three fresh sausages plants and their products, at different stages of manufacturing (27).
An important observation is that none of the samples of raw material were contaminated by L. monocytogenes, however, the pathogen was isolated in all samples from the final product demonstrating that raw material was not an important vehicle in the dissemination of this microorganism during processing. The contamination of meat products in processing plants mostly appears to be due to L. monocytogenes strains already present in the plant environment. There are some disagreements between the most important sources of L. monocytogenes contamination on processing lines. While many authors have emphasized the importance of equipment and environment as a source of contamination (2, 8, 15, 28), others highlighted the importance of raw material as contamination sources (11, 17).
Serotyping: 83 isolates of L. monocytogenes were investigated and 71.1% (59) belonged to serotype 1/2c, 12.1% (10) to serotype 1/2b, 2.4% (2) to serotype 4b, and 14.5% (12) to serogroup 1. Serotype 1/2c was isolated in all sampling points, with exception of the mixture ready for packing. Several studies have shown that L. monocytogenes strains isolated from meat processing environments are frequently of serotypes 1/2a, 1/2/b and 1/2c (6, 13, 29). This may be associated with enhanced capacity to L. monocytogenes serotype 1/2c to attach the stainless steel surfaces, commonly used in food-processing environments (19).
Despite the lack of information about human listeriose in Brazil, L. monocytogenes strains serotypes 4b, 1/2a and 1/2b are the most prevalent in samples of patients in this country (14, 16). This form, it is important to highlight that the isolation of the serotypes 4b and 1/2b in the final product is of great concern for public health. The presence of these strains may result in sporadic cases or an outbreak of listeriosis, particularly if this product is consumed by persons in the at-risk population or if properly cooked (15, 30).
Distribution of Listeria monocytogenes genotypes in the processing environment: PFGE discriminated the 83 isolates into 22 combined ApaI and AscI PFGE patterns (pulsotypes). DNAs digested with ApaI showed 10-16 fragments ranging from approximately 25.6 to 520.8kb (Figure 1), while 5-12 fragments of 30.8 to 626.6kb (Figure 2) were obtained following digestion using AscI.
Figure 1.
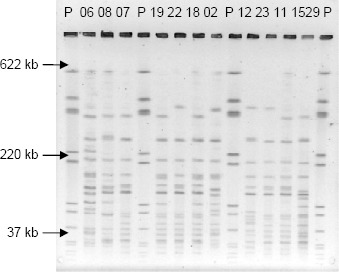
PFGE profiles obtained with the ApaI restriction enzime. P: L. monocytogenes H 2446 digested with AscI; 1529: L. monocytogenes from the outgroup; 06; 08; 07; 19; 22; 18; 02; 12; 23; 11: Isolates of L. monocytogenes digested with ApaI.
Figure 2.
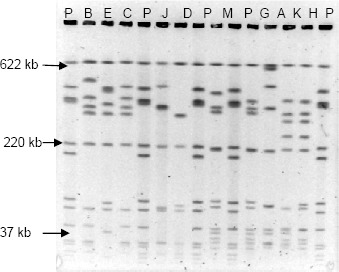
PFGE profiles obtained with the AscI restriction enzyme. P: L. monocytogenes H 2446 digested with AscI; B; E; C; J; D; M; G; A; K; H: isolates of L. monocytogenes digested with AscI.
The final product was the only one who made the highest number of pulsotypes (17). The majority of pulsotypes isolated of final product was unique (P5; P8; P9; P10; P11; P13; P15; P17; P18; P19; 20; P21; P22), in other words , they were not isolated from any other sample. These strains may be present in the processing plant in undetectable quantities to detect trough the conventional techniques of isolation, but once they contaminate the mass, they can multiply during the 24h period of storage in the cold room and be detected in the final product (7, 26).
The high genetic diversity of final product, without contamination of raw material demonstrate that the contamination occurred during the processing, and that raw meat seemed not to be an important factor in the contamination of fresh mixed sausage. Several studies, using different molecular techniques (AFLP, PFGE, RAPD, Ribotyping) have shown that the genotypes of L. monocytogenes strains collected from pork meat processing environment are extremely diverse (6, 20, 21, 28). Thévenot et al. (28) stated that high genotype diversity among strains collected during processing and processed products may indicate either a continuous source of inoculation by raw material or the persistence of several strains in spite of the cleaning and disinfection operations. The first hypothesis can’t be sustained in our study, since no L. monocytogenes was found in raw meat.
