Abstract
An enrichment culture technique was used to isolate bacterial strains responsible for the biodegradation of profenofos in a soil from Hubei province of central China. Two pure bacterial cultures, named W and Y, were isolated and subsequently characterized by sequencing of 16S rRNA genes and biochemical tests. Isolate W showed 96% similarity to the 16S rRNA gene of a Pseudomonas putida unlike Y which showed 99% similarity to the 16S rRNA gene of Burkholderia gladioli. Both strains grew well at pH 5.5-7.2 with a broad temperature profile ranging from 28° to 36 °C. Bioremediation of profenofos-contaminated soil was examined using soil treated with 200 ug g-1; profenofos resulted in a higher degradation rate than control soils without inoculation. In a mineral salt medium (FTW) reduction in profenofos concentration was 90% within 96 hours of incubation. A literature survey revealed that no data is available regarding the role of Burkholderia gladioli on pesticide biodegradation as well as on profenofos.
Keywords: Profenofos, Organophosphate pesticide, Gas Chromatography, Bioremediation
INTRODUCTION
Profenofos, a well known organophosphate pesticide has been in agricultural use over the last two decades for controlling Lepidopteron pests of cotton and tobacco. Profenofos has been classified as a moderately hazardous (Toxicity class II) pesticide by the World Health Organization (WHO) and it has a moderate order of acute toxicity following oral and dermal administration (32, 33). Profenofos is extremely toxic to fish and macro invertebrates (1). The acute toxic action of profenofos is the inhibition of the acetylcholinesterase activity (11) resulting in toxicity also in humans (8). Microbial degradation of organophosphate pesticides is of particular interest because of the high mammalian toxicity of such compounds and their widespread and extensive use. The most significant step in detoxifying organophosphate compounds is hydrolysis since that makes the compounds more vulnerable to further degradation (21). The enzyme responsible for catalyzing this reaction is referred as an esterase or phosphotriesterase. Research has found a wide range of microorganisms possessing the organophosphate hydrolase enzyme (9, 18, 24). The most well known examples of natural isolates able to degrade organophosphates are Pseudomonas diminuta MG and Flavobacterium ATCC 27551. They have been shown to possess the organophosphate hydrolase (OPH) enzyme (23). The effects of profenofos on total bacterial populations were supposed significant but studies showed that microbial populations decreased initially at concentrations of 100 to 300 μg g-1 but recovered rapidly to levels similar to those in the control (22) indicating that it can serve as Carbon source. Phototransformation of profenofos studies showed cleavage of ester bond (35). To our knowledge, degradation of profenofos by a soil bacterium has not been reported earlier. This paper describes the enrichment, isolation and identification of profenofos degrading bacteria.
MATERIALS AND METHODS
Reagents and Chemicals
Technical grade profenofos (99.5% pure) was purchased from Guangzhou Webber Chemical Co., Ltd. , HPLC grade acetone and hexanes were supplied by J.T Baker, USA and Mallinckrodt Baker Inc TEDIA, USA, respectively. Enzymes, marker and Buffer for PCR were obtained from Sangon Shanghai, China. All other chemicals, if not otherwise stated, used for media were purchased from Sinopharm Chemical Reagent Co. Ltd. (SCRC), Shanghai, China.
Sample Collection for Enrichment Studies
Two environmental samples were used in this study for the isolation of profenofos degrading microorganisms. Both soil samples (Table-1) were collected from different sites with a history of 6-7 years of profenofos applications in Hanchuan city of Hubei province, China. The coordinates of the sampling site were 30o 40’59.7” N, 113o 49’ 37.3” E. The collected samples were stored at 4°C.
Table 1.
