Abstract
The aim of this study was to characterize rhizobial isolates from Cratylia mollis Mart. ex Benth, Calliandra depauperata Benth. and Mimosa tenuiflora (Willd.) Poir. by means of rhizobial colonies morphology and restriction analysis of the 16S ribosomal gene (16S rDNA-ARDRA). Nodules were collected in the field and from plants cultivated in a greenhouse experiment using Caatinga soil samples. Sixty seven isolates were described by morphological analysis. Forty seven representative isolates were used for ARDRA analysis using seven restriction enzymes. We observed high diversity of both slow and fast-growing rhizobia that formed three morpho-physiological clusters. A few fast-growing isolates formed a group of strains of the Bradyrhizobium type; however, most of them diverged from the B. japonicum and B. elkanii species. Cratylia mollis nodule isolates were the most diverse, while all Mimosa tenuiflora isolates displayed fast growth with no pH change and were clustered into groups bearing 100% similarity, according to ARDRA results.
Keywords: Cratylia mollis, Mimosa tenuiflora, Calliandra depauperata, PCR-ARDRA, diversity
INTRODUCTION
The family Leguminosae consists of eighteen thousand species classified into around 650 genera widely spread around the globe (7). Distribution of these species over extreme environments has been attributed to the development of morpho-physiological mechanisms such as drought adaptation (6). The Caatinga biome is characterized by its shortage of water and by its edaphic and microclimate heterogeneity. All these features determine quite peculiar patterns of species distribution that allow for a large floristic diversity. Out of the 1041 species cataloged in the Caatinga, about 40% belong to the family Leguminosae, which has expressive relevance to the region, ecologically, socially and economically (10). Among these, the species Cratylia mollis Mart. ex Benth., Calliandra depauperata Benth. and Mimosa tenuiflora (Willd.) Poir. are used as forage for livestock, as firewood and as hedge (7, 11).
Most of species of this botanic family are capable of establishing symbiotic relationships with nitrogen-fixing bacteria in the Caatinga, helping to maintain soil fertility in the region (26). However, not much is known about the diversity of rhizobia that induces root nodules on C. mollis, C. depauperata and Mimosa tenuiflora.
In recent years, as molecular tools have been used in the studies about the diversity of rhizobia, new species have been described, and changes in the taxonomy of such group of microorganisms have been suggested, including other bacteria of the Burkholderiales order (4, 5, 15). The objective of this study was to characterize rhizobia indigenous from Caatinga region associated with C. mollis, C. depauperata and M. tenuiflora by applying morphological analysis of the rhizobia cultures and restriction analysis of the amplified ribosomal DNA (16S rDNA-ARDRA).
MATERIALS AND METHODS
Soil samples were collected from a Caatinga area (Petrolina, Pernambuco State, Brazil). The C. mollis and C. depauperata plants were grown in a pot with soil and sand (1:2, w:w), and kept in a greenhouse to the plant growth and development of the root nodules. The M. tenuiflora nodules and a portion of the C. mollis nodules were also field-collected in the same plot.
The nodules were washed with alcohol (70% v/v – 1 mim), to break the surface tension, superficially disinfected with hydrogen peroxide (H2O2 – 5 min) and washed five times with sterile distilled water. The surface disinfected nodules were crushed with claw on Petri dishes containing solid YMA medium with congo red indicator (27). The Petri dishes were incubated at 28°C for seven days with diary observation. The rhizobial isolates were transferred to Petri dishes containing YMA medium with bromothymol blue (27), purified and stored.
To the morphological characterization rhizobial isolates were grown in YMA medium (27), the parameter considered was growth period, pH alteration of culture medium, colony color, edge type, uniformity of the culture in the dish, colony diameter, and mucus consistency and elasticity (29).
Total DNA was extracted using Tris-EDTA-SDS buffer (50 mM Tris, 10 mM EDTA, 1% SDS, pH 8,0). The protein precipitation were carried out using phenol:chloroform:isoamylic alcohol (25:24:1), and chloroform:isoamylic alcohol (24:1). DNA precipitation was carried out using sodium acetate and isopropanol at -20°C. The 16S rDNA ribosomal gene was amplified using universal primers Y1 and Y3 (31). PCR reactions were prepared according to Zilli et al. (33), with initial denaturation at 95°C for 2 min, followed by 35 cycles of denaturation at 93°C for 45 s, annealing at 62°C for 45 s, and elongation at 72°C for 2 min, with a final elongation step of 5 min at 72°C.