It was verified that pulsotypes P1, P2, P4, P7 and P16 persisted in the environment of the processing plant, as they were isolated repeatedly during the sampling period (data not shown), suggesting that they are well adapted to the environment and able to colonize the production line. Similar results were found by Autio et al. (2), Berrang et al. (4), Lundén et al. (20) and Senczek et al. (26). In addition, pulsotypes P2 and P16 were found contaminating more than one sampling point during the same day.
Five samples (two from the final product, one from the mincer, one from the tieing equipment and one from the worker’s hands) presented contamination for more than one L. monocytogenes strain, which was evidenced by the presence of more than one pulsotype in these samples. The finding of several pulsotypes in a single sample may be due to contamination at many different sites. These results highlight that several isolates from a single sample should be typed in epidemiological and contamination studies, according also reported in other studies (2, 8).
Analyzing the dendrogram (Figure 3), it can be observed the existence of 7 distinct clusters. Cluster IA was the predominant one, grouping 8 pulsotypes and 34 of 83 strains analyzed. Profiles less genetically related formed separate clusters: IB, IC, ID and IIIA.
Figure 3.
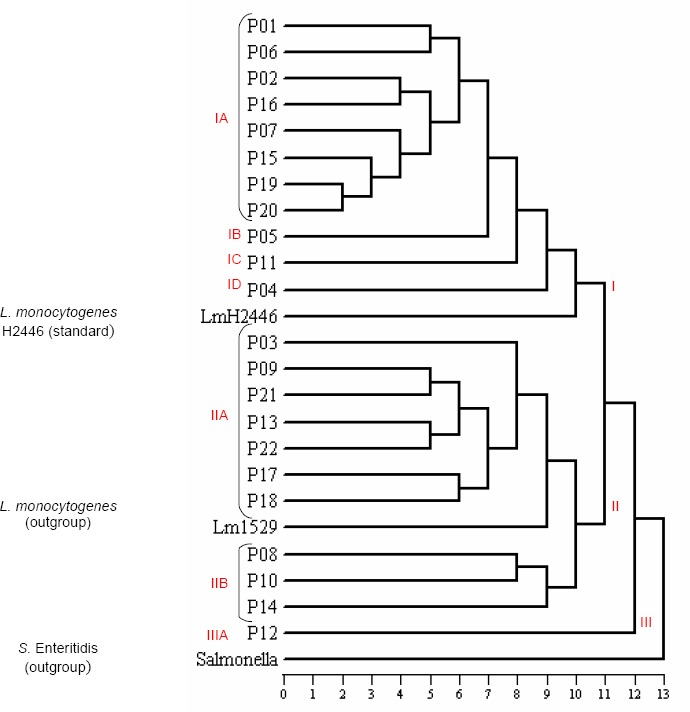
Representation of the genetic relationship among the 22 pulsotypes obtained from 83 L. monocytogenes isolates and L. monocytogenes H2446 (standard), L. monocytogenes 1529 and Salmonella Enteritidis chosen as outgroup.
The Table 1 demonstrates the persistence and the dissemination of some L. monocytogenes strains in the processing plant evaluated. Some strains (clusters IA, IIA, IIB e ID) were isolated repeatedly during the sampling period, suggesting their persistence in the environment of this industry. Moreover, some strains have more ability to contaminate and to multiply in the final product (IA, IIA, IB, IIB e IC), while others (ID e IIIA) were more adapted to the environment.
Table 1.
Distribution of clusters of L. monocytogenes on a processing line of fresh mixed sausage for sampling point and sampling day.
| Sampling | |||||
|---|---|---|---|---|---|
| Sampling Site and Sampling Source | 1 | 2 | 3 | 4 | 5 |
| Raw Material | - | - | - | - | - |
| Environmental and Surfaces | |||||
| Working tables | - | - | - | - | IDc |
| Boxes | IDc | - | - | - | - |
| Trolleys | IAb | - | IIIA | - | - |
| Drains | - | - | - | - | - |
| Equipments | |||||
| Disintegrator | - | - | - | - | - |
| Mincer | IAb | - | - | - | - |
| Mixer | IAb | - | - | - | - |
| Packaging equipment | - | IAb | - | - | - |
| Tieing equipment | - | IAb | - | - | IIBd |
| Handles | - | - | - | IAb | - |
| Mass ready to | IIAa | - | - | - | - |
| packing final | IB | IIAa | IIAa | IIAa | IIBd |
| product | IAb | IC | IAb | ||
| IIBd | |||||
L. monocytogenes not detected
a, b, c, d: The cluster was detected in more than one sampling point and/or more than one sampling day
Strains from clusters IIIA and ID appeared contaminating only the environment and surfaces (Table 1), suggesting that these strains are more adapted to these environments, more sensitive to spices and preservers used in the production of the fresh mixed sausage, or may be bad competitors, as they were not found in the final product. However, the clusters IA and IIB, probably are more important in cross contamination of the final product, once they contaminated the processing environment and, later, the fresh mixed sausage (Table 1).
