Abstract
Endophytic bacteria play an important role in agriculture by improving plant performance and adaptation against biotic and abiotic stresses. In the present study molecular methods were used for identifying Bacillus endophytic bacteria isolated from Brazilian sweet corn. SDS-PAGE of whole-cell protein extract of forty-two isolates revealed a high number of scrutinable bands. Twenty-four isolates were identified in nine different groups of duplicated bacteria and eighteen were identified as unique. Some high-accumulated polipeptides with variable length were observed in almost isolates. Partial sequencing of 16S ribosomal gene revealed that all isolates are Bacillus sp. and among thirteen isolates with similar protein profiles, two were different strains. Among the forty-two isolates identified by rDNA sequencing, Bacillus subitilis and B. pumilus were the most frequenty species (15 and 12 isolates, respectively) followed by B. licheniformes (7 isolates), B. cereus (5 isolates) and B. amiloliquefascens (3 isolates). According to present results, SDS-PAGE technique could be used as a fast and cheap first tool for identifying inter-specific variation in maize endophytic bacterial collections while rDNA sequencing could be applied for analyzing intra-specific variation among isolates with similar protein profile as well as for taxonomic studies.
Keywords: endophytic bacteria, Bacillus, sweet corn, SDS-PAGE, rDNA sequencing
INTRODUCTION
Endophytic bacteria are ubiquitous in virtually all plant on earth. Microbial endophytes, mainly bacteria and fungi, are defined as microorganisms that are detected after surface sterilization of a plant part (3,42) and are assumed to originate from the seeds, the roots surrounding environment and the aerial portions of plants (46). The soil, particularly the rhizosphere, is an important source of root endophytes (7,14). They are thought to enter the plant by local cellulose degradation or fractures in the root system (16). Endophytes inside a plant may either become localized at the point of entry or spread throughout the plant (14). Both gram-positive and gram-negative bacterial endophytes have been isolated from several tissue types in numerous plant species. Furthermore, several different bacterial species have been isolated from a single plant (28).
Traditionally, endophytes were assumed to be latent pathogens that did not trigger harmful reactions or disease symptoms and provided no benefit to the host plant (33). Nowadays, endophytes refer to symbiotic microorganisms colonizing the interior of plants without causing any pathogenic infection (4). A large number of experimental evidences demonstrate that bacterial endophytes support the plant growth, development and yielding by synthesizing different plant hormones (1,4,5,7,11,27,46). In some cases, bacterial endophytes can also accelerate seedling emergence and promote plant establishment under adverse condition (9). Moreover, several strains of endophytic bacteria can induce both biotic and abiotic stress tolerance of inoculated plant (19).
Pathogenic microorganisms affecting plant health are a major and chronic threat to food production and ecosystem stability worldwide (12). Bacterial endophytes are involved in natural plant protection against bacterial, fungal and viral diseases and may represent an important source of biocontrol agents. They produce high amounts of compounds with antimicrobial and insecticidal activity thus improving plant’s health (1,3,4,9,17,28,48,49). Diseases of fungal, bacterial or viral origin and in some instances even damage caused by insects and nematodes can be reduced following prior inoculation with endophytes (3,48,49). Erwinia carotovora, for example, is inhibited by numerous endophytic bacteria, including several Pseudomonas sp. strains (27), Curtobacterium luteum, and Pantoea agglomerans (48). Furthermore, Wilhelm and coworkers (54) demonstrated that Bacillus subtilis strains isolated from the xylem sap of healthy chestnut-trees exhibit antifungal effects against Cryphonectria parasitica causing chestnut blight.
Endophytic bacteria are also involved in the biological nitrogen fixation. Several N-fixing bacteria have been isolated from the rhizosphere of many crop plants (11). Endophytic diazotrophs, such as Acetobacter, Azoarcus, and Herbaspirillum, in gramineous plants have received special attention because of their occurrence mainly within plant tissues and evidence for significant nitrogen fixation (8,36,41). Therefore, endophytic bacteria-plant interaction has a potential role in developing sustainable systems of crop production (30,33,49).
Endophytic bacteria exert important influence in matter flux on earth (49). Endophytic methanotrophic bacteria are involved in the control of biogeochemical cycle on the efficient oxidation of methane, leading to highly effective in situ methane recycling to carbon dioxide, which is subsequently used for photosynthesis and fixed by plants into plant sterols (39,40). In wetland ecosystems both the efficient recycling of methane and the high organic carbon burial are explained by endophytic symbiosis (40).
The intensive and abusive use of agrochemical has leading to water and soil contamination. Several authors have investigated the role of bacteria to clean up environmental pollutant (35). Some pollutants, are not metabolised by plants and, thus, accumulate and cause phytotoxicity. Certain plant-bacterial associations increase polluent compounds degradation in soil indicating that, microorganisms play an important role in phytoremediation systems (15,33,35,43). Endophytic bacteria have been engineered to enhance their naturally ability to degrade pollutants as they pass through the plant (35) improving phytoremediation of water-soluble compounds as well as of xenobiotic organic contaminants (35,37,52,56). Engineered endophytic bacteria increase plant tolerance to toluene, and decrease the transpiration of toluene to the atmosphere (52).
Recently, endophytes are viewed as a new potential source of novel genes, proteins and natural biochemical compounds for medicine, agriculture, and industrial process (32, 47). The biotechnological potential of endophytic isolates assessed by their antagonistic activity or by the in vitro production of enzymes, antibiotics, siderophores, and plant growth hormones is high (47).
In spite of the great importance of microorganisms in agricultural ecosystems, only a very small part of the microbial diversity relevant to tropical agriculture was carefully described (3). The great amount of information regarding the key role of endophytic bacteria in agriculture, in addition to the constant substitution of local races of maize for improved varieties in tropical areas, clearly demonstrate the necessity to characterize the tropical maize endophytic bacterial collection. Microbial culture collections properly identified are valuable assets for conservation of tropical genetic resources, and the bioprospection of new molecules. Their taxonomic status represents the first relevant step for an adequate characterization and utilization of microbial germplasm. This work was carried out to obtain basic knowledge about the endophytic species of Bacillus associated with tropical Brazilian sweet corn.
