Abstract
The production of manganese peroxidase (MnP) from Bacillus pumilus and Paenibacillus sp. was studied under absence and presence of the inducers indulin AT, guayacol, veratryl alcohol, lignosulfonic acid and lignosulfonic acid desulfonated. Indulin AT increased the activity of B. pumilus MnP up to 31.66 U/L after 8 h, but no improve was observed for Paenibacillus sp., which reached maximum activity (12.22 U/L) after 20 h. Both MnPs produced by these microorganisms were purified in phenyl sepharose resin and the proteins from crude extracts were eluted in two fractions. However, only the first fraction of each extract exhibited MnP activities. Tests in different pH and temperature values, from pH 5.0 to pH 10.0 and 30 °C to 60 °C, respectively, were carried out with the purified MnP. The maximum activity reached for B. pumilus and Paenibacillus sp. MnPs were 4.3 U/L at pH 8.0 and 25 °C and 11.74 U/L at pH 9.0 and 35 °C, respectively. The molar masses determined by SDS-PAGE gel eletrophoresis were 25 kDa and 40 kDa, respectively, for the purified enzyme from B. pumilus and Paenibacillus sp.
Keywords: Bacillus pumilus, Paenibacillus sp., Manganese peroxidase, Purification, Characterization
INTRODUCTION
Plant cell walls have lignin in their structure as the most abundant component. Lignin is an aromatic and heterogeneous constituent that ensures strength and resistance towards microbial attack. White-rot basidiomycetes are the most common organisms known to efficiently degrade and mineralize lignin into CO2 and H2O, due to extracellular enzymes involved in lignin degradation, particularly the ligninase complex, formed by laccase, lignin peroxidase and manganese peroxidase (10, 11). Rarely these three enzymes are present in the same organism, and different combinations of them can operate. The ligninase complex is frequently produced during secondary metabolism but different species have particular responces to nutrients (19, 25).
Due to the important degradative potential of manganese peroxidase (MnP), there is a general interest in producing the enzyme biotechnologically. Manganese peroxidase, a glycosylated heme-containing enzyme, have been used besides biodegradation of lignin (13), in the biodegradation of polycyclic aromatic hydrocarbons (PAH) (24, 27), humic acids (28), synthetic dyes (12), and polychlorinated biphenyls (PCB) (2). MnP oxidizes Mn2+ to Mn3+ in an H2O2-dependent reaction and Mn3+ is stabilized by chelating dicarboxylic acids (8).
The optimum pH of almost all ligninolytic enzymes, including MnP, reported to date lies in the acidic range. However some industry activities, such as pulping and bleaching are mainly performed under highly alkaline conditions and the waste generated is also alkaline. Ligninolytic enzymes having acidic optimum pH values cannot be used under alkaline conditions (16).
Few reports on bacterial MnP are found in the literature. Two bacteria, B. pumilus, isolated from wood decomposition material by Duarte et al. (6) and Paenibacillus sp. isolated from paper mill effluent (22) were able to produce MnP in alkaline conditions. These enzymes were able to remove the color from paper mill effluent (23). In this study, we report on the purification and partial characterization of MnP from B. pumilus and Paenibacillus sp.
MATERIALS AND METHODS
Microorganisms
Bacillus pumilus CBMAI 0008 was isolated from wood decomposition material by Duarte et al. (6) and was maintained in a culture medium containing birchwood xylan (20).
Paenibacillus sp. CBMAI 868 was isolated from paper industry effluent, in a media containing 1% birchwood xylan (Sigma); 0.1% (NH4)2SO4; 50% paper mill effluent; and 2% agar-agar. After sterilization, nistatin (0,044 mg/mL) was added as an antifungal control. An aliquot of 0.3 mL from paper mill effluent was used to sow the surface medium in Petri plates. After incubation (37 °C, 48 h), the ability of colonies to grow and produce clear haloes of decolorization on the medium surface was verified. The colony that provided the greater clear haloe was purified and identified at Microbial Resources Divison – CPQBA/UNICAMP.
The subsequent assays were carried out at 45 °C for both bacteria, once this temperature is close to that used in several industrial processes.