Cluster IA (34 strains), was found on the food handler’s hands, in trolleys, in all equipment (with the exception of the disintegrator of frozen blocks) and in the final product. This demonstrated the capacity of strains belonging to this cluster to be disseminated throughout the plant and its potential involvement in cross contamination. These strains may be better adapted to the plant environment and are able to survive longer. This result is in agreement with the findings reported by Autio et al. (2), Berrang et al. (4), López et al. (17) and Lundén et al. (20), who stated that some strains can persist in a food processing facility, whereas other can´t. Some factors influencing the survival of L. monocytogenes strains in food processing plants are recognized: complexity of structure of processing machines and poor hygienic properties; strain-specific properties, such as differences in adherence to stainless steel surfaces; and susceptibility to disinfectants.
Cluster IIB (11 strains) was present in the final product and on the packing machine. Interesting, strains belonging to cluster IIA (26 strains) were found only in the mixture ready to be packaging and in the final product, therefore, it is believed that even though they were present at some stage prior to the mixture preparation, due the limitations of the conventional techniques of isolation and identification, it was not possible to detect them.
In conclusion, it was verified that L. monocytogenes seems ubiquitous in the processing plant evaluated. The analysis of clusters formed by PFGE patterns showed that some strains are adapted and persisted into the processing environment, thus, relevant for food contamination in this industry. It emphasizes the problem of the efficiency of cleaning and disinfection procedures used, to avoid in-plant colonization by L. monocytogenes. Samples from the final product were frequently contaminated by L. monocytogenes, and the wide genetic diversity among the isolates demonstrates the importance of cross contamination in the processing plant evaluated and the importance in food safety. The most prevalent serotype was 1/2c, however, the presence of isolates from serotype 4b and 1/2b in the final product indicate that consumption of fresh mixed sausage can be risky to susceptible population if not properly cooked.
ACKNOWLEDGEMENT
We are thankful to CAPES and to FAPERGS for financial support through scholarships; to Laboratório de Microbiologia Clínica (Departamento de Análises Clínicas e Toxicológicas, USP, SP) for the support for performing the PFGE analysis; to Dr. Ernesto Hofer, Laboratório de Zoonoses Bacterianas (FIOCRUZ, RJ) for serotyping of L. monocytogenes; and to Secretaria Estadual da Agricultura, Pecuária e Agronegócio do RS, for logistical supports.
RESUMO
Caracterização de Listeria monocytogenes isoladas em uma linha de processamento de lingüiça mista frescal em Pelotas-RS por PFGE
Listeria monocytogenes é uma bactéria com capacidade de formar biofilmes e de colonizar superfícies de equipamentos e utensílios. Esse microrganismo pode persistir por longos períodos em plantas de processamento de alimentos, sendo capaz de contaminar o produto final. O objetivo deste estudo foi traçar a rota de contaminação de L. monocytogenes em uma linha de processamento de lingüiça mista frescal, desde a matéria-prima até o produto final. Os isolados obtidos foram caracterizados por sorotipagem e por tipagem molecular, através de Pulsed Field Gel Electrophoresis (PFGE), usando as enzimas de restrição ApaI and AscI. L. monocytogenes foi detectada em 25% das amostras. Nenhuma amostra da matéria-prima apresentava o patógeno, entretanto, o microrganismo foi detectado em 21% das amostras ambientais (com e sem contato com o alimento), 20,8% dos equipamentos, 20% das amostras de mãos de manipuladores, 40% da massa pronta para o embutimento, e em todas as amostras do produto final. Isso demonstra que a contaminação do produto final ocorreu durante o processamento, e a importância da contaminação cruzada. PFGE produziu 22 pulsotipos e a sorotipagem produziu 3 sorotipos e 1 sorogrupo, entretanto, a presença dos sorotipos 4b e 1/2b no produto final é de grande importância para a saúde pública. Os perfis de macrorestrição mostraram que algumas cepas são adaptadas e persistiram no ambiente de processamento dessa indústria.