MATERIAL AND METHODS
Endophytic bacteria were isolated from randomly selected fresh health leaf of bulk population of sweet corn germplasm from Embrapa Milho e Sorgo (Maize and Sorghum Research Center, Sete Lagoas, MG, Brazil). The leaves were initially thoroughly washed in running tap water to remove soil debris and surface-disinfested by immersion in 70% ethanol for 1 min, 3% sodium hypochlorite for 4 min and rinsed 5 times in sterile distilled water. After surface disinfestation four leaf sections 2-3 cm long were excised with a sterile knife blade and were asseptically plated on each Petri dish containing D2 medium (25): (0,3g magnesium sulfate heptahydrate, 1g ammonium chloride, 5g lithium chloride, 10g glycose, 4g hydrolyzed casein, 2g yeast extract, 1,2g Tris, 15g agar and 1L water, pH 6.9). Plates were incubated at 28°C for 48–72 h and individual colonies were isolated and purified by successive plating in D2 medium. Isolates pathogenicity were evaluated in greenhouse conditions in maize and tobacco plants. Nonpathogenic isolates were reinoculated and recovered in maize and tobacco plants (6). Stock cultures were maintained on D2 agar slants and incorporated to the tropical maize microbial collection at the Embrapa Milho e Sorgo. In the present study, forty-two isolates were used to evaluate the usefulness of SDS-PAGE as a fast, simple and low cost method for preliminary bacterial identification and rDNA sequencing to validate SDS-PAGE results and taxonomic identification.
SDS-PAGE was performed according to Laemmli (31) and Jackman (24). An aliquote of 1.5 mL from 48 h old culture of each isolate was centrifuged at 20,800 x g for 5 min. Pellets were washed three times with 1 mL of TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA) and stored at –80°C until used. Bacterial mass (20 mg) was powdered with liquid nitrogen, using a pestle and a mortar, and transferred to a 1.5 mL microcentrifuge tubes containing 100 μL of sample buffer (62.5 mM Tris-HCl pH 8.0, 20% (v/v) glycerol, 2% SDS, 5% β-mercaptoethanol, 0.02% bromophenol blue). Samples heated for 10 min in boiled water were immediately placed on ice for five minutes and cooled. These samples were centrifuged for 10 min at 958 x g and 15 μL of supernatants were load onto a 12% acrylamide gel. Electrophoresis was performed in 10% Tris-Glycine buffer (0.025 M Tris base, 0.192 M glycine, 0.1% SDS pH 6,8) with Protean II minigel electrophoretic system (Bio-Rad Laboratories, Mississauga, ON, Can) at 60v for 1h. After electrophoresis, protein were visualized by coomassie blue staining method (31) and photographed with Eagle Eye System (Stratagene, La Jolla, CA). The fingerprints were compared visually with the overview gels.
Genomic DNA extraction was performed according to modified method of Gürtler and Stanisich (18). Fifty mL from 48 h liquid culture of each isolate were centrifuged at 958 x g, washed two times with TE, centrifuged and pellets were powered in liquid nitrogen using a mortar and pestle. The macerate was transferred to 50 mL propylene tubes containing 5 mL of extraction buffer (0.1 M Tris-HCl pH 8.0, 0.2 M NaCl, 0.02 M EDTA, 1,0% SDS, 0.1% β-mercaptoethanol). Each tube was vigorously agitated for obtaining an uniform suspension without lumps and then incubated for 15 min at room temperature. Afterwards, an equal volume of chloroform:isoamyl alcohol (24:1 v/v) was added to each sample, vigorously agitated, and incubated 10 min at room temperature. Cell debris was removed by centrifugation at 4°C at 6,810 x g for 10 minutes. Aliquots of 5 mL of the supernatant layer was transferred to 50 mL propylene tube and equal volume of ice cold ethanol was added to each sample and gently inverted several times to precipitate nucleic acids. After centrifugation at 20,800 x g for 15 min, nucleic acids were washed with 70% ethanol, air dried and dissolved with 0,5 mL of TE buffer containing 40 μg/mL RNAse H. The quality of DNA was checked by spectrophotometer (OD 260/280) and by electrophoresis in 1% agarose gel. The final concentration of DNA was adjusted to 25 ng/ μL.
The 16S rDNA was amplified with the 16F27 (forward) (5’-AGAGTTTGATCCTGGCTCAG-3’) and 16R1542 (reverse) (5’-AAGGAGGTGATCCAGCCGCA-3’) universal primers (18). PCR reactions were performed with 25 ng of bacterial genomic DNA plus 2.5 μL 10X PCR buffer (20 mM Tris-HCl pH 8.4, 50 mM KCl), 2.0 μM of each primer, 25 mM dNTP, 2,5 mM MgCl2, and 1 U Taq DNA polymerase (Phoneutria, Belo Horizonte, Brazil) in a total volume of 25 μL. PCR was performed in a model PTC-100 thermalcycler machine (MJ Research, MS, USA) with the following conditions: one cycle for denaturation of DNA samples at 94°C for 1 min, 30 cycles of 1 min at 94°C, 1 min at 50°C (annealing) and 2 min at 72°C (extension). Finally, reactions were incubated for 10 min at 72°C. The Amplified DNA were analyzed by horizontal gel electrophoresis at 6 V/cm2 in 1.0 % agarose gel (wt/v) in 1X TAE buffer (0.04M Tris-acetate, 0.001M EDTA, pH 8.0) containing ethidium bromide (0.5 mg/L). Gels were visualized under UV light, photographed and the fingerprints were compared visually with the overview gels. Gel slices containing the amplified DNA fragments were cut off from gels and DNA were purified with the GeneClean kit II (BIO 101, Vista, CA, USA).