Properties of the soil samples
| Sample | pH | Organic matter (g/kg) | sand (%) | Silt (%) | Clay (%) |
|---|---|---|---|---|---|
| 1 | 6.58 | 28.3 | 80.61 | 6.64 | 12.75 |
| 2 | 6.76 | 26.7 | 84.00 | 9.00 | 7.00 |
Isolation of profenofos7degrading bacteria by enrichment culture
Medium for isolation
Salt medium FTW (15) with the following composition was used: (g L-1 deionized water) K2HPO4 0.225; KH2PO4 0.225; (NH4)2SO4 0.225; MgSO4J7H2O 0.05; CaCO3 0.005; and FeCl2J4H2O 0.005, blended with 1 mL of Focht trace elements solution (10). The Focht trace element solution contained (mg L-1): MnSO4JH2O 169; ZnSO4J7H2O 288; CuSO4J5H2O 250; NiSO4J6H2O 26; CoSO4 28; and Na2MoO4J2H2O 24; pH- 7.2±0.2.
The medium for isolation was composed of FTW and profenofos. FTW was autoclaved and then supplemented with 100 mgL-1 profenofos. Plates for isolation were prepared by adding 20 g agar per liter of isolation medium.
Luria-Bertani (LB) Medium composition was: (g L-1 deionized water) NaCl 10.0; Tryptone 10.0; Yeast extract 5.0; pH- 7.2±0.2 (28).
Isolation procedure
The isolation method of Lansing M. Prescott, 2002, was followed with some modifications. The soil samples (Table 1) were sieved through a 90 mesh sieve to remove stones and plant material. Then 10 g of soil was placed in a 250 mL conical flask containing 100 mL FTW media and incubated at 30o C in a rotary shaker at 150 rpm for two days. The flasks were then left for a few hours to allow the soil particles to settle, and the suspension containing microorganisms was then used to inoculate fresh sterilized FTW media containing 100 mgL-1 profenofos and incubated for two days.
To obtain pure cultures of single strains, 5 mL aliquots of enrichment cultures were centrifuged at 3000 rpm for 5 min (TGL-16A centrifuge, Changsha Pingfan instrument Ltd) and the cell pellets were resuspended in 2 mL sterile media. Aliquots of this suspension were streaked on FTW profenofos agar plates. Inoculated plates were incubated under aerobic conditions at 30°C and discrete colonies were isolated.
The resulting colonies were repeatedly subcultured in the same medium (FTW with 100 mgL-1 of profenofos) to confirm their profenofos utilizing ability. All colonies were transferred into fresh sterile medium to obtain a pure culture. Two strains, designated Y and W, which possessed the highest degradation capability, were selected for further investigation (26).
Identification of the isolates
The morphological, physiological and biochemical tests of isolates W and Y, were performed by using standard methods. The bacterial strains were taxonomically identified from Bergey’s Manual of systematic bacteriology (12) as Pseudomonas sp. and further confirmation was made by sequencing 16S rRNA gene.
Genomic DNA isolation and sequencing of 16S rRNA gene
Genomic DNA was isolated using standard bacterial procedures (14). The following primers were used for PCR amplification of the gene encoding 16S rRNA: 63f (5’ AGGCCTAACACATGCAAG TC 3’), 1387r (5’ GGGCGGAGTGTACAAGGC 3’) . The PCR mixtures (50 μl) contained 10μM of each primer, PCR buffer, 5 U of Taq DNA polymerase, BSA 10μM and 2μl of DNA. The thermocycling conditions consisted of a denaturation step at 94°C for 3 min, 28 amplification cycles of 94°C for 20 s, 58°C for 40 s and 72°C for 1 min and a final polymerization for 3 min 30 s with a MJ Research Thermalcycler (MJ Research, PTC-100, USA). PCR products were visualized on 1.0% agarose gels with Gel Doc 2000 (Bio-Rad USA). The purified PCR products were then cloned into the pMD 18 T vector (TaKaRa, Dalian, China) and sequenced in Sangon Shanghai, China. Nucleotide sequence similarities were determined using BLAST (National Center for Biotechnology Information databases).
Biodegradation studies Inoculums preparation for degradation studies
The isolated bacterial cells were pre cultured in LB medium at 30°C with 150 rpm shaking, harvested by centrifugation at 6000 rpm for 10 min and washed three times with sterilized water. For all experiments, cells were used at a concentration of 20 mg dry weight per 50 mL media and for 50 g soil 40 mg dry weight cells were used. If not otherwise stated the incubation conditions were 30°C with 150 rpm shaking.