Palavras-chave: Listeria monocytogenes; PFGE; sorotipagem, cepas persistentes
REFERENCES
- 1.APHA. Compendium of Methods for Microbiological Examination of Foods. 3rd ed. Washington D.C.: American Public Health Association.; 1992. [Google Scholar]
- 2.Autio T., Hielm S., Miettinen M., Sjöberg A.M., Aarnisalo K., Björkroth J., Mattila-Sandholm T., Korkeala H. Sources of Listeria monocytogenes contamination in a cold-smoked rainbow trout processing plant detected by pulsed-field gel electrophoresis typing. Appl. Environ. Microbiol. 1999;65:150–155. doi: 10.1128/aem.65.1.150-155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barros M.A.F., Nero L.A., Manoel A.V., d’Ovídio L., Silva L.C., Franco B.D.G. M., Beloti V. Listeria spp. associated to different levels of autochthonous microbiota in meat, meat products and processing plants. Braz. J. Microbiol. 2007;38:603–609. [Google Scholar]
- 4.Berrang M.E, Meinersmann, R.J., Frank J.F., Smith D.P., Genzlinger L.L. Distribution of Listeria monocytogenes subtypes within a poultry further processing plant. J. Food Prot. 2005;68:980–985. doi: 10.4315/0362-028x-68.5.980. [DOI] [PubMed] [Google Scholar]
- 5.Carminatia D., Perronea A., Giraffaa G., Nevianib E., Mucchetti G. Characterization of Listeria monocytogenes strains isolated from Gorgonzola cheese rinds. Food Microbiol. 2004;21:801–807. [Google Scholar]
- 6.Chasseignaux E., Toquin M.T., Ragimbeau C., Salvat G., Colin P., Ermel G. Molecular epidemiology of Listeria monocytogenes isolates collected from the environment, raw meat and raw products in two poultry- and pork-processing plants. J. Appl. Microbiol. 2001;91:888–899. doi: 10.1046/j.1365-2672.2001.01445.x. [DOI] [PubMed] [Google Scholar]
- 7.Cruz C.D., Silvestre F.A., Kinoshita E.M., Landgraf M., Franco B.D.G.M., Destro M.T. Epidemiological survey of Listeria monocytogenes in a gravlax salmon processing line. Braz. J. Microbiol. 2008;39:375–383. doi: 10.1590/S1517-838220080002000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dauphin G., Ragimbeau C, Malle P. Use of PFGE for tracing contamination with Listeria monocytogenes in three cold-smoked salmon processing plants. Int. J. Food Microbiol. 2001;64:54–61. doi: 10.1016/s0168-1605(00)00442-6. [DOI] [PubMed] [Google Scholar]
- 9.Destro M.T., Leitão M.F.F., Farber J.M. Use of molecular typing methods to trace the dissemination of Listeria monocytogenes in a shrimp processing plant. Appl. Environ. Microbiol. 1996;62:705–711. doi: 10.1128/aem.62.2.705-711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farber J.M., Warburton D.W., Babiuk T. Government of Canada HPB Method, Quebec (Canada).: Polyscience Publications.; 1994. Isolation of Listeria monocytogenes from all food and environmental samples. [Google Scholar]
- 11.Giovannacci I., Ragimbeau C., Queguiner S., Salvat G., Vendeuvre J.L., Carlier V., Ermel G. Listeria monocytogenes in pork slaughtering and cutting plants. Use of RAPD, PFGE, and PCR-REA for tracing and molecular epidemiology. Int J Food Microbiol. 1999;53:127–140. doi: 10.1016/s0168-1605(99)00141-5. [DOI] [PubMed] [Google Scholar]
- 12.Graves L.M., Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and Pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2001;65:55–62. doi: 10.1016/s0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 13.Jay J. Prevalence of Listeria spp. in meat and poultry products. Food Control. 1996;7(415):209–214. [Google Scholar]
- 14.Hofer E., Ribeiro R., Feitosa D.P. Species and serovars of the genus Listeria isolated from different sources in Brazil from 1971 to 1997. Mem. Inst. Oswaldo Cruz. 2000;95(5):615–620. doi: 10.1590/s0074-02762000000500005. [DOI] [PubMed] [Google Scholar]