Inocula preparation
The isolates were individually transferred to 125 mL Erlenmeyer flasks, containing 12.5 mL of the liquid media (20), and incubated at 45 °C in a shaker (250 rpm) for 20 h. The culture was centrifuged in aseptic conditions and the cells were ressuspended in 40 mL of the media. Further steps of MnP production were carried out using inoculum at 8 % (v/v).
Effect of inducers on the MnP production
For cinetic studies, inocula were prepared as described above and the fermentation was carried out in 250 mL Erlenmeyer flasks, containing 50 mL of the liquid media, and incubated at 45 °C in a shaker (250 rpm) during 32 h. At regular periods, samples were collected for MnP activity measurement. MnP production was also studied in the presence of veratryl alcohol, industrial lignin (indulin AT), guayacol, lignosulfonic acid and lignosulfonic acid desulfonated at 0.1% (w/v) at the same conditions. The fermented media was centrifuged for 15 min at 12000 x g for the activity assays.
Enzyme activity assay
Manganese peroxidase activity was assayed spectrophotometrically according to Kuwahara (17). The reaction mixture contained 0.1 mL of 0.25 M sodium lactate, 0.05 mL of 2 mM MnSO4, 0.2 mL of 0.5% serum albumin bovine, 0.1 mL of 0.1% phenol red, 0.5 mL enzyme and 0.05 mL of 2 mM H2O2 in 0.2 M sodim phosphate buffer (pH 8.0). The mixture was left at room temperature for 5 min and the reaction was ended with 0.04 mL NaOH 2 N. The absorbance was read at 610 nm and the activity expressed in U/L. One activity unit was defined as amount of enzyme necessary to oxidize 1 M mol of substrate per minute.
Proteins measurement
Protein concentration was measured by the Bradford method (3). Bovin serum albumin was used as a standard. The results were used to calculate specific activity.
Culture conditions for crude extracts production aim purification and characterization
The culture conditions for crude extracts production aim purification and characterization were determined after inducers studies. The bacteria were cultured as described above for inoculum production and subsequently inoculated in the medium containing xylan (20) enriched with indulin AT (B. pumilus) or without inducer (Paenibacillus sp.), and incubated at 45 °C in a shaker (250 rpm) for 8 h or 20 h, respectively. The cultures were centrifuged during 15 min at 12000 x g prior the purification assays.
MnP purification and partial characterization
All purification procedures were done at room temperature. The enzyme activity and protein concentrations were determined in all steps. The supernatant obtained from the crude broth was filtered (Whatman 0.45 μm) and futher concentrated in a Pellicon™ ultrafiltration system using 10 kDa Biomax 10™ membrane. The concentrated solution was loaded to a Pharmacia KX-26 40/26 column packed with Phenyl Sepharose hydrophobic interaction chromatography (HIC) resin coupled to a Pharmacia FPLC, previously equilibrated with 1.7 M (NH4)2SO4 (9). The flow rate was 3 mL/min and proteins were eluted in a linear (NH4)2SO4-gradient from 1.7 to 0 M in sodium phosphate buffer, pH 7.0, 50 mM. The peaks containing MnP activity were pooled, concentrated and dialyzed against the same buffer. The HIC-separated MnP was further loaded to a Pharmacia KX-16 16/2 column packed with Q-Sepharose anion exchange chromatography resin coupled to a Pharmacia FPLC, previously equilibrated with sodium phosphate 50 mM. The flow rate was 2 mL/min and proteins were eluted with sodium phosphate 50 mM in a linear NaCl-gradient from 0 to 1 M, pH 7.0. The peaks were all collected in a Red Frak™ (Pharmacia) system, monitored at 280 nm.
Electrophoresis – SDS PAGE
Purity and apparent molecular weights of the peaks were examined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as described by Laemmli (18) using a Mini-Protean II system (Biorad). Molecular weight markers (Promega) were included in 10% gels, followed by Comassie Blue R-250 staining.
Effect of pH and temperature on MnP activity
The effect of pH on purified MnP activity from B. pumilus and Paenibacillus sp. was studied in the following buffers (200 mM): citrate phosphate, pH 5.0 and pH 6.0; sodium phosphate, pH 7.0 and pH 8.0; and glycine-NaOH, pH 9.0 and pH 10.0. The effect of temperature was determined in range from 25 °C to 60 °C with 5 °C intervals and the incubation was according to Kuwahara et al. method (17).