Kinetics of biodegradation
The degrading ability of the isolated strains was assayed with batch cultures in mineral salts medium (FTW) supplemented with profenofos (100 mgL-1) in triplicates. The cultures were inoculated with the isolated strain and incubated. Aliquots of 2 mL were taken at suitable interval between 0 to 4 days and were subjected to GC analysis after extraction of profenofos residues. Triplicate sets of each composition without inoculation were kept as controls.
Degradation studies in soil
Air-dried sieved (< 2 mm) soil samples, used for isolation (Table 1), were placed in 50 g portions in pre sterilized 100 mL Erlenmeyer flasks and were moistened with sufficient water to provide 60% water holding capacity. The soil samples were amended with 200 mg/L of profenofos and inoculated with the isolates separately and incubated in duplicate. The duplicate sets of controls consisting of sterilized soil with identical amount of profenofos but without the isolate were run simultaneously under identical conditions. At different time points between 0 to 25 d., aliquots of 3 g soil were removed in duplicate from the microcosms and subjected to extraction of profenofos residues in soil samples by acetone hexane (20:80) mixture (17) and then analyzed by GC.
Gas chromatography analysis
Extraction of profenofos residues: The samples (aliquots of liquid culture as well as soil) were subjected to organic solvent extraction three times with acetone: hexane (20:80) mixture. The extract was filtered and the organic solvents were evaporated to near dryness on a rotary thin film evaporator. The residues were dissolved in 5 ml hexane and stored at 4°C until analysis. Efficiency of extraction and estimation of profenofos was over 90%.
GC-ECD : Profenofos residues, extracted in hexane from salt media or from the soil samples, were analyzed in Agilent Gas chromatograph (Model 6890 Series USA) equipped with a Ni 63 electron capture detector with HP-5MS column (length 30 m, diameter 0.25 mm). The operating conditions: initial temperature 120 °C, then heated at 7°C/min to a final temperature of 250°C. The total run time was 33.07 min. The splitless mode was used for the injection. The injector temperature was at 240°C and the detector temperature was at 300°C. Nitrogen gas (99.999%) was used as the carrier gas with a gas flow at 23.3 cm sec-1 linear velocity.
RESULTS
Isolation and Identification of profenofos degrading bacteria
Four different microorganisms were isolated from the profenofos-exposed soil samples by above mentioned method. All isolates were found to possess the ability to degrade the pesticide. Strains named as W and Y degraded profenofos were subejected for further studies and are presented in this study. Theses strains were a rod-shaped, gram negative, motile bacterium (Table 1).
By sequencing the 16S rRNA gene of W and Y and comparing them with previously published 16S rRNA gene sequences, the strains were classified as a member of the genus Pseudomonas and Burkholderia respectively. The sequence of strain W displayed the highest identity (96%) with the 16S rRNA gene of a Pseudomonas putida (FJ865579) and strain Y showed (99%) similarities with Burkholderia gladioli (FJ865580).
Biodegradation of profenofos
Degradation in liquid culture media
Under aerobic conditions, profenofos degradation by both bacterial isolates was monitored by GC for a period of 96 h (Fig. 1a1 & a2) in liquid culture media.
Figure 1.