- 15.Kathariou S. Listeria monocytogenes virulence and pathogenicity: a food safety perspective. J. Food Prot. 2002;65:1811–1829. doi: 10.4315/0362-028x-65.11.1811. [DOI] [PubMed] [Google Scholar]
- 16.Lemes-Marques E.G., Cruz C.D., Destro M.T. Pheno- and genotypic characterization of Listeria monocytogenes clinical isolates from the southwestern region of the state of São Paulo, Brazil. Braz. J. Microbiol. 2007;38:287–292. [Google Scholar]
- 17.López V., Villatoro D., Ortiz S., López P., Navas J., Dávila J.C., Martínez Suárez, J. Molecular tracking of Listeria monocytogenes in an Iberian pig abattoir and processing plant. Meat Sci. 2008;78:130–134. doi: 10.1016/j.meatsci.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Lundén J.M. Persistent Listeria monocytogenes contamination in food processing plants. Helsinki, Finland: 2004. p. 60. (Academic Dissertation. Faculty of Veterinary Medicine, University of Helsinki). [DOI] [PubMed] [Google Scholar]
- 19.Lundén J., Miettinen M.K., Autio T.J., Korkeala H.J. Persistent Listeria monocytogenes strains show enhanced adherence to food contact surfaces after short contact time. J. Food Prot. 2000;63:1204–1207. doi: 10.4315/0362-028x-63.9.1204. [DOI] [PubMed] [Google Scholar]
- 20.Lundén J.M., Autio T.J., Sjoberg A.M., Korkeala H.J. Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 2003;66:2062–2069. doi: 10.4315/0362-028x-66.11.2062. [DOI] [PubMed] [Google Scholar]
- 21.Martinez I., Rorvik L.M., Brox V., Lassen J., Seppola M., Gram L., Fonnesbech-Vogel B. Genetic variability among isolates of Listeria monocytogenes from food products, clinical samples and processing environments, estimated by RAPD typing. Int. J. Food Microbiol. 2003;4:285–297. doi: 10.1016/s0168-1605(02)00423-3. [DOI] [PubMed] [Google Scholar]
- 22.Neves E., Lourenco A., Silva A. C., Coutinho R., Brito L. Pulsed-field gel electrophoresis (PFGE) analysis of Listeria monocytogenes isolates from different sources and geographical origins and representative of the twelve serovars. Syst. Appl. Microbiol. 2008;31:387–392. doi: 10.1016/j.syapm.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Rohlf F.J. Setauked, NY.: Exeter Publishing, Ltd.; 2002. NTSYSpc: Numerical Taxonomy System, ver. 2.1. [Google Scholar]
- 24.Ryser E.T., Marth E.H. Listeria, Listeriosis, and Food Safety. 3rd ed. Boca Raton, Florida: CRC Press; 2007. p. 873. [Google Scholar]
- 25.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 26.Senczek D., Stephan R., Untermann F. Pulsed-field gel electrophoresis (PFGE) typing of Listeria strains isolated from a meat processing plant over a 2-years period. Int. J. Food Microbiol. 2000;2:155–159. doi: 10.1016/s0168-1605(00)00395-0. [DOI] [PubMed] [Google Scholar]
- 27.Silva W.P., Lima A.S., Gandra E.A., Araújo M.R., Macedo M.R.P., Duval E.H. Listeria spp. no processamento de lingüiça frescal em frigoríficos de Pelotas, RS, Brasil. Ciência Rural. 2004;34(3):911–916. [Google Scholar]
- 28.Thévenot D., Delignette-Muller M.L., Christieans S., Leroy S., Kodjo A., Vernozy-Rozand C. Serological and molecular ecology of Listeria monocytogenes isolates collected from 13 French pork meat salting–curing plants and their products. Int. J. Food Microbiol. 2006;112:153–161. doi: 10.1016/j.ijfoodmicro.2006.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Thévenot D., Delignette-Muller M.L., Christieans S., Vernozy-Rozand C. Prevalence of Listeria monocytogenes in 13 dried sausage processing plants and their products. Int. J. Food Microbiol. 2005;102:85–94. doi: 10.1016/j.ijfoodmicro.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Thévenot D., Dernburg A., Vernozy-Rozand C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J. Appl. Microbiol. 2006;101:7–17. doi: 10.1111/j.1365-2672.2006.02962.x. [DOI] [PubMed] [Google Scholar]