The amplification product was digested separately using seven restriction endonuclease (AluI, HaeIII, HhaI, HinfI, MspI, RsaI and TaqI) following the manufacturer’s recommendations. The fragments produced were separated by electrophoresis for 3 hours at 85V in ultrapure agarose gel (3% w/v). In this analysis, six known strains were also used: BR 29 (Bradyrhizobium elkani), BR 111 (Bradyrhizobium japonicum), BR 10026 (Rhizobium etli), BR 10016 (Rhizobium tropici), BR 112 (Sinorhizobium fredii) and BR 5410 (Azorhizobium caulinodans). The strains were obtained from the diazotroph culture collection of Empraba Agrobiologia (www.cnpab.embrapa.br).
Morphological characterization data and ARDRA profiles were converted into an absence/presence binary matrix (0, 1). Simple Matching Coefficient and the UPGMA clustering method, available in the NTSYS program, version 2.1 (18), were used to build dendrograms.
RESULTS AND DISCUSSION
Sixty seven isolates from the three species were obtained and morphologically characterized. Thirty nine out of these were isolated from field-collected nodules and 28 from nodules acquired in a greenhouse. Forty isolates (59.7%) were extracted from the C. mollis nodules, 23 were derived from the field and 17 from greenhouse experiment. Eleven C. depauperata isolates were collected from root nodules grown in a greenhouse, while 16 M. tenuiflora isolates were obtained from nodules collected from the field.
In relation to the growth period, 6 isolates were characterized as slow-growing and 61 isolates as fast-growing (Figure 1). It was observed that all slow growing isolates were obtained from Cratylia mollis. For this specie the proportion between fast and slow growing rhizobia were similar to isolates obtained from field collected nodules and nodules obtained from greenhouse experiment. It suggests that the sampling to obtain rhizobial isolates from Cratylia mollis with different growth rates may be conducted at field or greenhouse conditions. Therefore, rhizobial isolates from Calliandra depauperata and M. tenuiflora did not presented slow growth rate.
Figure 1.
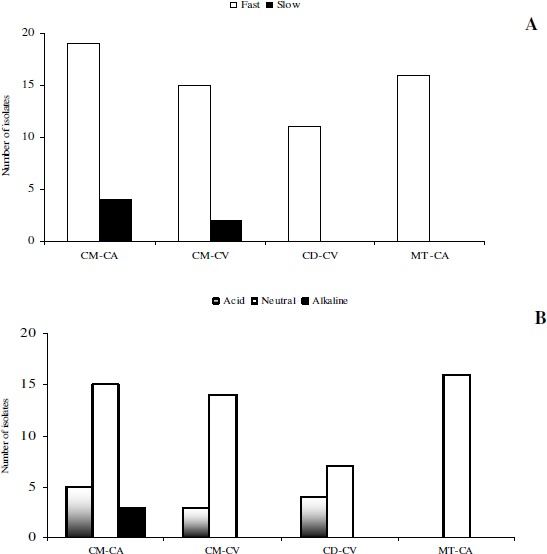
Organization of the isolates according to the species studied and the method of collecting nodules. Classification based on (A) growth period of the colonies and on (B) pH alteration of the culture medium. CM – C. mollis, CD – C. depauperata, MT – M. tenuiflora, CA – field and CV – greenhouse.
Regarding the type of pH change in the culture medium, 12 isolates were able to acidify the medium, 3 alkalized the medium, and 52 did not alter the pH of the medium (Figure 1). The fact that fast-growing rhizobia have been often found in arid regions reflects a survival strategy in adverse conditions based on the reduction of the generational interval, as suggested by Sprent (23). Martins et al. (12) and Santos et al. (20) obtained similar results with cowpea root nodules and peanut root nodules, respectively.
