Abstract
External pH constitutes one of the most important environmental factors that control growth, metabolism and differentiation in microorganisms, including fungi. We have analyzed the effect of external pH on sterigmatocystin biosynthesis in Aspergillus nidulans. It was observed in repeated experiments that alkaline pH, in opposition to acid pH, increased sterigmatocystin production and the transcript levels of aflR, the master gene that regulates expression of the sterigmatocystin cluster in A. nidulans. It is known that pH effects in fungi operate mostly through the Pal/Pac signaling pathway, originally described in Aspergillus nidulans. Accordingly, we studied the role of this signaling pathway in ST biosynthesis. It was observed that aflR transcript levels were increased in the “alkalinity mimicking” mutant pacCc14 and were minimal in the “acidity mimicking” mutant palA1. No sterigmatocystin was produced by palA1 or pacC- mutants at neither acid or alkaline pH of incubation. Finally, fluG and flbA, genes known to regulate both conidiation and sterigmatocystin synthesis upstream in the regulatory cascade, were up-regulated at alkaline pH.
Keywords: Aspergillus nidulans, pH, sterigmatocystin, pacC
INTRODUCTION
Mycotoxins are a group of toxic secondary metabolites produced by fungi, mainly from the genera Aspergillus, Penicillium and Fusarium (22). They are frequently found contaminating a wide variety of food and feed, representing a serious threat to human and animal health and causing severe economic losses (8). Aflatoxins (AF) are among the most studied mycotoxins. Aflatoxins are produced mainly by Aspergillus flavus and A. parasiticus. Aflatoxin B1 is not only toxic but also teratogenic and mutagenic; being the most potent carcinogenic compound found in nature (15).
Studies using the model organism A. nidulans, which produces the AF precursor sterigmatocystin (ST), have led to a better understanding of the biosynthetic steps involved in AF production (4). The fact that the mycotoxin gene clusters from A. nidulans and A. parasiticus have high homology (25) has also been helpful in the study of AF/ST synthesis and regulation.
Among the most important physiological determinants that regulate AF/ST production in Aspergilli are the carbon and nitrogen sources, and the pH of the medium. Simple carbohydrates, such as glucose and sucrose, favor AF biosynthesis in A. parasiticus and ST biosynthesis in A. nidulans (19). It is also known that while ammonium favors AF biosynthesis in A. parasiticus, it inhibits ST production in A. nidulans (13). However, studies on the effect of pH on mycotoxin synthesis have often produced complex and at times contradictory results; e.g. data in the literature have reported either a positive (9, 11, 18) or no effect (5) of acid pH in aflatoxin biosynthesis by A. flavus and/or A. parasiticus, but no information exists on the mechanism involved (for a discussion see (7)). It has also been reported that ST biosynthesis was stimulated by acid pH in A. nidulans (18).
Regulation of fungal metabolism by pH is mostly effected through the Pal/Pac signaling pathway, described for the first time in A. nidulans (6). PacC is a transcription factor activated at alkaline pH by the products of six pal genes that sense external pH and transduces the activating signal. PacC activation may lead to repression of genes expressed preferentially under acidic growth conditions, and up-regulation of genes transcribed preferentially under alkaline growth conditions, including pacC itself (23). Loss of function mutations in any of the pal genes or pacC, lead to an “acidity mimicking” phenotype; i.e., display a pattern of gene expression similar to that of the wild-type strains grown under acidic conditions, independently of ambient pH. In contrast, pacC constitutive mutants show an “alkalinity mimicking” phenotype, displaying a gene expression pattern similar to that of the wild-type strains grown under alkaline conditions, irrespective of ambient pH (see (20, 21) for helpful reviews). A pacC null mutant from A. nidulans displays an “acidity mimicking” phenotype, and exhibits poor growth and almost null conidiation, leading to the hypothesis that PacC might also be involved in development (23). Interestingly, Guzman-de-Peña and Ruiz-Herrera (12) found a correlation between AF biosynthesis and sporulation in A. parasiticus; and between both sexual and asexual sporulation and ST production in A. nidulans (13), thus linking mycotoxin synthesis and differentiation in Aspergilli. Further on, several reports that proposed a model G-protein/cAMP/PKA signaling pathway that connected growth, conidiation and ST production in A. nidulans have been published (3, 14).