Partial sequencing of 16S PCR-amplified rDNA were made with one of the following universal primers (18): 16S518F (5´-CAGCAGCCGCGGTAATAC-3´) or 16S928R (5´-CCCTCAATTCCTTTGAGTTT-3´). Sequencing reactions were performed in a total volume of 25 μL containing 200-300 ng of amplified rDNA, 20 pmol of primer and 8.0 μL reaction premix (Applied Biosystems, Lincoln Centre Drive Foster City, USA). Reaction conditions were established with an initial step of DNA denaturation at 96°C for 30 s, followed by 25 cycles of 30 s at 96°C, annealing for 15 s at 50°C and extension for 4 min at 60°C. The reaction products were precipitated with 2,5 μL of 3 M sodium acetate, pH 4.6 plus 50 μL 95% cold ethanol for 10 min on ice, centrifuged for 30 min at 27,239 x g, and washed with 250 μL of 70% ethanol. DNA sequencing was performed in an automatic sequencer (ABI-377, Applied Biosystems, Lincoln Centre Drive Foster City, USA) and repeated at least three times. 16S rDNA sequences were aligned using the Clustal multiple-alignment program (Clustal W) (51). Bacterial 16S rDNA partial sequences generated in the present study were deposited in EMBL/GenBank/DDBJ nucleotide sequence data libraries and their respective accession numbers are shown in Table 1. The DNA sequences were analyzed in the GenBank database using the algorithm BLASTN (2) and CLUSTAL W (51) to identify the most similar 16S rDNA sequences (table 1).
Table 1.
DNA sequence identity of bacterial rDNA 16S partial sequencing among endophytic bacteria isolated from sweet corn and GeneBank 16S DNA sequences.
| Isolate (CNPMS) | GenBank Accession Number | Identity (16S rDNA sequences) | Accession Number (GeneBank) | Max Score | Max Identity (%) |
|---|---|---|---|---|---|
| Endo 01 | EU795010 | B. subtilis strain B43 | gi|158323764|EU169188.1| | 722 | 99 |
| Endo 02 | EU795011 | B. subtilis strain HDYM-23 | gi|151935459|EF428247.2| | 704 | 98 |
| Endo 03 | EU795012 | B. pumilus isolate 6 | gi|94442977|AM260977.1| | 704 | 98 |
| Endo 04 | EU795013 | B. pumilus strain FO-033 | gi|7107438|AF234851.1| | 681 | 97 |
| Endo 05 | EU795014 | B. amyloliquefaciens strain TPL13 | gi|171191186|EU373386.1| | 708 | 98 |
| Endo 06 | EU795015 | B. amyloliquefaciens strain HNR20 | gi|171191187|EU373387.1| | 744 | 100 |
| Endo 07 | EU795016 | B. pumilus isolate zyj 1-1 | gi|163311394|gb|EU302128.1| | 717 | 98 |
| Endo 08 | EU795017 | B. subtilis strain C4-1 | gi|159145578|EU257444.1| | 706 | 98 |
| Endo 09 | EU795018 | B. pumilus strain FO-033 | gi|7107438|dbj |AF234851.1| | 742 | 100 |
| Endo 10 | EU795019 | B. subtilis strain GH29. | gi| 150372731 |AB301003.1| | 704 | 98 |
| Endo 11 | EU795020 | B. subtilis strain C4-1 | gi|159145578|EU257444.1| | 690 | 97 |
| Endo 12 | EU795021 | B. licheniformis strain 3EC4A9 | gi|7107449|AF234862.1| | 737 | 99 |
| Endo 13 | EU795022 | B. pumilus strain FO-033 | gi|7107438|AF234851.1| | 729 | 99 |
| Endo 14 | EU795023 | B. pumilus strain FO-033 | gi|7107438|AF234851.1| | 708 | 98 |
| Endo 15 | EU795024 | B. pumilus strain S8-09 | gi|171191236|EU373436.1| | 720 | 99 |
| Endo 16 | EU795025 | B. pumilus strain FO-033 | gi|7107438|gb|AF234851.1| | 664 | 96 |
| Endo 17 | EU795026 | B. pumilus TPR18 | gi|171191236|EU373436.1| | 720 | 99 |
| Endo 18 | EU795027 | B. subtilis strain F198 | gi|78498901|DQ234847.1| | 700 | 98 |
| Endo 19 | EU795028 | B. pumilus strain FO-033 | gi|7107438|gb|AF234851.1| | 693 | 97 |
| Endo 20 | EU795029 | B. subtilis strain GB13 | gi|118574032|EF101728.1| | 710 | 98 |
| Endo 21 | EU795030 | B. pumilus strain FO-033 | gi|7107438|AF234851.1| | 699 | 97 |
| Endo 22 | EU795031 | B. pumilus strain FO-033 | gi|7107438|AF234851.1| | 699 | 97 |
| Endo 23 | EU795032 | Low G+C Gram-positive bacterium | gi|18149254|AB074701.1| | 708 | 98 |
| B. cereus strain FM-4 | gi| 193794800|gb|EU794727.1| | 700 | 98 | ||
| Endo 24 | EU795033 | Unidentified bacteria 16S RNA | gi|2209053|AB004761.1| | 715 | 99 |
| B. cereus strain FM-4 | gi| 193794800|gb|EU794727.1| | 710 | 98 | ||
| Endo 25 | EU795034 | B. licheniformis strain FO-085 | gi|7107449|AF234862.1| | 731 | 99 |
| Endo 26 | EU795035 | B. subtilis strain FO-029a | gi|78498901|DQ234847.1| | 700 | 98 |
| Endo 27 | EU795036 | B. cereus strain Ag 13 | gi|163960970|gb|EU327888.1| | ||
| Endo 28 | EU795037 | Unidentified bacteria 16S RNA | gi|2209053|AB004761.1| | 740 | 100 |
| B. cereus strain FM-4 | gi| 193794800|gb|EU794727.1| | 735 | 99 | ||
| Endo 29 | EU795038 | B. licheniformis strain FO-085 | gi|7107449|AF234862.1| | 733 | 99 |