The diversity of this microorganism group may be influenced by the different chemical composition of the root exudates, as well as by the land use system, among other factors (8, 24). Because of the symbiotic relation with legumes, it is suggested that rhizobia diversity is strictly related to the variety of legume species in the location, pointing evolutional adaptations of both symbiotic partners (23).
Regarding other morphological features, there was a small variation among the isolates: most presented white coloring (97%), smooth colony edge (97%) and large quantity of mucus production (70%). Batista et al. (3) noted an increasing tendency of Bradyrhizobium isolates to produce mucus as a reflection of the adaptation to the acid soils of the Brazilian tropical Savannah known as Cerrado.
According to the clustering based on the morphological characteristics of the colonies, we could notice the formation of three main groups (Figure 2). Group A consisted of slow-growing isolates that alkalized the medium, a feature of the Bradyrhizobium genus. Group B consisted of most fast-growing isolates, which divided in several subgroups. One of these subgroups (jp isolates) gathered most of the M. tenuiflora isolates, which displayed neutral and fast growing, viscosity, and 2 mm diameter colonies. Isolates from Group C did not display mucus elasticity, unlike most isolates from preceding groups. Group C presented the only isolates that displayed root like border colonies.
Figure 2.
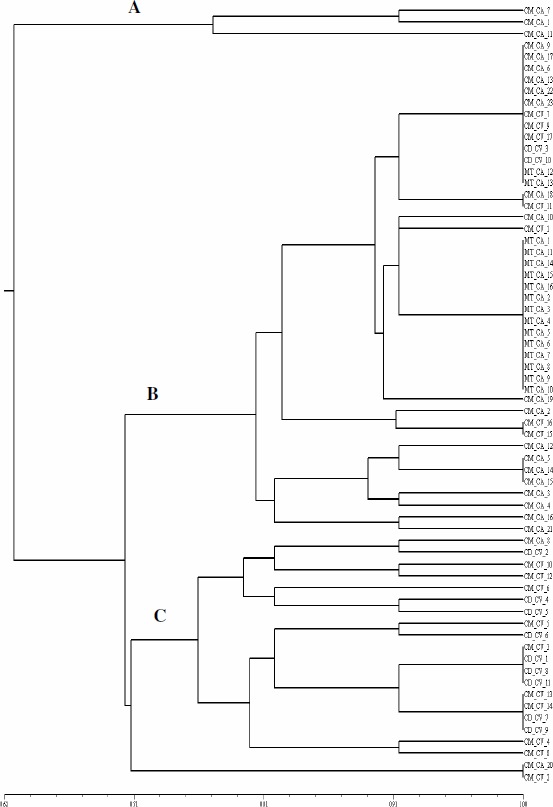
Similarity dendrogram created from the morphophysiological characterization data using the Simple Matching similarity index and the UPGMA clustering method. CM – C. mollis, CD – C. depauperata, MT – M. tenuiflora, CA – field and CV – greenhouse.
Another study detected a high diversity of rhizobia using Arachis hypogaea (20) and Vigna unguiculata L. Walp. (12) as trap host to obtain rhizobial from Caatinga soil. Lammel et al. (9) observed that the morphological features of rhizobia isolated from legumes grown in araucaria region were different from those of standard strains (Rhizobium tropici, Bradyrhizobium elkanii and Burkholderia sp).
Forty seven (70%) morphological clusters representative isolates were selected for the restriction analysis of the 16S rDNA gene. It were observed a variation from 10 (MspI) to 18 (HinfI) bands generated by each restriction enzyme. A total of 95 bands were generated analyzing all endonucleases and isolates studied. The dendrogram in Figure 3 shows three main groups (A, B and C).
Figure 3.