Taking into consideration all these data, we proceeded to analyze the regulation of ST production by ambient pH in A. nidulans, and whether the Pal/Pac signaling pathway was or not involved in this process.
MATERIALS AND METHODS
Strains and Growth Media and Conditions
The strains used for these studies were Aspergillus nidulans FGSC26 (biA1, veA1), A. nidulans MAD002 (biA1, veA1), A. nidulans MAD134 (biA1, vea1, palA1, wA3), A. nidulans MAD135 [biA1,veA1, pacCc14 (5-492)] and MAD812 (pantoB100, veA1, pacC+/-209). A. nidulans FGSC26 and A. nidulans MAD002 are both good ST producers. A. nidulans MAD134 is a mutant strain unable to process the inactive form of PacC, and therefore mimicks growth in acidic conditions regardless of the pH of the medium (“acidity mimicking” phenotype). On the other hand, the A. nidulans mutant strain MAD135 produces only a pH-independent activated form of PacC, mimicking growth in alkaline conditions, also regardless of the pH of the medium (“alkalinity mimicking” phenotype); and strain MAD812 has a stringent loss of function mutation. All MAD strains were kindly donated by Miguel A. Peñalva (Centro de Investigaciones Biológicas, Madrid, Spain).
All strains were kept at 4 °C as silica stocks. For conidia production, A. nidulans strains FGSC26, MAD002, MAD135, and MAD812 aliquots from the stocks were transferred to solid Kafer medium (16) with sodium nitrate as the sole nitrogen source and supplemented with 5% sucrose as carbon source, and grown for 5 days at 37 °C. A. nidulans MAD134 was grown at 28 °C on solid Kafer medium with ammonium tartrate and 5% sucrose (acidic growth conditions) in order to avoid extragenic suppressors of the palA mutation (M. A. Peñalva, personal communication).
Spore suspensions
Conidia were harvested by adding 10 ml of a Triton X-100 solution (0.01%) to each Petri dish, counted with a hemocytometer and adjusted to the desired inoculum concentration with sterile distilled water. Before inoculation conidia suspensions were kept overnight at 4 °C to ensure an even germination.
Growth conditions
Conidia were inoculated into 100 x 15 mm Petri dishes containing 50 ml of liquid (1x107) or solid (1x106) Kafer medium (see Results) supplemented with 5% sucrose and 70 mM sodium nitrate as carbon and nitrogen sources respectively, and incubated under static conditions at 37 °C for variable periods of time. In some experiments, pH of the media was adjusted with 100 mM citrate buffer.
At the end of the incubation period, in liquid cultures the medium was separated from the mycelium by filtration. Mycelia were transferred to a Petri dish, washed with 10 ml of distilled water containing 0.01% Triton X-100, and gently shaken to release the spores. This procedure was repeated twice. Mycelia were then dried at 75 °C for 24 h, stored at room temperature in desiccators with silica gel as drying agent for another 24 h, and weighed. The residual growth medium was processed immediately or stored at 4 °C until used. When mycelia were used for RNA extraction, they were collected by vacuum filtration, frozen with liquid nitrogen and stored at -70 °C until used. When solid medium was used, after spore recovery ST was extracted as described below, agar was melted in a microwave oven, mycelia were recovered with the use of tweezers, washed with hot water, dried and weighed as above.
ST quantitation
ST was extracted from the mixture of culture medium, dry mycelium (see above) and conidia, with 50 ml acetone for 30 min, followed by 50 ml chloroform by further 30 min (17). The organic phase was separated, filtered through anhydrous sodium sulfate and evaporated in a fume hood in a water boiling bath. The residue was resuspended in 500 µL HPLC grade methanol and filtered through C-18 columns (Alltech).
Analysis and quantitation were performed by HPLC as previously described (13). The detection limit was 3 ng ST in 20 µl samples.