RESULTS AND DISCUSSION
Effect of inducers on the production of MnP
The production of MnP was determined in the crude extract of B. pumilus and Paenibacillus sp., in the absence and presence of inducers. The maximum activity reached for the B. pumilus MnP without inducers was 6.41 U/L after 16 h (Figure 1-A). When the inducers were added in the culture medium, the activity increased to 31.66 U/L in the presence of indulin AT after 8 h (Figure 1-B), followed by lignosulfonic acid (15.6 U/L) and lignosulfonic acid desulfonated (8.6 U/L) after 6 h (Figures 1-D and 1-E, respectively). The maximum MnP production by Paenibacillus sp. was 13.76 U/L in the presence of veratryl alcohol after 28 h (Figure 1-C). The addition of lignosulfonic acid desulfonated resulted in a maximum activity of 12.78 U/L at the same time (Figure 1-D), while maximum activity in the presence of guayacol was 12.20 U/L after 20 h (Figure 1-F). These results did not show improve in the Paenibacillus sp. MnP activity since the activity without inducers was 12.33 U/L after 20 h (Figure 1.-A). No data was found in the literature about the use of inducers in the MnP production from bacteria. The use of Polyfon H as inducer in the MnP production by the fungus Irpex flavus allowed reaching 0.08 U/mL. Inducers absence in the media was the best conditions for MnP production by Dichomitus squalens (0.29 U/mL) and Polyporus sanguineus (0.43 U/mL) (7). Further study revelead that indulin AT, Polyfon H, Reax 80, Orzan S, veratryl alcohol and guayacol did not increase MnP activity by Phlebia floridensis, a White-rot fungus (1).
Figure 1.
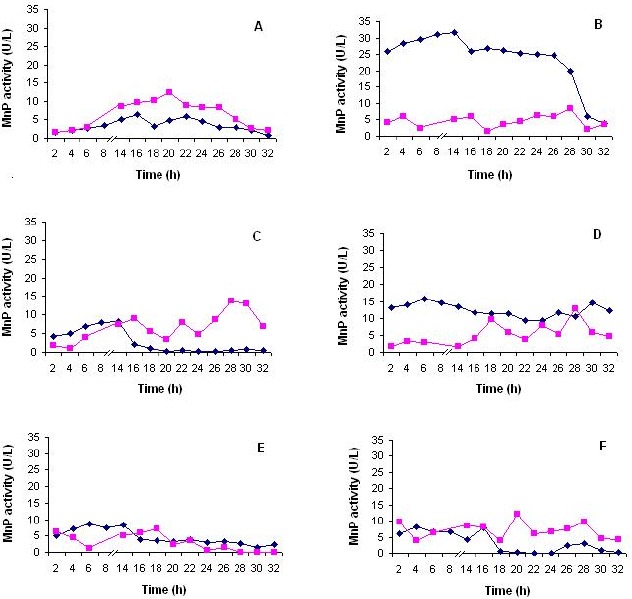
MnP production by B. pumilus CBMAI 0008 ( ) and Paenibacillus sp. CBMAI 868 (
) and Paenibacillus sp. CBMAI 868 ( ). A – no inducers; B – indulin AT; C – veratryl alcohol; D – dissulf lignin acid; E – lignin acid and F – guayacol.
). A – no inducers; B – indulin AT; C – veratryl alcohol; D – dissulf lignin acid; E – lignin acid and F – guayacol.
Enyme purification
The crude extract produced by B. pumilus and Paenibacillus sp. was first taken through the hydrophobic interaction resin (phenyl sepharose) obtaining two peaks as shown in Figures 2-A and 2-B. Subsequently, the eluted peaks were submitted to ultrafiltration and further purification in Q-Sepharose anion exchange chromatography resin, confirming the presence of only one proteic fraction. In all purification steps, MnP activity and total proteins were determined. A summary of the purification steps is shown in Table 1.
Figure 2.