| Endo 30 | EU795039 | B. licheniformis | gi|7107449|AF234862.1| | 726 | 99 |
| Endo 31 | EU795040 | B. licheniformis strain FO-085 | gi|7107449|AF234862.1| | 720 | 99 |
| Endo 32 | EU795041 | B. subtilis | gi|193804895|gb|EU790487.1| | 729 | 99 |
| Endo 33 | EU795042 | B. subtilis | gi| 166012621 |gb|EU3663 85.1| | 690 | 97 |
| Endo 34 | EU795043 | B. licheniformis strain FO-085 | gi|7107449|AF234862.1| | 719 | 99 |
| Endo 35 | EU795044 | B. amyloliquefaciens strain TPL13 | gi|171191186|EU373386.1| | 744 | 100 |
| Endo 36 | EU795045 | B. subtilis strain A184 | gi|17646567|AF447803.1| | 677 | 96 |
| Endo 37 | EU795046 | B. subtilis strain DA7 | gi| 152218451 |EU000054.1| | 708 | 98 |
| Endo 38 | EU795047 | B. subtilis | gi|193804895|gb|EU790487.1| | 729 | 99 |
| Endo 39 | EU795048 | B. subtilis strain B43 | gi| 158323764|EU 169188.1| | 708 | 98 |
| Endo 40 | EU795049 | B. licheniformis strain FO-036 | gi|7107449|AF234862.1| | 722 | 99 |
| Endo 41 | EU795050 | B. subtilis strain DA7 | gi| 152218451 |gb|EU000054.1| | 693 | 97 |
| Endo 42 | EU795051 | B. cereus strain FM-4 | gi| 193794800|gb|EU794727.1| | 731 | 99 |
RESULTS
In the present study, SDS-PAGE technique was used as a first-step procedure for identifying endophytic Bacillus isolated from tropical sweet maize and rDNA sequencing was used for taxonomic information. SDS-PAGE of whole-cell protein extract of forty-two bacterial isolates showed a high heterogeneous profile (Figure 1). The main difference in protein pattern was related to some high-accumulated polypeptides with different molecular weight present in almost isolates. Protein profile allowed the comparision of the forty-two isolates wich were distributed into six groups of duplicated bacteria: a) Endo 1 and Endo 2; b) Endo 5 with Endo 6; c) Endo 3, Endo 7, Endo 9, Endo 13, Endo14, Endo 15, Endo 16, Endo 17, Endo 19, Endo 21 and Endo 22; d) Endo 29 with Endo 30; e) Endo 31 and Endo 34, f) Endo 32 with Endo 36, Endo 37, Endo 38 and Endo 41. The remaining eighteen isolates were considered as unique.
Figure 1.
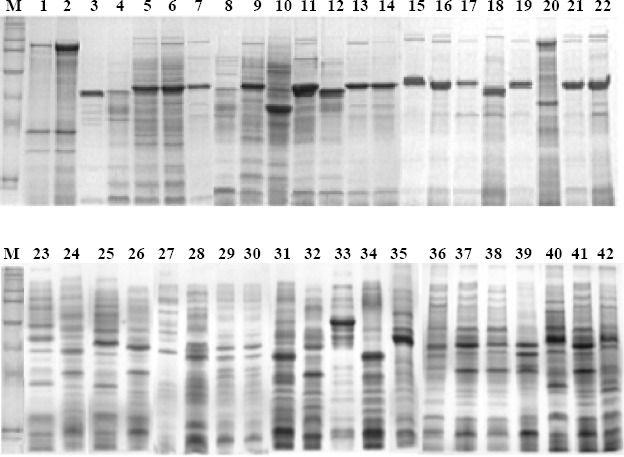
Electrophoretic profile (SDS-PAGE) of whole protein extract of forty-two endophytic bacteria isolated from tropical sweet corn. Numbers 1 to 42 indicate bacterial isolates 1 to 42, respectively. M = Protein Molecular weight markers (Rainbow, New England Biolabs, Ipswich, MA).
In order to gain insights about the bacterial identity, an accumulated polypeptide of 42-kDa present in twenty-one isolates was electroeluted from the SDS gel and the amino acid sequence for 27 amino acid residues at the N-termini was determined (data not shown). Amino acid sequency comparison in GeneBank revealed a high identity with flagellin H, a protein found in the Bacillus sp bacterial flagellum. The identity with Bacillus subtilis flagelin was 100% and identity with B. amyloliquefaciens was 96%. The identity among the 42-kDa protein with flagelin and other Bacillus species (B. amiloliquefascens, B. licheniformes, B. pumilus, B. Licheniformis, B. pumilus and Oceanobacillus iheyensis) ranged from 88% to 76%, but it was still high.
In the present study, the partial DNA sequencing of 16S rRNA gene was performed in order to validate SDS-PAGE results. The 16S rRNA gene was amplified by PCR using 16F27 and 16R1542 universal primer (18). All amplified products produced a single band with approximately 1500 base pair in length and differences among them were not visible in 1 % agarose gel (data not shown). Two universal primers (16S518F and 16S928R) were used for partial sequencing of the amplified 16S rDNA. All the fouty-two bacterial isolates were Bacillus spp. with B. subitilis been the most prevalent (15 isolates) (Table 1). The other Bacillus isolates were close to B. pumilus (12 isolates), B. licheniformes (7 isolates), B. cereus (5 isolates) and B. amiloliquefascens (3 isolates). One bacterial isolate (Endo 23) showed a high score with an unidentified bacterium with low G+C content associated with the gut bacterial flora from pea aphid intracellular symbiont (22). Data generated by DNA sequencing of rRNA genes confirmed the twenty-one Bacillus isolates as revealed by the partial amino acid sequencing (data not shown) of the 42-kDa polipeptide corresponding to flagellin H of Bacillus species. Although two isolates (Endo 24 and Endo 28) are close to B. cereus (98 and 99% identity), they showed high identity with unidentified bacteria (99 and 100% identity).