RT-PCR analysis of the expression of genes encoding proteins involved in growth and ST synthesis
Total RNA extraction was performed using TriZOL (Invitrogen). RNA purification was achieved using RNeasy mini Kit (Qiagen). RNA integrity was confirmed by electrophoresis in a 1.5% agarose gel in TAE 1X. The expression levels of the genes fluG, flbA, laeA, pacC and aflR, were evaluated semi quantitatively by RT-PCR. First-strand cDNA was obtained using 1 µg of total RNA, adding 1 µl Oligo dT (Invitrogen), 1 µl dNTP mix (10 µM each) and enough RNase-free water to adjust to 12 µl, heating the mix at 65 °C for 15 min and transferring it immediately to ice. 4 µl of 5x RT Buffer, 2 µl of DTT 0.1 M and 1 µl RNase OUT (Invitrogen) were added to each reaction, gently mixed and incubated at 42 °C for 2 min. After that, 1 µl Superscript II Reverse Transcriptase (Invitrogen) was added to each reaction for a final volume of 20 µl. RT conditions were: an initial incubation at 42 °C for 50 min, followed by 15 min at 70 °C. The cDNA derived from 0.1 µg of total RNA was used as a template. PCR conditions were as follows: After an initial incubation at 94 °C for 3 min, 28 cycles of 94 °C for 45 s, 54 °C for 1 min and 72 °C for 1 min were performed, followed by a final incubation at 72 °C for 7 min.
The PCR primers for each gene were as follows: 5’-ACCCTAATGTTTATTTGGAT-3’ (forward) and 5’-TGGATAGGTCTGGTATAAGG-3’ (reverse) for fluG; 5’-, TCCCTCAAATTCTCTCAATCGAACCGG-3’ (forward) and 5’-GTAGAATGACAGGTTTTCTTCGCAGA-3’ (reverse) for flbA; 5’-GGTGACGATTTGTATAGTCC-3’ (forward) and 5’-CTCTTCATGAAACTGGTTTC-3’ (reverse) for laeA; 5’-GACTGACGGTATGACTTCTG-3’ (forward) and 5’-GTTGGCAATGTAGTTACGTA-3’ (reverse) for pacC; 5’-GCCATCCTGTCTCCGAATAC-3’ (forward) and 5’-CGAACCTCTACGACTGTCTTG-3’ (reverse) for aflR; and 5’-CCAAGGCCAACCGCGAGAAGATGAC-3 (forward) and 5’-AGGGTACATGGTGGTGCCGCCAGAC-3’ (reverse) for γ-actin (control gene). Transcription levels were normalized by comparing the UV absorption intensity of the bands to that of γ-actin using ImageJ software (http://rsb.info.nih.gov/ij/).
Statistical analyses
Each experiment was repeated three to five times, and values presented in this paper are means of these replicates. Differences between means were evaluated by Tukey’s Test (p=0.05) using SAS (version 6.12; SAS Institute, Cary, NC, USA). Coefficients of determination (r2) and regression data are included in the corresponding results.
RESULTS
Relationship between initial pH, growth and ST production
A. nidulans MAD002 (wild-type strain) was grown in Kafer medium at three different initial pH values for 72 h. Growth, conidiation and ST synthesis were evaluated approximately every 12 h. When initial pH was 4.61, it was observed that only basal levels of ST were produced until 58 hours of incubation, ST synthesis being activated after this time (Table 1). Activation of ST synthesis occurred earlier when the initial pH value was raised (58 h at initial pH= 5.22, and 46 h at pH= 6.90) (Table 1). As it may be seen in this table, time of activation of ST synthesis occurred when pH reached a value above 7.5. The amount of ST produced and mycelial growth at 72 h of incubation were also linearly increased as initial pH value of the medium increased (r2 = 0.977 and r2 = 0.993, respectively).
Table 1.