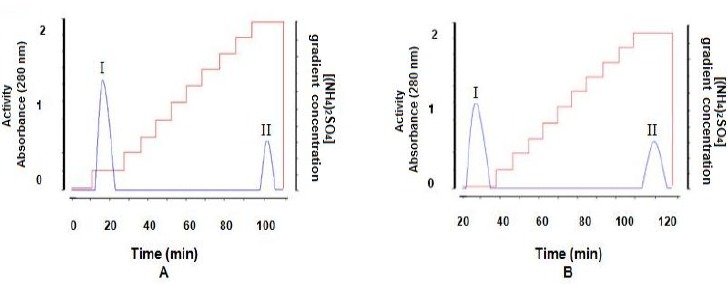
FPLC chromatogram in a Fenil Sepharose hydrophobic interaction resin. A – B. pumilus CBMAI 0008 MnP elution characterized in peak I. B – Paenibacillus sp. CBMAI 868 MnP elution characterized in peak I. Activity ( ) and gradient concentration (
) and gradient concentration ( ).
).
Table 1.
Purification steps of MnP from B. pumilus CBMAI 0008 and Paenibacillus sp. CBMAI 868.
| Purification step | Total volume (mL) | MnP activity (U/L) | Total protein (mg/mL) | Specific activity (U/mg) | Yield (%) | Fold |
|---|---|---|---|---|---|---|
| B. pumilus | ||||||
| Fenil Sepharose | 140 | 0.43 | 0.080 | 5.37 | 100.00 | 1.00 |
| Ultraf./Dial. | 14 | 4.20 | 0.138 | 30.39 | 976.74 | 5.66 |
| Q-Sepharose | 80 | 0.38 | 0.013 | 29.23 | 88.37 | 5.44 |
| Ultraf./Dial. | 8 | 3.45 | 0.110 | 31.36 | 802.33 | 5.84 |
| Paenibacillus sp. | ||||||
| Fenil Sepharose | 110 | 0.35 | 0.053 | 6.60 | 100.00 | 1.00 |
| Ultraf./Dial. | 11 | 3.20 | 0.317 | 10.09 | 914.29 | 1.53 |
| Q-Sepharose | 70 | 0.28 | 0.044 | 6.36 | 80.00 | 0.96 |
| Ultraf./Dial. | 7 | 2.40 | 0.284 | 8.45 | 685.71 | 1.28 |
Electrophoresis – SDS-pAGE
The purified MnP from B. pumilus and Paenibacillus sp. appeared as single bands on 10% SDS-pAGE and presented molecular weights of 25 kDa and 40 kDa, respectively (Figure 3). MnP-PGY and MnP-GY produced by fungus Pleorotus ostreatus (15), and a purified MnP from Trametes versicolor (4) revelead molecular weights of 42 kDa close to the molecular weight found for Paenibacillus sp. in the present study. Also Hoshino et al. (14) verified a similar molecular weight of 40 kDa for MnP produced by fungus Lenzites betulinus. According to this study, the purified MnP from B. pumilus showed inferior molecular weights comparing to those produced by fungus reported in the literature.
Figure 3.
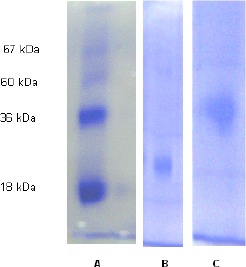
SDS electrophoresis: A – molecular weight standards; B – purified MnP from B. pumilus CBMAI 0008; C – purified MnP from Paenibacillus sp. CBMAI 868. Coomassie Brilliant Blue staining.
Effect of pH and temperature on MnP activity
The results of enzymatic activity in different pH and temperature values for purified MnP from B. pumilus and Paenibacillus sp. are shown in Figures 4-A and 4-B. The activity of B. pumilus MnP increased according to pH from 5.0 to 8.0, where occurred the maximum activity (4.3 U/L), and decreased at superior pH values (Figure 4.-A). Optimum activity for Paenibacillus sp. was observed at pH 9.0 (5.65 U/L). MnP produced by this bacteria showed greater stability in different pH values than MnP produced by B. pumilus, since the lowest activity was at pH 6.0 (3.56 U/L) (Figure 4.-A). Since the isolation conditions of these microorganisms were carried out in alkaline media, these results confirmed the maximum activities of purified enzymes in these conditions. Tests aiming the determination of optimum temperature values were done at best pH conditions for both microorganisms. Higher MnP activity occurred at 25 °C for B. pumilus (4.3 U/L) and 35 °C for Paenibacillus sp.(11.74 U/L), (Figure 4.-B). MnP activity was not detected at 55 °C and above (Figure 4.-B).