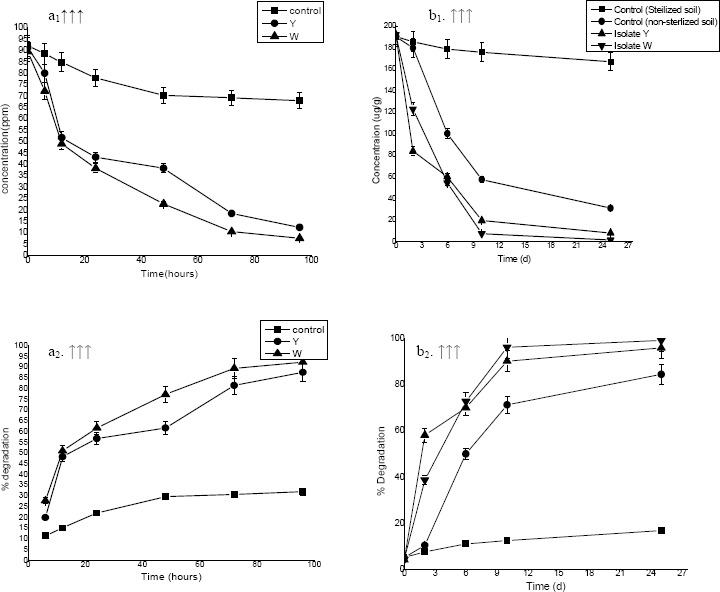
Degradation kinetics of profenofos by bacterial strains; (a1) degradation in liquid batch culture media (FTW amended with profenofos) at different time interval; ■ control, ● Y, ▲ W; (a2) % degradation of profenofos at different time interval in liquid batch culture media; ■ control, ● Y strain , ▲ W strain; (b1) degradation in soil sample supplemented with profenofos at different days; ■ control (sterilized soil), ● Control (non sterilized soil), ▲ Isolate Y, ▼ Isolate W ; (b2) % degradation of profenofos at different time interval in soil sample; ; ■ control (sterilized soil), ● Control (non sterilized soil), ▲ Isolate Y ▼ Isolate W
Both Pseudomonas putida W and Burkholderia gladioli effected very rapid degradation of profenofos, added to the mineral salts medium as a sole carbon source.
Rapid degradation began shortly after 6 hours of incubation under aerobic conditions, suggesting that a constitutively expressed enzyme(s) was involved in the degradation of profenofos. The degradation of profenofos supported cell growth, indicating that isolated strains could utilize profenofos as a carbon source.
After 6 h, 27.77% and 19.92 % of pesticide had rapidly removed by Pseudomonas putida W and Burkholderia gladioli respectively followed by a slower decrease of profenofos with longer incubation times. After 96 hrs 92.37% pesticide was removed by Pseudomonas putida showing its ability to degrade op compounds in liquid media while in case of Burkholderia gladioli results were 87.58%.
Degradation of profenofos in soils
The soil sample, previously used to isolate profenofos degrading bacteria, was used in this study. The soils characteristics are given in the Table 2. The standard error was within 5% of the mean.
Table 2.
Morphological and Biochemical characteristics of profenofos degrading isolated bacterial strains
| Biochemical Test and staining | Isolate W | Isolate Y |
|---|---|---|
| Gram straining | Negative | Negative |
| Oxidase | Positive | Negative |
| Catalase | Positive | Positive |
| Indole test | Positive | Negative |
| Methyl red | Negative | Negative |
| V oges-Proskauer | Positive | Positive |
| Citrate | Positive | Negative |
| H2S production | Positive | Negative |
| Urease | Negative | Negative |
| Glucose , Sucrose | Positive | Positive |
| Lactose | Positive | Negative |
| Manitol | Negative | Positive |
| Methanol, Ethanol, Butanol, Isobutanol | Negative | Positive |
| Denitrification | Positive | Negative |
| Cell type (shape) | rods | Curve rods |
| Color(colony in FTW media) | white | Yellow |
| Elevation | flat (irregular) | convex (entire) |
| Surface | Not smooth | Smooth |
The addition of isolated strains to soils resulted in a more rapid rate of profenofos degradation than that by control (non sterilized soil and sterilized) soils. Degradation of profenofos in control sterilized soils (without inoculation) was minimal where less than 16% of the applied concentration was degraded in 25-day incubation studies (Fig. 1b2). Degradation of profenofos was significant in control non sterilized (without inoculation) soil where 84% degradation was recorded in 25 days. Fig. 1b1 & b2 demonstrates that approximately 70% profenofos was degraded within 5 days but the degradation reaches 96.06 and 99.37within 25 days by Pseudomonas putida and Burkholderia gladioli respectively. This suggested the same fashion of degradation by both strains in soil and as well as in liquid media such as in first phase rapid steep decline in concentration of the profenofos followed by slower degradation.