Rate of growth and ST synthesis in A. nidulans MAD002 (wild-type).
| Initial pH | Incubation Time (h) | Final pH | Growth (mg DW) | Total Sterigmatocystin (ng) |
|---|---|---|---|---|
| 4.61 | 22 | 5.4 | 45.9 A | 0 |
| 34 | 6.2 | 62.8 A | 35 A | |
| 46 | 6.8 | 104.8 A | 38 A | |
| 58 | 7.2 | 211.2 | 43 A | |
| 72 | 7.7 | 483.9 | 1,972 | |
| 5.22 | 11 | 5.4 | 13.9 A | 65 A |
| 23 | 5.9 | 26.5 A | 83 A | |
| 34 | 6.3 | 49.4 A | 38 A | |
| 46 | 6.9 | 93.1 A | 201 | |
| 58 | 8.3 | 190.7 | 2,247 | |
| 72 | 8.4 | 525.3 | 12,779 | |
| 6.90 | 10 | 6.9 | 23.2 A | 275 B |
| 22 | 7.0 | 38.9 A | 26 B | |
| 34 | 7.4 | 80.3 A | 49 B | |
| 46 | 7.6 | 232.5 | 5,896 A | |
| 58 | 8.8 | 364.2 | 3,699 A | |
| 72 | 9.0 | 605.0 | 28,727 |
Cultures were obtained in unbuffered liquid Kafer medium incubated under static conditions, and growth and ST formation were measured as described in Materials and methods. Statistical analysis of samples (n=5) proceeded by the Tukey test. Values in the same experimental block (same initial pH) marked with the same letter in the same column are statistically similar (p= 0.05).
Transcription of regulatory genes through time of incubation
The previous experiment suggested a correlation between pH and ST production. Taking into consideration that both phenomena are regulated in Aspergilli by known pathways (6, 25), we decided to investigate the correlation existing between the drift in external pH during incubation, and the expression of the corresponding regulatory genes. A. nidulans MAD002 was incubated in Kafer medium pH = 6.60 for 83 h. Mycelia were collected at different intervals, and levels of transcripts of the regulatory gene aflR was evaluated (Fig. 1). aflR expression levels increased through time linearly (r2 = 0.922) as pH of the medium was increased, raising the hypothesis that alkaline pH promotes ST biosynthesis.
Figure 1.
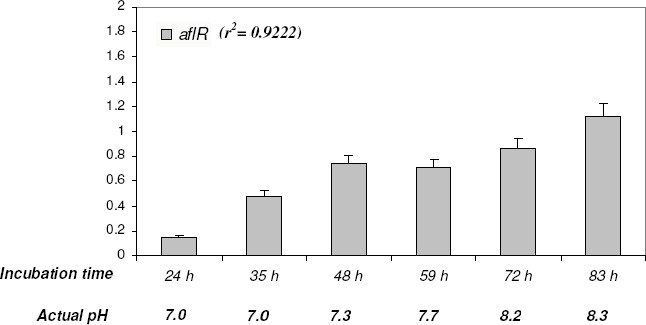
Effect of pH on the transcription of the regulatory gene aflR in the wild-type strain MAD002 through incubation time. Cultures were incubated in large Petri dishes containing liquid pH= 6.5 unbuffered Kafer medium under static conditions at 37 ºC. At intervals, sets of dishes were recovered, RNA was extracted and transcription levels were measured by semiquantitative RT-PCR. γ-actin was used as an endogenous reference. The amount of each mRNA was normalized to the amount of γ-actin mRNA in each sample. Data are means ± SD (n= 3). Time of incubation and actual pH of the different samples are indicated.
Determination of the optimum pH for growth and ST production in the wild-type and mutant strains affected in the Pal/Pac pathway
A. nidulans mutants affected in the Pal/Pac pathway were included in these experiments because of the general observation on the role of this pathway in the regulation of different phenomena by pH in Aspergilli. All strains were grown in Kafer (liquid or solid) medium containing 100 mM citrate buffer at different pH values. It was observed that mycelial growth of wild-type strains (FGSC26 and MAD002) was higher at alkaline pH. ST levels were about 30-70-fold higher at alkaline pH as compared to acidic or neutral conditions in both strains. In contrast to these results, the A. nidulans “acidity mimicking” mutant MAD134 (palA1) grew poorly at these three different pH values compared to the wild-type strains grown at similar pH values and time. Interestingly, this mutant was unable to produce ST at any of the pH values tested. On the other hand, the MAD135 (pacCc14) mutant produced ST regardless of the pH of the medium. Nevertheless, as previously reported (18), its levels of ST production were well below those from the wild-type strains. These results are shown in Table 2. Finally, contrary to the behavior of strain MAD135, a pacC negative mutant (MAD812) was unable to synthesize ST at neither acid nor alkaline pH (Table 3).