Figure 4.
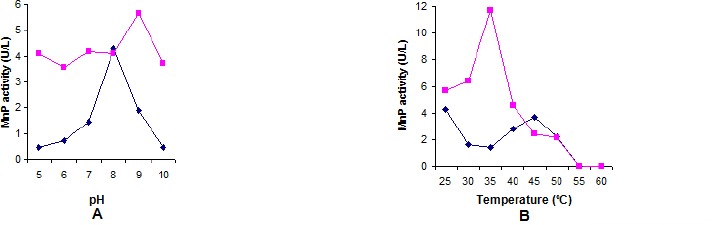
MnP activity from B. pumilus CBMAI 0008 ( ) and Paenibacillus sp. CBMAI 868 (
) and Paenibacillus sp. CBMAI 868 ( ). A – Different pH values, buffers (0.2M): citrate phosphate, pH 5.0 and pH 6.0; sodium phosphate, pH 7.0 and pH 8.0; glycine-NaOH, pH 9.0 and pH 10.0. B-Different temperature values: B. pumilus, at pH 8.0-sodium phosphate buffer 0.2 M and Paenibacillus sp., at pH 9.0 glycine-NaOH buffer 0.2 M.
). A – Different pH values, buffers (0.2M): citrate phosphate, pH 5.0 and pH 6.0; sodium phosphate, pH 7.0 and pH 8.0; glycine-NaOH, pH 9.0 and pH 10.0. B-Different temperature values: B. pumilus, at pH 8.0-sodium phosphate buffer 0.2 M and Paenibacillus sp., at pH 9.0 glycine-NaOH buffer 0.2 M.
The MnP optimum temperature is variable according to microorganism, as verified in previous studies. Purified MnP from Aspergillus terreus LD-1 showed maximum activity at 37 °C (16), while the optimum temperature for the enzymes produced by Dichomitus squalens, Irpex flavus and Polyporus sanguineus was 30 °C. In these cases, the enzyme was not detectable at 35 °C, except for MnP from Polyporus sanguineus (7). In the present study, the optimum temperature for purified MnP from B. pumilus and Paenibacillus sp. is the same verified, respectively, for Phebia floridensis (1) and Schizophyllum sp. F17 (5).
We concluded that MnP activity obtained from the crude extract from B. pumilus (31.66 U/L) was almost three times above that found for Paenibacillus sp. (12.33 U/L).
Recent researches indicate multiples biotechnological applications for fungi MnP. A few mentions on bacterial MnP are found in the literature. A summary on MnP fungi production comparing with the results obtained in the present study is shown in Table 2. The data show that some fungi spend a longer time (around 8 days) for enzymes production than observed for the bacteria studied in the present work. Therefore, this suggests that may be possible to reach the same activities if we cultured the bacteria during the same period.
Table 2.
MnP activity found in the literature from different microorganisms.
| Microorganisms | Culture and enzyme production conditions | Activity (U/L) | References |
|---|---|---|---|
| Dichomitus squalens | Malt extract broth, pH 5.5 at 25 °C, after 8 days | 290 | |
| Irpex flavus | Mineral salts broth (MSB) with rice straw, pH 5.5 at 25 °C, after 8 days | 340 | 7 |
| Polyporus sanguineus | Malt extract broth, pH 5.5 at 25 °C, after 8 days | 430 | |
| Pleurotus ostreatus | Yeast extract medium, peptone/glucose, pH 7.5 at 28 °C, after 8 days | 740 | 15 |
| Phlebia floridensis | N-limited MSB broth, pH 4.5 at 25 °C, after 4 days | 60 | 1 |
| Ganoderma sp. | Wheat bran, yeast extract, glucose and ammonium chloride, at 30 °C, after 7 days | 7.8 | 26 |
| Trametes versicolor | Glucose, pH 6.0 at 25 °C, after 8 days | 44 | 21 |
| Bacillus pumilus | Xylan medium with indulin AT, pH 9.0 at 45 °C, after 8h | 31.66 | |
| Bacillus pumilus | Purified MnP, pH 8.0 at 25 °C | 4.3 | |
| Paenibacillus sp. | Xylan medium, pH 9.0 at 45 °C, after 20 h | 12.33 | PRESENT STUDY |
| Paenibacillus sp. | Purified MnP, pH 9.0 at 35 °C | 11.74 |
In this study, we chose the alkaline effluent from paper industry to cultivate and isolate microorganisms which could grow and secretes useful alkaline lignin-degrading enzymes. Thus, we have purified two alkaline MnPs, respectively from B. pumilus and Paenibacillus sp., and we believe that this is the first report on the bacterial ligninolytic enzymes. Subsequent tests carried out aimed the color removal from paper mill effluent showed a decrease in the compounds responsible for the colour and confirmed that the compounds present in the paper effluent were depolymerized during the treatment (23), indicating an important application area for these enzymes.