DISCUSSION
Although SDS-PAGE of bacterial whole-cell protein extracts is shown to be very sensitive to taxonomic differences its use is still limited in some bacterial species (20,21,26,38). The high level of protein polymorphism observed in maize endophytic bacteria, indicates that protein profile is an effective method for endophytic bacterial fingerprinting when a high number of isolates are necessary to be identified. Furthermore, SDS-PAGE could be an inexpensive and fast procedure allowing the rational use of microorganism collections. After exhausting search in specialized literature we concluded that the present study was the first report using SDS-PAGE technique for endophytic bacterial identification. However, the development of powerful molecular methods like rDNA sequencing, although more expensive, have been widely used for strain identification and taxonomic information.
In general, results obtained with SDS-PAGE technique show a high correlation with those obtained from nucleic acid hybridization (24). Comparison of rDNA sequencing data with SDS-PAGE profile results showed that SDS-PAGE duplicates (Endo 1-Endo 2 and Endo 5-Endo 6) are different strains of Bacillus subitilis and B. amyloliquefaciens, respectively. The other three groups were identified as B. licheniformes (Endo 29 and Endo 30), B. subtilis (Endo 32, Endo 36, Endo 37, Endo 38 and Endo 41) and B. pumilus (Endo 3, Endo 7, Endo 9, Endo 13, Endo 14, Endo 15, Endo 16, Endo 17, Endo 19, Endo 21 and Endo 22). Two bacterial isolates with similar protein pattern (Endo 31 and Endo 34) and three with completely different profile (Endo 12, Endo 25, and Endo 40), fit in B. licheniformis specie. Interestingly, two bacterial isolates with different protein profile (Endo 1 and Endo 39) showed a high identity with the strain B43 of B. subtilis. Likewise, eigth isolates (Endo 4, Endo 9, Endo 13, Endo 14, Endo 16, Endo 19, Endo 21 and Endo 22) showed high similarity with the strain FO-033 of B. pumilus isolated from spacecraft (53).
The present result with sweet corn is in accordance with previous study concerning to bacterial community present in 14 maize Chinese cultivars (14). In that study, Bacillus spp. was the endophytic bacterium with a higher frequency in roots with eight species been identified (B. subtilis, B. megaterium, B. cereus, B. licheniformis, B. anthracis, B. mycoides, B. pumilus and B. circulans). Other endophytic bacteria isolated in that study were Enterobacter spp., Serratia spp., Pseudomonas spp., Xanthomonas spp., Clavibacter spp. (14). However, McInroy & Kloepper (34) found that endophytic bacterial community in sweet corn (stems and roots) was represented mainly by the class Proteobacteria (gamma-proteobacteria) within Enterobacter spp. is the prevalecent, followed by members of the beta-proteobacterial (Burkholderia spp.). Likewise, study performed with maize comercial varieties found that Enterobacter agglomerans, Klebsiella terrigena, Pseudomonas corrugata, P. fluorescens, P. marginalis e Vibrio sp. were the predominant species in the maize stems (10,13). In another study, Chelius & Triplett (10) performed a comparative study on diversity of bacteria and Archaea associating on the surface and interior of maize roots using two different techniques: culture collection and clonal analysis. Only four bacterial divisions were found in the culture collection, which represented 27 phylotypes, whereas 6 divisions were identified in the clonal analysis, comprising 74 phylotypes. The predominant group in the culture collection was the actinobacteria. The population of maize-associated proteobacteria resembled the proteobacterial population of a typical soil community, which resided a subset of specific plant-associated bacteria, such as Rhizobium- and Herbaspirillum-related phylotypes (10). The representation of phylotypes within other divisions suggested that maize plants support a distinct bacterial community.
Both, gram-positive and gram-negative bacterial endophytes have been isolated from several tissue types in numerous plant species. Furthermore, several different bacterial species have been isolated from a single plant (28). Similarly, significant variations appear to exist in the types of endophytic bacteria isolated from maize. Several factors may explain these differences, including host specificity, geographical distribution, plant age, and tissue type (28). Likewise, the biodiversity and population dynamic of bacterial endophytes in Brassica napus are highly influenced by genetic background, growth periods and environmental conditions (55). The abundance and diversity of bacteria isolated from different tissues of field grown potato revealed a high heterogeneity of community composition suggesting the existence of microenvironment-specific communities’ (29). In soybean, significant differences were observed in bacterial population densities in relation to season, growth phase and the tissues from which the endophytes were obtained (30). In Medicago spp., the addition of ethylene decreased endophytic colonization and ethylene-mediated inhibition was reversed by addition of the ethylene action inhibitor, 1-methylcyclopropene (23). In addition, most studies concerning to endophytic community biodiversity are cultivation-dependent and growth requirements are unknown for many bacterial species (50). Therefore, cultivation-dependent biodiversity studies of the endophytic community are somewhat limited and biodiversity studies rescue only about 48% of the bacterial diversity retrieved by cultivation-independent techniques (10,44). Different media used for bacterial isolation could be another factor affecting bacterial community diversity recovered from maize tissues. In the present study, the D2 medium (25) was used for bacterial isolation. Unfortunately, culture media used by another workers for endophytic bacterial isolation from maize tissue were not described. Finally, agricultural practices like agrochemicals usage is another factor that significantly influence bulks soil microbial community and also affect the root endophytic community (45). In conjunction, differences in the bacterial biodiversity among maize bacterial endophytes observed in previous studies (10,13,14,34) as well as those observed in the present study could be explained by one or more different factors and indicated that maize plants support a high diversity of distinct endophytic bacterial community. In addition, fast and slow growing bacteria require different times of incubation. In the present study, only fast growing Bacillus species were isolated in 48-72h of explant incubation. Studies on maize endophytic bacterial communities showed that the time of incubation as well as medium composition are very important factors affecting recovering of bacterial diversity (10,13,14, 34). Fast and slow growing bacteria were isolated with time of explant incubation from 48 to 72h in different media composition (medium R2A, for oligotrophic bacteria; TSA for culturable heterotrophic bacteria; and medium SC, to support the growth of fastidious organisms) (34) as well as with explant incubation higher than 72h (7-10 days) (10,13,14). In conjunction, all these aspects are very relevant and might be considered for the screening and the diversity preservation of microbial germplasm.