Table 2.
Growth and ST synthesis in A. nidulansstrains grown in buffered medium.
| Strain | Initial pH | Final pH | Growtha (mg mycelium) | Total Sterigmatocystin (ng) |
|---|---|---|---|---|
| FGSC26 (wt) | 3.0 | 3.2 | 175 A | 7,683 A |
| 6.5 | 6.4 | 186 A | 7,477 A | |
| 8.0 | 8.7 | 406 | 508,799 | |
| MAD002 (wt) | 3.3 | 4.1 | 515 A | 4,249 A |
| 6.2 | 7.4 | 356 | 2,158 A | |
| 8.0 | 8.6 | 562 A | 141,097 | |
| MAD134 (palA1) | 3.3 | 3.5 | 70.4 | 0 |
| 6.1 | 6.1 | 57.3 A | 0 | |
| 7.9 | 7.6 | 35.6 A | 0 | |
| MAD135 | 3.3 | 3.5 | 130.5 A | 463 A |
| (pacCc14) | 6.1 | 6.2 | 76.6 A | 229 A |
| 7.9 | 8.2 | 298.4 | 325 A |
Cultures were obtained in buffered (100 mM citrate) liquid Kafer medium incubated under static conditions, and growth and ST formation were measured as described in Materials and methods. Statistical analysis of samples (n=5) proceeded by the Tukey test (p=0.05). Statistical significance of the differences in the same experimental block (same strain and medium) were treated as described for Table 1.
Table 3.
Growth and ST synthesis of A. nidulans MAD002 and MAD812 strains grown in solid buffered medium.
| Strain | Initial pH | Final pH | Growth (mg1) | Total ST |
|---|---|---|---|---|
| MAD002 (wt) | 3.1 | 4.4 | 726.9 A | 29,076 |
| 7.8 | 8.9 | 747.0 A | 2’530,700 | |
| MAD812 (pacC+/209) | 3.1 | 4.0 | 707.3 | 0 |
| 7.8 | 8.3 | 260.6 | 0 |
Cultures were obtained in buffered (100 mM citrate) solid Kafer medium, and growth and ST formation were measured as described in Materials and methods. Statistical analysis of samples (n=5) proceeded by the Tukey test (p=0.05). Statistical significance of the differences in the same experimental block (same strain and medium) were treated as described for Table 1.
Determination of the expression of regulatory genes in wild-type and mutants affected in the Pal/Pac pathway
In order to analyze the effect of pH on the expression of the regulatory genes (pacC and aflR) in wild-type and Pal/Pac affected mutants, we used buffered medium of different pH values as described above. It was observed that expression levels of pacC and aflR in strain FGSC26 increased linearly as pH of the medium increased (r2 = 0.981 and r2 = 0.997, respectively). Expression levels of pacC in strain MAD134 (palA1) remained low and constant at all pH values. aflR levels were also basal, in agreement with the absence of ST production at any pH value. Transcript levels from strain MAD135 (pacCc14) contrasted with those of strain MAD134 (palA1). Levels of pacC transcript in pacCc14 were twice higher than those from palA1 mutant, and its aflR transcript levels were high regardless of the pH of the medium, correlating with ST production in this strain (these results are shown in Fig. 2).
Figure 2.