Acknowledgments
The first author is thankful to CNPq scholarship.
REFERENCES
- 1.Arora D.S., Gill P.K. Production of ligninolytic enzymes by Phlebia floridensis. W. J. Microbiol. Biotechnol. 2005;21:1021–1028. [Google Scholar]
- 2.Beaudette L.E., Davies S., Fedorak P.M., Ward O.P., Pickard M.A. Comparison of biodegradation and mineralization as methods for measuring loss of selected polycholorinated biphenyl congeners in cultures of four white-rot fungi. Appl. Environ. Microbiol. 1998;64:2020–2025. doi: 10.1128/aem.64.6.2020-2025.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M.M. A rapid and sensitive method for quantification of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Champagne P.P., Ramsay J.A. Contribution of manganese peroxidase and laccase to dye decoloration by Trametes versicolor. Appl. Microbiol. Biotechnol. 2005;69:276–285. doi: 10.1007/s00253-005-1964-8. [DOI] [PubMed] [Google Scholar]
- 5.Cheng X., Jia R., Li P., Tu S., Zhu Q., Tang W., Li X. Purification of a new manganese peroxidase of the white-rot fungus Schizophyllum sp. F17, and decolorization of azo dyes by the enzyme. Enz. Microb. Technol. 2007;41(3):258–264. [Google Scholar]
- 6.Duarte M.C.T., Portugal E.P., Ponezi A.N., Bim M.A., Tagliari C.V., Franco T.T. Production and purification of alkaline xylanases. Bioresour. Technol. 1997;68:49–53. [Google Scholar]
- 7.Gill P.K., Arora D.S. Effect of culture conditions on manganese peroxidase production and activity by some white-rot fungi. J. Indust. Microbiol. Biotechnol. 2003;30(1):28–33. doi: 10.1007/s10295-002-0002-4. [DOI] [PubMed] [Google Scholar]
- 8.Gold M.H., Youngs H.L., Sollewijn Gelpke M.D. Manganese peroxidase. In: Sigel A., Sigel H., editors. Metal ions in biological systems. New York, USA: Marcel Dekker; 2000. pp. 559–586. [PubMed] [Google Scholar]
- 9.Hakala T.K., Lundell T., Galkin S., Maijala P., Kalkkinen N., Hatakka A. Manganese peroxidases, laccases and oxalic acid from the selective white-rot fungus Physisporinus rivulosus grown on spruce wood chips. Enz. Microb. Technol. 2005;36:461–468. [Google Scholar]
- 10.Hatakka A. Biodegradation of lignin. In: Hofrichter M., Steinbuchel A., editors. Biopolymers. Vol. 1. Germany: Wiley VCH, Weinheim; 2001. pp. 129–180. Lignin, humic substances and coal. [Google Scholar]
- 11.Hatakka A. Lignin modifying enzymes from selected white-rot fungi: production and role in lignin degradation. FEMS Microbial Rev. 1994;13:125–135. [Google Scholar]
- 12.Heinfling A., Martinez M.J., Martinez A.T., Bergbauer M., Szewzyk U. Purification and characterization of peroxidases from the dye-decolorizing fungus Bjerkandera adusta. FEMS Microbiol Lett. 1998;165:43–50. doi: 10.1111/j.1574-6968.1998.tb13125.x. [DOI] [PubMed] [Google Scholar]
- 13.Hilden L., Johansson G., Pettersson G., Li J., Ljungquist P., Henrikson G. Do the extracellular enzymes cellobiose dehydrogenase and manganese peroxidase form a pathway in lignin biodegradation? FEBS Lett. 2000;477:79–83. doi: 10.1016/s0014-5793(00)01757-9. [DOI] [PubMed] [Google Scholar]