DISCUSSIONS
Isolation of bacterial strain through enrichment technique has been extensively used for the study of biodegradation of pesticides (31). The results obtained in this study were in agreement with earlier reports that indicated the involvement of pseudomonas putida in the degradation of organophosphorus insecticides like chlorpyrifos (4), methylparathion (27), phosphonate (7) parathion (13), ethoprophos (19) and glyphosate (34).
Members of the genus Burkholderia are versatile organisms that occupy a wide range of ecological niches. These bacteria are exploited for biocontrol, bioremediation and plant growth promotion purposes, but safety issues regarding human infections, especially in cystic fibrosis patients, have not been solved. Many members of genus Burkholderia have been discovered as pesticide degrader, For example, the species B. kururiensis (36), B. sacchari (5), B. phenoliruptrix 11(6), B. terrae (16) and B. cepacia (2) but there have been only a few previous reports on the ability of biodegradation of Burkholderia gladioli.
Degradation of pentachlorophenol (30) and glyphosate (20) has been early reported. To our knowledge this is very first report on ability of Burkholderia gladioli to degrade profenofos.
OPH gene in Burkholderia sp. Strain NF100 has been extensively studied (29), similarly esterase gene was found in Burkholderia gladioli (25), as previous reports clearly stated that esterase gene can be responsible for the breakdown of ester bond in organophosphate pesticide which may be the reason for biodegradation of profenofos.
In the natural environment, the competition for carbon sources is immense and the utilization of profenofos as an energy source by this bacterium provides it with a substantial competitive advantage over other microorganisms. Successful removal of pesticides by the addition of bacteria (bioaugmentation) had been reported earlier for many compounds, including parathion (37), coumaphos (3) and ethoprop (19).
ACKNOWLEDGEMENT
This work was supported by both the National Key Technology R & D Program of China (2008BADA7B03) and the City Key Technology R & D Program of Wuhan in China (200720422150). The authors are grateful to Mr. Shamba Chatterjee, Prof. S. H. Qi, Dr. G. L.Sheng and their students for their valuable suggestions as well as technical support. The authors would like to thank to Dr.Julia Ellis Burnet for her helpful comments on the manuscript.
REFERENCES
- 1.Akerblom N. Uppsala: Sveriges Lantbruks Univ.; 2004. Agricultural pesticide toxicity to aquatic organisms: a literature review. p. 31. [Google Scholar]
- 2.Anthony R., Smith W., Carol A.B. Induction of enzymes of 2, 4 dichlorophenoxyacetate degradation in Burkholderia cepacia 2a and toxicity of metabolic intermediates. Biodegradation. 2008;19:669–681. doi: 10.1007/s10532-007-9172-0. [DOI] [PubMed] [Google Scholar]
- 3.Barles R.W., Daughton G.C., Hsieh D.P.H. Accelerated parathion degradation in soil inoculated with acclimated bacteria under field conditions. Arch. Environ. Contam. Toxicol. 1979;8:647–660. doi: 10.1007/BF01054867. [DOI] [PubMed] [Google Scholar]
- 4.Brajesh K.S., Allan W., Alun W., Denis J.W. Biodegradation of Chlorpyrifos by Enterobacter Strain B-14 and Its Use in Bioremediation of Contaminated Soils, American Society for Microbiology. Appl. Envir. Microbiol. 2004;27:4855–4863. doi: 10.1128/AEM.70.8.4855-4863.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramer C.O., Andamme P.V., da Silva L.F., Gomez J.G., Steinbuchel A. Burkholderia sacchari sp. nov. a polyhydroxyalkanoate accumulating bacterium isolated from soil of a sugar cane plantation in Brazil. Int. J. Syst. Evol. Microbiol. 2001;51:1709–1713. doi: 10.1099/00207713-51-5-1709. [DOI] [PubMed] [Google Scholar]