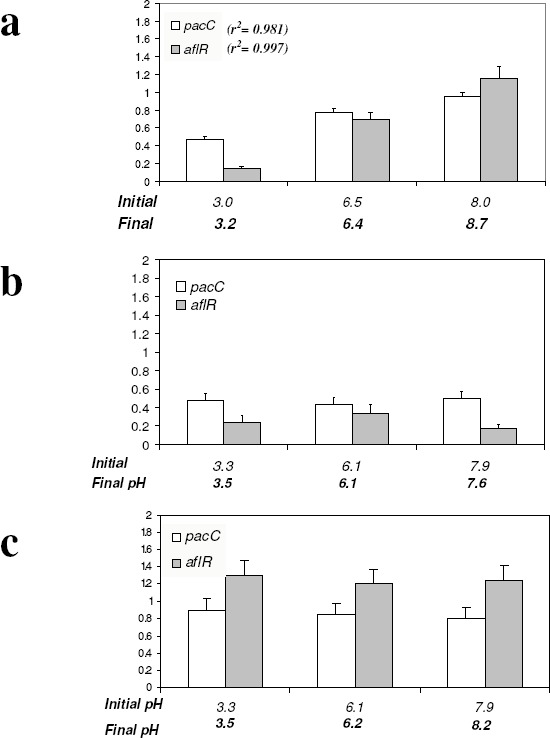
Effect of pH on the transcription of pacC and aflR genes in wild-type and mutant strains of A. nidulans grown at three different pH values. Culture conditions proceeded as described for Fig. 1, but medium was added of 100 mM citrate buffer at the indicated pH values, and incubation lasted for 72 h. Gene transcription levels were analyzed as described for Fig. 1. Data are means ± SD (n= 3). a) wild-type strain FGSC26; b) “acidity mimicking” mutant MAD134 (palA1); c) “alkalinity mimicking” mutant MAD135 (pacCc14). Initial and final pH values of the different samples are indicated.
Effect of pH on the regulatory mechanism of ST synthesis
We further proceeded to determine the probable level at which external pH affects ST production. It has been suggested that fluG and flbA encoded proteins have a positive role in ST production (17). In this model, laeA transcript product also activates ST formation (3). Transcript levels of these genes were evaluated in FGSC26 and MAD134 (palA1) strains grown at three different pH values in citrate buffered medium for 72 h (Fig. 3). It was noted that fluG (r2= 0.998 in wild-type and r2= 0.971 in MAD134 strains) and flbA (r2= 0.889 and r2= 0.976, respectively) were preferentially expressed at alkaline pH in both strains. On the other hand transcript levels of laeA were higher at neutral and alkaline pH in the wild-type strain (Fig. 3), but in the MAD134 (palA1) strain an opposite trend in expression was observed, decreasing with an increase in pH (data not shown).
Figure 3.
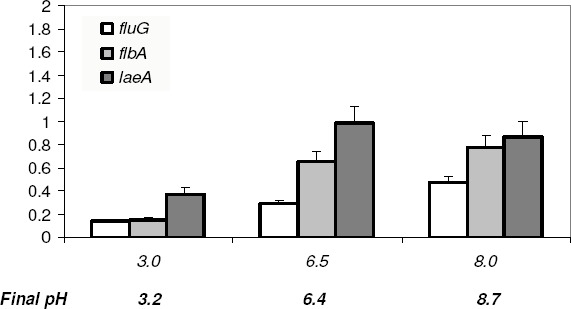
Effect of pH on the transcription of fluG, flbA and laeA genes in the wild-type strain FGSC26 grown at three different pH values. Culture conditions proceeded as described for Fig. 2, and transcription levels were analyzed as described for Fig. 1. Data are means ± SD (n= 3).
DISCUSSION
The formation of natural products resulting from the secondary metabolism of plants and microorganisms is subjected to different environmental cues, such as temperature, humidity, light, pH, nutrient source, etc. Aflatoxins (AF), probably the most important and studied mycotoxins, do not escape to this rule. Accordingly, it has been described that temperature and nitrogen source affect AF production in A. parasiticus and ST synthesis in A. nidulans, but in different ways (11). It was demonstrated that nitrate as the sole nitrogen source promoted ST synthesis in A. nidulans, while ammonium promoted AF production in A. parasiticus. It was also observed that AF production by A. parasiticus was best at 27 °C, while ST was maximally produced by A. nidulans when incubated at 37 °C (11).
pH is also an important factor in the production of aflatoxins. Acid pH has been described to stimulate AF synthesis in A. parasiticus (10, 18). According to the results described in this paper, we may conclude that pH of the medium does have an effect on ST synthesis in A. nidulans, with alkaline pH promoting mycotoxin production. These data are in contradiction with a previous communication reporting stimulation of ST biosynthesis at acid pH (18). The reasons for this discrepancy are difficult to pin point. Possibly, conditions of growth, time of incubation and the determination only of ST present in the mycelium (contrary to our protocols) may be partially responsible for this behavior.