- 14.Hoshino F., Kajino T., Sugiyama H., Asami O., Takahashi H. Thermally stable and hydrogen peroxide tolerant manganese peroxidase (MnP) from Lenzites betulinus. FEBS Letters. 2002;530:249–252. doi: 10.1016/s0014-5793(02)03454-3. [DOI] [PubMed] [Google Scholar]
- 15.Kamitsuji H., Honda Y., Watanabe T., Kuwahara M. Production and induction of manganese peroxidase isozymas in a white-rot fungus Pleurotus ostreatus. Appl. Microbiol. Biotechnol. 2004;65:287–294. doi: 10.1007/s00253-003-1543-9. [DOI] [PubMed] [Google Scholar]
- 16.Kanayama N., Suzuki T., Kawai K. Purification and characterization of an alkaline manganese peroxidase from Aspergillus terreus LD-1. J. of Biosci. Bioeng. 2002;93(4):405–410. [PubMed] [Google Scholar]
- 17.Kuwahara M., Glenn J.K., Morgan M.A., Gold M.H. Separation and characterization of two extracellular H2O2 dependent oxidases from lignolytic cultures of Phaerochaete chrysosporium. FEBS Letter. 1984;169:247–250. [Google Scholar]
- 18.Laemmli U.K. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Li D., Alic M., Gold M.H. Nitrogen regulation of lignin peroxidase gene transcription. Appl. Environ. Microbiol. 1994;60:3447–3449. doi: 10.1128/aem.60.9.3447-3449.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mandels N., Stenberg D. Recent advances in cellulase technology. J. Fermentation Technol. 1976;54:267–286. [Google Scholar]
- 21.Mikiashvili N., Elisashbili V., Wasser S., Nevo E. Carbon and nitrogen sources influence the ligninolytic enzyme activity of Trametes versicolor. Biotech. Lett. 2005;27:955–959. doi: 10.1007/s10529-005-7662-x. [DOI] [PubMed] [Google Scholar]
- 22.Oliveira P.L. Purificação e caracterização bioquímica de manganês peroxidase de Bacillus pumilus e Paenibacillus sp. e sua atuação na remoção da cor do efluente da indústria papeleira. São Paulo, Brasil: (M.Sc. Dissertation. Faculdade de Engenharia de Alimentos. UNICAMP); 2008. p. 69. [Google Scholar]
- 23.Oliveira P.L., Duarte M.C.T., Ponezi A.N., Durrant L.R. Use of Bacillus pumilus CBMAI 0008 and Paenibacillus sp. 868 for colour removal from paper mill effluent. Brazil. J. Microbiol. 2009;40:354–357. doi: 10.1590/S1517-838220090002000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickard M.A., Roman R., Tinoco R., Vazquez Duhalt R. Polycyclic aromatic hydrocarbon metabolism by white-rot fungi and oxidation by Coriolopsis gallica UAMH 8260 laccase. Appl. Environ. Microbiol. 1999;65:3805–3809. doi: 10.1128/aem.65.9.3805-3809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy C.A., D’Souza T.M. Physiology and molecular biology of the lignin peroxidases of Phanerochaete chrysosporium. FEMS Microbial Rev. 1994;13:137–152. doi: 10.1111/j.1574-6976.1994.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 26.Silva C.M.M.S., Melo I.S., Oliveira P.R. Ligninolytic enzyme production by Ganoderma spp. Enz. Microbial Technol. 2005;37:324–329. [Google Scholar]
- 27.Wariishi H., Valli K., Gold M.H. Manganese (II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. J. Biol. Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 28.Ziegenhagen D., Hofrichter M. Degradation of humic acids by manganese peroxidase from the white-rot fungus Clitocybula dusenii. J. Basic Microbiol. 1998;38:289–299. [PubMed] [Google Scholar]