- 6.Coenye T.D., Henry D.P., Speert Vandamme P. Burkholderia phenoliruptrix sp. nov. to accommodate the 2,4,5 trichlorophenoxyacetic acid andhalophenol degrading strain AC1100 System. Appl. Microbiol. 2004;27:623–627. doi: 10.1078/0723202042369992. [DOI] [PubMed] [Google Scholar]
- 7.Cook M.A., Christian G., Daughton Martin A. Phosphonate Utilization by Bacteria. J. Bacteriol. 1978;1:85–90. doi: 10.1128/jb.133.1.85-90.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa L.G., Cole T.B, Jansen K.L., Furlong C.E. Paraoxonase (pon1) and Organophosphate toxicity: Springer; pp. 209–220. chap 13. [Google Scholar]
- 9.Dave K.I., Miller C.E., Wild J.R. Characterization of organophosphorus hydrolase and the genetic manipulation of the phosphotriesterase from Pseudomonas diminuta. Chem. Biol. Interact. 1993;87:55–68. doi: 10.1016/0009-2797(93)90025-t. [DOI] [PubMed] [Google Scholar]
- 10.Focht D.D. Weaver R.W., et al., editors. Microbiological procedures for biodegradation research. Methods of soil analysis. Part 2. SSSA. Book Ser. 1994;5:407–426. [Google Scholar]
- 11.Fukuto T.R. Mechanism of action of organophosphorus and carbamate insecticides. Environ. Health Perspect. 1990;87:245–254. doi: 10.1289/ehp.9087245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.George M.G. Gammaproteobacteria. Bergey’s Manual of Systematic Bacteriology. In: Brenner Don J., Krieg Noel R, Staley James T., editors. Vol. 2. Springer USA: 2005. pp. 323–378. [Google Scholar]
- 13.Gilbert E.S., Walker A.W., Keasling J.D. A constructed microbial consortium for biodegradation of the organophosphorus insecticide parathion. Appl Microbiol Biotechnol. 2003;61:77–81. doi: 10.1007/s00253-002-1203-5. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg J.B., Ohman D.E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J. Bacteriolo. 1984;158:1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herman D.C., Frankenberger W.T.Jr. Bacterial reduction of perchlorate and nitrate in water. J. Environ. Qual. 1999;28:1018–1024. [Google Scholar]
- 16.Hiroaki I., Kazuya A., Yoshie H. Isolationand characterization of a new2, 4 dinitrophenol degrading bacterium Burkholderia sp. strainKU 46 and its degradation pathway. FEMS Microbiol. Lett. 2007;274:112–117. doi: 10.1111/j.1574-6968.2007.00816.x. [DOI] [PubMed] [Google Scholar]
- 17.Horne I., Harcourt R.L., Sutherland T.D., Russel R.J., Oakeshott J.G. Isolation of a Pseudomonas monteilli strain with a novel phosphotriesterase. FEMS Microbiol. Lett. 2002;206:51–55. doi: 10.1111/j.1574-6968.2002.tb10985.x. [DOI] [PubMed] [Google Scholar]
- 18.Horne I., Sutherland T.D., Harcourt R.L., Russell R.J., Oakeshott J.G. Identification of an opd (organophosphate degradation) gene in an Agrobacterium isolate. Appl. Environ. Microbiol. 2002;68:3371–3376. doi: 10.1128/AEM.68.7.3371-3376.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpouzas D.G., Walker A. Factors influencing the ability of Pseudomonas putida strains epI and II to degrade the organophosphate ethoprophos. J. Appl. Microbiol. 2000;89:40–48. doi: 10.1046/j.1365-2672.2000.01080.x. [DOI] [PubMed] [Google Scholar]
- 20.Kuklinsky S.J., Luiz A. W., Mendes R., Pizzirani I., Kleiner A.A., Azevedo J.L. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant and soil. 2005;273:91–99. [Google Scholar]
- 21.Kumar S., Mukerji K.G., Lal. Molecular aspects of pesticide degradation by microorganisms. Crit. Rev. Microbiol. 1996;22:1–26. doi: 10.3109/10408419609106454. [DOI] [PubMed] [Google Scholar]