Among our data that support the conclusion that it is alkaline pH the one that promotes ST production, we may cite the following: 1) the repeated observation that higher levels of ST are produced at alkaline pH in the wild-type strains; 2) the observation that expression levels of aflR (the transcriptional regulator of the gene cluster encoding proteins involved in ST synthesis (24)) in the wild-type strain of A. nidulans were high under alkaline conditions, and very low at acidic pH; 3) the observation that an “acidity mimicking” palA mutant did not produce ST at any pH value; 4) the observation that a PacC constitutive “alkalinity mimicking” mutant produced similar levels of ST at all the pH values tested, although as reported (18), at lower levels than the wild-type strain; 5) the fact that a pacC negative mutant did not form ST at any pH value. These last results also meet the criteria for recognizing gene regulation by pH through the Pal/Pac signaling pathway (1); and finally 6) it is also important to point out that the regulation of other genes involved in the common control of ST biosynthesis (laeA, fluG and flbA) are in agreement with a positive regulation by alkaline pH.
It is accepted that LaeA is a global regulator of secondary metabolism in A. nidulans (2). Levels of transcription of laeA gene were higher at neutral to alkaline growth conditions in the wild-type strain.
fluG and flbA are known to be involved in a G-protein/cAMP/PKA signaling pathway that connects ST synthesis and development in A. nidulans (3, 14). The observation that transcription of both genes was stimulated by alkaline pH of the medium agrees with the general observation that sterigmatocystin production is stimulated at alkaline pH.
In summary, based on our results, we may suggest that alkaline pH regulates the signaling pathway that controls ST synthesis in A. nidulans at the level of aflR by the Pal/Pac signaling pathway.
ACKNOWLEDGEMENT
We wish to thank Prof. Miguel A. Peñalva for providing us all of the A. nidulans MAD strains, and to Prof. José Ruiz Herrera for his helpful comments and suggestions to this manuscript. We acknowledge technical support to Gloria Laura Anguiano-Ruvalcaba and Yolanda Rodriguez-Aza. FDV Ph.D fellowship (181787), was supported by CONACYT, México
REFERENCES
- 1.Arst H.N. Jr, Peñalva M.A. Recognizing gene regulation by ambient pH. Fungal Genet. Biol. 2003;40:1–3. doi: 10.1016/s1087-1845(03)00077-x. [DOI] [PubMed] [Google Scholar]
- 2.Bok J.W., Keller N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryotic cell. 2004;3:527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodhagen M., Keller N.P. Signaling pathways connecting mycotoxin production and sporulation. Mol. Plant Pathol. 2006;7:285–301. doi: 10.1111/j.1364-3703.2006.00338.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown D.W., Yu J.H., Kelkar H.S., Fernandes M., Nesbitt T.C., Keller N.P., Adams T.H., Leonard T.J. Proc. Natl. Acad. Sci. Vol. 93. USA: 1996. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans; pp. 1418–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan R.L., Ayres J.C. Effect of initial pH on aflatoxin production. Appl Microbiol. 1975;30:1050–1051. doi: 10.1128/am.30.6.1050-1051.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caddick M.X., Brownlee A.G., Arst H.N.Jr. Regulation of gene expression by pH of the growth medium in Aspergillus nidulans. Mol. Gen. Genet. 1986;203:346–353. doi: 10.1007/BF00333978. [DOI] [PubMed] [Google Scholar]
- 7.Calvo A.M., Wilson R.A., Bok J.W., Keller N.P. Relationship between secondary metabolism and fungal development. Microbiol. Mol. Biol. Rev. 2002;66:447–459. doi: 10.1128/MMBR.66.3.447-459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardwell K.F., Desjardins A., Henry H.S., Munkvold G., Robens J. Mycotoxins: The cost of achieving food security and food quality. APSnet.org. 2001. Available at: http://apsnet.org/online/feature/mycotoxin/top.html. Accessed 13 Feb 2009.