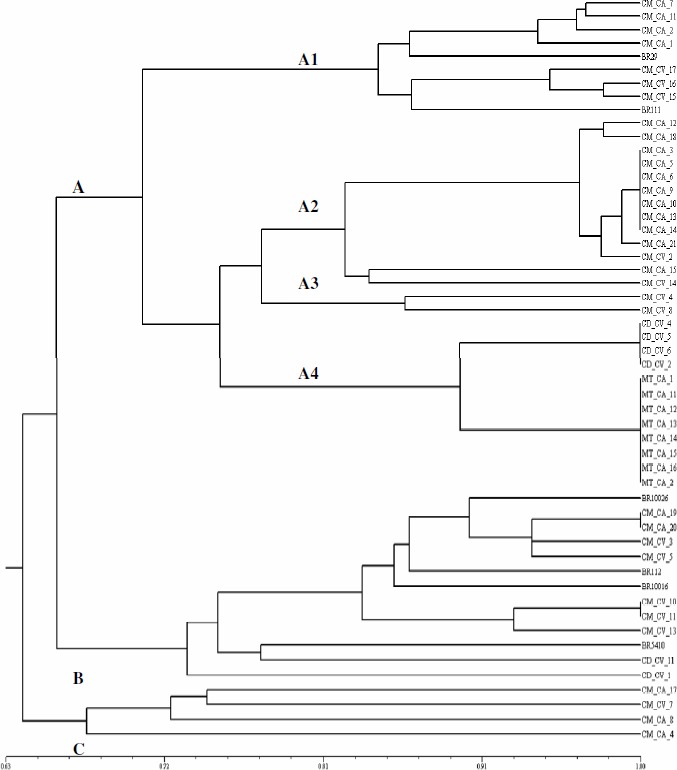
Similarity dendrogram created from the restriction profiles of the 16S rDNA gene of the isolates using the Simple Matching similarity index and the UPGMA clustering method. CM – C. mollis, CD – C. depauperata, MT – M. tenuiflora, CA – field and CV – greenhouse.
In group A, four subgroups with 80% of the similarity were formed (Figure 3). The first (A1) consisted of slow-growing C. mollis isolates having neutral or alkaline pH change. In this clustering there were strains of the Bradyrhizobium genus (BR29 e BR111). The second subgroup (A2) also consisted of C. mollis isolates, being 11 isolates from nodules collected in the field and two from nodules collected in a greenhouse; all isolates were fast-growing and only one was able to acidify the culture medium. Subgroup three (A3), which consisted of two C. mollis isolates collected in a greenhouse, displayed fast-growing isolates with no pH change in the medium. The last subgroup (A4) had two clusters with isolates that presented 100% similarity: the first cluster presented four C. depauperata isolates that acidified the medium and did not display mucus elasticity; and the second had eight M. tenuiflora isolates that did not change the pH of the medium culture, and displayed mucus elasticity and viscosity.
M. tenuiflora isolates displayed a high level of similarity according to the phenotypic analysis (Figure 2), and clustered in identical groups according to the genotypic analysis (Figure 3), indicating a possible model for the evolutional process of the legume-rhizobium. Recent works have showed that nodule isolates from different species of Mimosa belong to the β-rhizobium group of the Burkholderia genus (2, 4, 5).
Due to the fact that group A comprised the B. japonicum and B. elkanii strains (Figure 3), which displayed three subgroups of fast-growing isolates that differed from the Rhizobium and Sinorhizobium strains, it is suggested that the Bradyrhizobium genus is the most diverse genus observed so far, confirming data from previous studies (30, 25). Such knowledge reinforces that changes in the rhizobia taxonomy and new species description or reclassification are still current (32).
Group B, having nine isolates, displayed one subgroup comprising most isolates that clustered with strains of the Rhizobium and Sinorhizobium genera (BR10026, BR10016 e BR 112). These isolates originated from C. mollis nodules, although, unlike group A, most of these isolates (five out of seven) were obtained from nodules collected in the greenhouse experiment. Furthermore, this cluster displayed one strain of the Azorhizobium genus (BR5410) and two C. depauperata isolates, which were found in farther subgroups (Figure 3).
By studying the diversity of soybean and common bean Alberton et al. (1) observed how the type of sample affected the diversity of the isolates collected. In this study, the common bean isolates that were obtained from nodules of plants inoculated with a 10–4soil dilution showed to be more diverse than the isolates obtained from nodules collected in the field, whereas in the soybean culture there was a decrease in diversity.