ACKNOWLEDGEMENT
This work was supported by grants of the State of Minas Gerais Research Foundation (FAPEMIG - Fundação de Amparo à Pesquisa do Estado de Minas Gerais), (www.fapemig.br).
RESUMO
Análise molecular de bactérias endofíticas do gênero Bacillus isoladas de milho tropical (Zea mays L.)
Bactérias endofíticas desempenham papel importante na agricultura, melhorando a performance e adaptação de plantas contra estresses bióticos e abióticos. No presente estudo, métodos moleculares foram empregados para identificar bactérias endofíticas do gênero Bacillus isoladas de cultivares de milho doce brasileiro. SDS-PAGE de extratos protéicos totais de quarenta e dois isolados revelaram elevado número de bandas escrutináveis. Vinte e quatro isolados formaram nove grupos diferentes de réplicas bactérianas e dezoito foram considerados como únicos. Entre os isolados, alguns polipeptídios, de tamanhos variados, foram altamente acumulados. Seqüenciamento parcial do gene ribosomal 16S revelou que todos os isolados pertencem ao gênero Bacillus e que, entre treze isolados com padrão protéico similar, dois eram linhagens diferentes. Entre os quarenta e dois isolados identificados por seqüenciamento de rDNA, Bacillus subtilis e B. pumilus foram mais frequentes (15 e 12 isolados, respectivamente), seguido por, B. licheniformes (7 isolados), B. cereus (5 isolados) e B. amiloliquefascens (3 isolados). Baseado nos resultados, conclui-se que a técnica de SDS-PAGE poderá ser usada como primeiro procedimento, rápido e barato, para identificar variação inter-específica em coleções de bactérias endofíticas isoladas do milho, enquanto o método de seqüenciamento de rDNA poderá ser aplicado para analisar variações intraespecífica entre isolados com padões similares de proteínas e estudos de taxonomia.
Palavras-chave: Bactéria endofítica, Bacillus, milho doce, SDS-PAGE, seqüenciamento de rDNA
REFERENCES
- 1.Adhikari T.B., Joseph C.M., Yang G., Phillips D.A., Nelson L.M. Evaluation of bacteria isolated from rice for plant growth promotion and biological control of seedling disease of rice. Can. J. Microbiol. 2001;47(10):916–924. doi: 10.1139/w01-097. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search program. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azevedo J.L., MaCCheroni W., Araújo W.L., Pereira J.O. Endophytic microorganisms: A review on insect control and recent advances on tropical plants. Electron. J. Biotechnol. 2000;3(2):40–65. [Google Scholar]
- 4.Bai Y., Zhou X., Smith D.L. Enhanced soybean plant growth resulting from coinoculation of Bacillus strains with Bradyrhizobium japonicum. Crop Sci. 2003;43:1774–1781. [Google Scholar]
- 5.Bottini R., Cassán F., Piccoli P. Gibberellin production by bacteria and its involvement in plant growth promotion and yield increase. Appl. Microbiol. Biotechnol. 2004;65(5):497–503. doi: 10.1007/s00253-004-1696-1. [DOI] [PubMed] [Google Scholar]
- 6.Bressan W., Borges M.T. Delivery methods for introducing endophytic bacteria into maize. BioControl. 2004;49(3):315–322. [Google Scholar]
- 7.Castro-Sowinski S., Herschkovitz Y., Okon Y., Jurkevitch E. Effects of inoculation with plant growth-promoting rhizobacteria on resident rhizosphere microorganisms. FEMS Microbiol. Lett. 2007;276(1):1–11. doi: 10.1111/j.1574-6968.2007.00878.x. [DOI] [PubMed] [Google Scholar]
- 8.Cavalcante V.A., Dobereiner J. A new acid-tolerant nitrogen-fixing bacterium associated with sugarcane. Plant Soil. 1998;108(1):23–31. [Google Scholar]
- 9.Chanway C.P., Shishido M., Nairn J., Jungwirth S., Markham J., Xiao G., Holl F.B. Endophytic colonization and field responses of hybrid spruce seedlings after inoculation with plant growth-promoting rhizobacteria. For. Ecol. Manage. 2000;133(1-2):2–88. [Google Scholar]
- 10.Chelius M.K., Triplett E.W. The diversity of archaea and bacteria in association with the roots of Zea mays L.Microbiol. Ecol. 2001;41(3):252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 11.Chi F., Shi-Hua S., Hai-Ping C., Yu-Xiang J., Yanni Y.G., Dazzo F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005;71(11):7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Compant S., Duffy B., Nowak J., Clément C., Barka E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005;75(9):4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher P.J., Petrini O., Scott H.M.L. The distribution of some fungal and bacterial endophytes in maize (Zea mays L.). New Phytol. 1992;122:299–305. doi: 10.1111/j.1469-8137.1992.tb04234.x. [DOI] [PubMed] [Google Scholar]
- 14.Gao Z., Zhuang J., Chen J., Liu X., Tang S. Population of endophytic bacteria in maize roots and its dynamic analysis. Ying Yong Sheng Tai Xue Bao. 2004;15(8):1344–1348. [PubMed] [Google Scholar]
- 15.Germaine K., Liu X., Cabellos G., Hogan J., Ryan D., Dowling D.N. Bacterial endophyte-enhanced phyto-remediation of the organochlorine herbicide 2,4-dichlorophenoxyacetic acid. FEMS Microbiol. Ecol. 2006;57(2):302–310. doi: 10.1111/j.1574-6941.2006.00121.x. [DOI] [PubMed] [Google Scholar]