- 9.Cotty P.J. Aflatoxin and sclerotial production by Aspergillus flavus: Influence of pH. Phytopathology. 1988;78:1250–1253. [Google Scholar]
- 10.Detroy R.W., Hesseltine C.W. Net synthesis of 14C-labeled lipids and aflatoxins in resting cells of Aspergillus parasiticus. Dev. Ind. Microbiol. 1969;10:127–133. [Google Scholar]
- 11.Feng G.H., Leonard T.H. Culture conditions control expression of the genes for aflatoxin and sterigmatocystin biosynthesis in Aspergillus parasiticus and A. nidulans. Appl. Environ. Microbiol. 1998;64:2275–2277. doi: 10.1128/aem.64.6.2275-2277.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.uzman-de-Peña D., Ruiz-Herrera J. Relationship between aflatoxin biosynthesis and sporulation in Aspergillus parasiticus. Fungal Gen. Biol. 1997;21:198–205. doi: 10.1006/fgbi.1996.0945. [DOI] [PubMed] [Google Scholar]
- 13.Guzman-de-Peña D., Aguirre J., Ruiz-Herrrera J. Vol. 73. Antonie Van Leeuwenhock.; 1998. Corrrelation between the regulation of sterigmatocystin biosynthesis and asexual and sexual sporulation in Emericella nidulans. pp. 199–205. [DOI] [PubMed] [Google Scholar]
- 14.Hicks J.K., Yu J.H., Keller N.P., Adams T.H. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Ga protein-dependent signaling pathway. EMBO J. 1997;16:4916–4923. doi: 10.1093/emboj/16.16.4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.International Agency for Research on Cancer (IARC). Aflatoxins (Naturally occurring mixtures). Monogr. Eval. Carcinog. Risks. Hum. 2002;82 Available at http://monographs.iarc.fr. Accessed 13 Feb 2009. [Google Scholar]
- 16.Kafer E. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 1977;19:131–133. doi: 10.1016/s0065-2660(08)60245-x. [DOI] [PubMed] [Google Scholar]
- 17.Keller N.P., Kantz N.J., Adams T.H. Aspergillus nidulans veA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 1994;60:1444–1450. doi: 10.1128/aem.60.5.1444-1450.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller N.P., Nesbitt C., Sarr B., Phillips T.D., Burow G.B. pH regulation of Sterigmatocystin and Aflatoxin Biosynthesis in Aspergillus spp. Phytopathology. 1997;87:643–648. doi: 10.1094/PHYTO.1997.87.6.643. [DOI] [PubMed] [Google Scholar]
- 19.Payne G.A., Brown M.P. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 1998;36:329–362. doi: 10.1146/annurev.phyto.36.1.329. [DOI] [PubMed] [Google Scholar]
- 20.Peñalva M.A., Arst H.N.Jr. Regulation of gene expresión by ambient pH in filamentous fungi and yeasts. Microbiol. Mol. Biol. Rev. 2002;66:426–446. doi: 10.1128/MMBR.66.3.426-446.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peñalva M.A., Arst H.N.Jr. Recent advances in the characterization of ambient pH regulation of gene expression in filamentous fungi and yeasts. Ann. Rev. Microbiol. 2004;58:425–451. doi: 10.1146/annurev.micro.58.030603.123715. [DOI] [PubMed] [Google Scholar]
- 22.Task Force Report. Mycotoxins: Risks in plant, animal, and human systems. Iowa, USA.: the Council for Agricultural Science and Technology. CAST, Ames; 2003. Fungal growth and mycotoxin development by major mycotoxigenic fungi. pp. 129–135. [Google Scholar]
- 23.Tilburn J., Sarkar S., Widdick D.A., Espeso E.A., Orejas M., Mungroso J., Peñalva M.A., Arst H.N.Jr. The Aspergillus PacC zinc finger transcription factor mediates regulation of both acid- and alkaline-expressed genes by ambient pH. EMBO J. 1995;14:779–790. doi: 10.1002/j.1460-2075.1995.tb07056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woloshuk C.P., Foutz K.R., Brewer J.F., Bathnagar D., Cleveland T.E., Payne G.A. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 1994;60:2408–2414. doi: 10.1128/aem.60.7.2408-2414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu J., Chang P.K., Erlich K.C., Cary J.W., Bhatnagar D., Cleveland T.E., Payne G.A., Linz J.E., Woloshuk C.P., Bennet J.W. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 2004;70:1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