- 22.Martinez T., Salmeron M.V., Gonzalez V.L. Effect of an organophosphorus insecticide, profenofos, on agricultural soil microflora. Chemosphere. 1992;24:71–80. [Google Scholar]
- 23.Mulbry W. Characterization of a novel organophosphorus hydrolase from Nocardiodes simplex NRRL B 24074. Microbiol. Res. 2000;154:285–288. doi: 10.1016/S0944-5013(00)80001-4. [DOI] [PubMed] [Google Scholar]
- 24.Mulbry W., Karns W., Kearney J.C., Nelson P.C., McDaniel J.O., Wild C.S. dentification of plasmid borne parathion hydrolase gene from Flavobacterium sp. by southern hybridization with opd from Pseudomonas diminuta. Appl. Environ. Microbiol. 1986;51:926–930. doi: 10.1128/aem.51.5.926-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petersen E.I., Valinger G., Solkner B., Stubenrauch G., Schwab H. A novel esterase from Burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to β lactamases and DD peptidases. J. Biotechnol., 9. 2001;1:11–25. doi: 10.1016/s0168-1656(01)00284-x. [DOI] [PubMed] [Google Scholar]
- 26.Prescott L.M., Harley, Klein Microbial Nutrition. Microbiology. (5th ed.) :101–110. [Google Scholar]
- 27.Rani N.L., Lalitha Kumari D. Degradation of methyl parathion by Pseudomonas putida. Can. J. Microbiol. 1994;15:984–987. doi: 10.1139/m94-160. [DOI] [PubMed] [Google Scholar]
- 28.Ronald M. CRC Press USA.: 2000. Atlas Handbook of microbiological media 2000; p. 905. [Google Scholar]
- 29.Tago K., Yoenezawa S., Ohkouchi T., Ninomiya T., Hashimoto M., Hayatsu M. A Novel Organophosphorus Pesticide Hydrolase Gene Encoded on a Plasmid in Burkholderia sp. Strain NF100. Microbes Environ. 2006;21:53–57. [Google Scholar]
- 30.Takashi N., Takayuki M., Yoshikatsu S., Isamu Y. Biotransformation of pentachlorophenol by Chinese chive and a recombinant derivative of its rhizosphere competent microorganism. Pseudomonas gladioli M-2196 Soil Biology and Biochemistry. 2004;36:787–795. [Google Scholar]
- 31.Tatiane M.S., Maria I.S., André M.M., Fabiana D.A., Sônia A.V.P., Paulo R. F., Marcelo D.C., Emanuel C.B., Carneiro Marcos P. Degradation of 2, 4 D herbicide by microorganisms isolated from Brazilian contaminated soil. Brazil. J. Microbiol. 2007;38:522–525. [Google Scholar]
- 32.US EPA 2000 The HED chapter of the registration eligibility decision document (RED) for profenofos EPA 738-F-00-005. [Google Scholar]
- 33.WHO Geneva, Switzerland.: World Health Organization; 2004. The WHO recommended classification of pesticides by hazard and guidelines to classification 2004. WHO/PCS/90.1. [Google Scholar]
- 34.Yi Cheng S., Yan Cheng C., Zhe Xian T., Feng M.L., Xin Yue W., Jing Zhang H., Hanada S., Shigematsu T.K., Shibuya Y., Kamagata T., Kanagawa & Kurane R. Burkholderia kururiensis sp. nov. a trichloroethylene (TCE) degrading bacterium isolated from aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 2000;50:743–749. doi: 10.1099/00207713-50-2-743. [DOI] [PubMed] [Google Scholar]
- 35.Zamy C., Mazellier P., Legube B. Phototransformation of selected organophosphorus pesticides in dilute aqueous solutions. Water Research. 2004;38:2305–2314. doi: 10.1016/j.watres.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z.L., Xiao M.L., Niamh G., David N., Dowling, Yi Ping W. Novel AroA with High Tolerance to Glyphosate, Encoded by a Geneof Pseudomonas putida 4G 1 Isolated from an Extremely Polluted Environment in China. App. Env. Microbiol., 71. 2005;8:4771–4776. doi: 10.1128/AEM.71.8.4771-4776.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhongli C., Shunpeng L., Guoping F. Isolation of methyl parathion degrading strain M6 and cloning of the methyl parathion hydrolase gene. Appl. Environ. Microbiol. 2001;67:4922–492. doi: 10.1128/AEM.67.10.4922-4925.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