- 16.Gough C., Galera C., Vasse J., Webster G., Cocking E.C., Denarie J. Specific flavonoids promote intercellular root colonization of Arabidopsis thaliana by Azorhizobium caulinodans ORS571. Mol. Plant-Microbe Interact. 1997;10(5):560–570.. doi: 10.1094/MPMI.1997.10.5.560. [DOI] [PubMed] [Google Scholar]
- 17.Gunatilaka A.A.L. Natural products from plant-associated microorganisms: distribution, structural diversity, bioactivity, and implications of their occurrence. J. Nat. Prod. 2006;69(3):509–526. doi: 10.1021/np058128n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gürtler V., Stanisich V.A. New approaches to typing and identification of bacteria by using the 16S-23S rDNA spacer region. Microbiology. 1996;142:3–16. doi: 10.1099/13500872-142-1-3. [DOI] [PubMed] [Google Scholar]
- 19.Hallmann J., Quadt-Hallmann A., Mahaffee W.F., Kloepper J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997;43:895–914. [Google Scholar]
- 20.Hantula J.H., Korhonen T.K., Bamford D.H. Determination of taxonomic resolution power of SDS-polyacrylamide gel electrophoresis of total cellular proteins using Enterobacteriaceae. FEMS Microbiol. Lett. 1990;70:325–330. doi: 10.1111/j.1574-6968.1990.tb13998.x. [DOI] [PubMed] [Google Scholar]
- 21.Hantula J.H., Kurki A., Vuoriranta P., Bamford D.H. Rapid classification of bacterial strains by SDS-polyacrylamide gel electrophoresis: population dynamics of the dominant dispersed phase bacteria of activated sludge. Appl. Microbiol. Biothecnol. 1991;34(4):551–555. [Google Scholar]
- 22.Harada H., Oyaizu H., Ishikawa H. A consideration about the origin of aphid intracellular symbiont in connection with gut bacterial flora. J. Gen. Appl. Microbiol. 1996;42(1):17–26. [Google Scholar]
- 23.Iniguez A.L., Dong Y., Carter H.D., Ahmer B.M., Stone J.M., Triplett E.W. Regulation of enteric endophytic bacterial colonization by plant defenses. Mol. Plant-Microbe Interact. 2005;18(2):169–178. doi: 10.1094/MPMI-18-0169. [DOI] [PubMed] [Google Scholar]
- 24.Jackman P.J.H. Bacterial taxonomy based on electrophoretic whole-cell protein patterns. In: Goodfellow M., Minnikin D.E., editors. Chemical Methods in Bacterial Systematics. London, UK: Academic Press; 1985. pp. 115–129. [Google Scholar]
- 25.Kado C.I., Heskett M.G. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopatology. 1970;60(6):969–976. doi: 10.1094/phyto-60-969. [DOI] [PubMed] [Google Scholar]
- 26.Kersters K., DeLey J. Classification and identification of bacteria by electrophoresis of their proteins. In: Goodfellow M., Board R.G., editors. Microbiological classification and identification. London, UK: Academic Press; 1980. p. 273. [PubMed] [Google Scholar]
- 27.Kloepper J.W. Effect of seed pieces inoculation with plant-growth promoting rhizobacteria on populations of Erwinia carotovora on potato roots and daughter tubers. Phytopathology. 1983;73(2):217–219. [Google Scholar]
- 28.Kobayashi D.Y., Palumbo J.D. Bacterial endophytes and their effects on plants and uses in agriculture. In: Bacon C.W., White J.F., editors. Microbial endophytes. New York, USA: Marcel Dekker; 2000. pp. 199–233. [Google Scholar]
- 29.Krechel A., Faupel A., Hallmann J., Ulrich A., Berg G. Potato-associated bacteria and their antagonistic potential towards plant-pathogenic fungi and the plant-parasitic nematode Meloidogyne incognita (Kofoid & White) Chitwood. Can. J. Microbiol. 2002;48(9):772–786. doi: 10.1139/w02-071. [DOI] [PubMed] [Google Scholar]
- 30.Kuklinsky-Sobral J., Araujo W.L., Mendes R., Geraldi I.O., Pizzirani-Kleiner A. A., Azevedo J.L. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004;6(12):1244–1251. doi: 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli U.K. Cleavage of structural protein during the assembly of the head of bactheriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Lima A.O., Quecine M.C., Fungaro M.H., Andreote F.D., MacCheroni W.JR., Araujo W.L., Silva-Filho M.C., Pizzirani-Kleiner A.A., Azevedo J.L. Molecular characterization of a beta-1,4-endoglucanase from an endophytic Bacillus pumilus strain. Appl. Microbiol. Biotechnol. 2005;68(1):57–65. doi: 10.1007/s00253-004-1740-1. [DOI] [PubMed] [Google Scholar]
- 33.Lodewyckx C., Vangronsveld J., Porteous F., Moore E.R.B., Taghavi S., Mezgeay M., Van der Lelie D. Endophytic bacteria and their potential applications. Crit. Rev. Plant Sci. 2002;21(6):583–606. [Google Scholar]
- 34.McInroy J.A., Kloepper J.W. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil. 1995;173(2):337–342. [Google Scholar]
- 35.Moore F.P., Barac T., Borremans B., Oeyen L., Vangronsveld J., van der Lelie D., Campbell C.D., Moore E.R.B. Endophytic bacterial diversity in poplar trees growing on a BTEX-contaminated site: the characterisation of isolates with potential to enhance phytoremediation. Syst. Appl. Microbiol. 2006;29(7):539–556. doi: 10.1016/j.syapm.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 36.Muthukumarasamy R., Revathi G., Seshadri S., Lakshminarasimhan C. Gluconacetobacter diazotrophicus (syn. Acetobacter diazotrophicus), a promising diazotrophic endophyte in tropics. Curr. Sci. 2002;83(2):137–145. [Google Scholar]
- 37.Newman L.A., Reynolds C.M. Bacteria and phytoremediation: new uses for endophytic bacteria in plants. Trends Biotechnol. 2005;23(1):6–8. doi: 10.1016/j.tibtech.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Owen R.J., Jackman P.J.H. The similarities between Pseudomonas paucimobilis and allied bacteria derived from analysis of deoxyribonucleic acids and electrophoretic protein patterns. J. Gen. Microbiol. 1982;128(2):2945–2954. doi: 10.1099/00221287-128-12-2945. [DOI] [PubMed] [Google Scholar]
- 39.Raghoebarsing A.A., Smolders A.J., Schmid M.C., Rijpstra W.I., Wolters-Arts M., Derksen J., Jetten M.S., Schouten S., Sinninghe D.J.S., Lamers L.P., Roelofs J.G., Op den Camp H.J., Strous M. Methanotrophic symbionts provide carbon for photosynthesis in peat bogs. Nature. 2005;436(7054):1153–1156. doi: 10.1038/nature03802. [DOI] [PubMed] [Google Scholar]
- 40.Raghoebarsing A.A., Pol A., van der Pas-Schoomen K.T., Smolders A.J., Ettwig K.F., Rijpstra W.I., Schouten S., Damsté J.S., Op den Camp H.J., Jetten M.S., Strous M. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature. 2006;440(7086):878–870. doi: 10.1038/nature04617. [DOI] [PubMed] [Google Scholar]
- 41.Reinhold–Hurek B., Maes T., Gemmer S., Van Montagu M., Hurek T. An endoglucanase is involved in infection of rice roots by the not-cellulose-metabolizing endophyte Azoarcus sp. strain BH72. Mol. Plant-Microbe Interact. 2006;19(2):181–188. doi: 10.1094/MPMI-19-0181. [DOI] [PubMed] [Google Scholar]
- 42.Rosenblueth M., Martínez-Romero E. Bacterial endophytes and their interactions with hosts. Mol. Plant-Microbe Interact. 2006;19(8):827–837. doi: 10.1094/MPMI-19-0827. [DOI] [PubMed] [Google Scholar]
- 43.Ryan R.P., Germaine K., Franks A., Ryan D.J., Dowling D.N. Bacterial endophytes: recent developments and applications. FEMS Microbial Lett. 2008;278:1–9. doi: 10.1111/j.1574-6968.2007.00918.x. [DOI] [PubMed] [Google Scholar]
- 44.Sandhers C.J., Yu S.Y., Moore D.A., Williams R., Gewirtz A.T. Humoral immune response to flagellin requires T cells and activation of innate immunity. J. Immunol. 2006;177:2810–2818. doi: 10.4049/jimmunol.177.5.2810. [DOI] [PubMed] [Google Scholar]
- 45.Seghers D., Wittebolle L., Top E.M., Verstraete W., Siciliano S.D. Impact of agricultural practices on the Zea mays L. endophytic community. Appl. Environ. Microbiol. 2004;70(3):1475–1482. doi: 10.1128/AEM.70.3.1475-1482.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sessitsch A., Reiter B., Berg G. Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can. J. Microbiol. 2004;50(4):239–249. doi: 10.1139/w03-118. [DOI] [PubMed] [Google Scholar]
- 47.Strobel G.A. Harnessing endophytes for industrial microbiology. Curr. Opin. Microbiol. 2006;9(3):240–244. doi: 10.1016/j.mib.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 48.Sturz A.V., Christie B.R., Matheson B.G., Arsenault W.J., Buchanan N.A. Endophytic bacterial communities in the periderm of potato tubers and their potential to improve resistance to soil-borne plant pathogens. Plant Pathol. 1999;48(3):360–369. [Google Scholar]
- 49.Sturz A.V., Christie B.R., Nowak J. Bacterial endophytes: potential role in developing sustainable systems of crop production. Crit. Rev. Plant Sci. 2000;19(1):1–30. [Google Scholar]
- 50.Tholozan J.L., Cappelier J.M., Tissier J.P., Delattre G., Federighi M. Physiological characterization of viable-but-nonculturable Campylobacter jejuni cells. Appl. Environ. Microbiol. 1999;65(3):1110–1116. doi: 10.1128/aem.65.3.1110-1116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.van Aken B., Peres C.M., Doty S.L., Yoon J.M., Schnoor J.L. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoides x nigra DN34). Int. J. Syst. Evol. Microbiol. 2004;54:1191–1196. doi: 10.1099/ijs.0.02796-0. [DOI] [PubMed] [Google Scholar]
- 53.Venkateswaran K., Satomi M., Chung S., Kern R., Koukol R., Basic C., White D. Molecular microbial diversity of a spacecraft assembly facility. Syst. Appl. Microbiol. 2001;24(2):311–320. doi: 10.1078/0723-2020-00018. [DOI] [PubMed] [Google Scholar]
- 54.Wilhelm E., Arthofer W., Schafleitner R. Bacillus subtilis, an endophyte of chestnut (Castanea sativa), as antagonist against chestnut blight (Cryphonectria parasitica). In: Cassells A.C., editor. Pathogen and microbial contamination management in micropropagation. The Netherlands: Kluwer Academic Publishers, Dortrecht; 1997. pp. 331–337. [Google Scholar]
- 55.Yang R.X., Sun G.Y., Zhang R., Chen L.J. 16S rDNA RFLP analysis of endophytic bacteria from Brassica napus. Wei Sheng Wu Xue Bao. 2005;45(4):606–609. [PubMed] [Google Scholar]
- 56.Yoon J.M., van Aken B., Schnoor J.L. Leaching of contaminated leaves following uptake and phytoremediation of RDX, HMX, and TNT by poplar. Int. J. Phytorem. 2006;8(1):81–94. doi: 10.1080/15226510500507128. [DOI] [PubMed] [Google Scholar]