Group C displayed four C. mollis isolates that were about 67% similar among them and 64% similar to the other isolates evaluated. The bacteria group collected from C. mollis nodules presented fast-growing and slow-growing bacteria, close to the standard strains of the Bradyrhizobium genus, placing the rhizobia that form nodules on this legume within the tropical rhizobia group. This tropical rhizobial group or cowpea group has been classified as rhizobia with a wide range of hosts (19). Another species of such genus, Cratylia argentea, has also been depicted as capable of forming nodules with rhizobial strains of the cowpea group (28).
Among the species studied, C. mollis presented the highest isolate diversity, since isolates of this species were present in all clustering of the both 16S rDNA and morphological-based similarity dendrogram. Cratylia mollis showed to be the specie that recovered higher rhizobial diversity. Isolates from Calliandra depauperata were distributed into two clusters through the ARDRA analysis, showing to belong to two different rhizobial genotypes. Therefore, M. tenuiflora presented the lowest rhizobial diversity, among the three plant species studied, once its isolates belonged to a single clusters with 100% of simple matching similarity index, in the ARDRA analisys (Figure 3). Recent results have been shown that Mimosa genera are able to form nodules with β-rhizobium (2, 4, 5).
Soares et al. (21, 22) had not found relations between the genotypic clusters, based on total-protein profile, and the morphological culture characterization of common bean and cowpea rhizobial isolates. In present study, both characterization approaches (genetic and morphologic) showed similar clustering patterns. Slow-growing isolates that alkalinize the culture medium clustered in a small group, not only in the morphologic analysis, but also in the restriction analysis of the 16S rDNA gene (Figures 2 and 3). M. tenuiflora isolates formed a consistent cluster both morphological and ARDRA analysis (Figures 2 and 3). Several isolates present in subgroups of cluster B through the morphological (Figure 2) formed consistent clusters through ARDRA analysis (Figure 3).
In recent years the classification of these microorganisms had been carried out with polyphasic taxonomic approaches, which aggregates phenotypic, genotypic and phylogenetic information (14, 17, 30, 33, 34). The studies that involve phenotypic and genetic characterization of rhizobial strains are important to elucidate relationships among these strains.
The results demonstrated the wide diversity of rhizobia present in the Caatinga, not only on a morphological level but also on a genetic level, as based on the three host legumes Cratylia mollis, Mimosa tenuiflora and Calliandra depauperata, which are typical of plants well adapted to the semi-arid climate and that until now had not been extensively studied in relation to the diversity of associated rhizobia. The knowledge about the ecology of Caatinga indigenous rhizobia must reveal parameters related to strain adaptability and symbiotic specificity, which are useful when selecting efficient strains to new inoculant formulations development (13, 16).
ACKNOWLEDGEMENTS
The authors would like to thank Dr. Sérgio M. de Faria for revision of the manuscript, Dr. Lindete Míria Vieira Martins for the technical support, PBV (UFRJ), CPGA-CS (UFRuralRJ), CAPES, Embrapa-CNPAB and CNPq for financial support.
REFERENCES
- 1.Alberton O., Kaschuk G., Hungria M. Sampling effects on the assessment of genetic diversity of rhizobia associated with soybean and common bean. Soil Biol. Biochem. 2006;38:1298–1307. [Google Scholar]
- 2.Barrett C.F., Parker M.A. Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. System. Appli. Microbiol. 2005;28:57–65. doi: 10.1016/j.syapm.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Batista J.S.S., Hungria M., Barcellos F.G., Ferreira M.C., Mendes I.C. Variability in Bradyrhizobium japonicum and B. elkanii seven years after introduction of both the exotic microsymbiont and the soybean host in a Cerrados soil. Microb. Ecol. 2007;53:270–284. doi: 10.1007/s00248-006-9149-2. [DOI] [PubMed] [Google Scholar]
- 4.Chen W.M., De faria S.M., Straliotto R., Pitard R.M., Simões-Araújo J.L., Chou J.H., Chou Y.J., Barrios E., Prescott A.R., Elliott G.N., Sprent J.I., Young J.P.W., James E.K. Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 2005;71(11):7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen W.M., De Faria S.M., James E.K., Elliott G.N., Lin K.Y., Chou J.H., Sheu S.Y., Cnockaert M., Sprent J.I., Vandamme P. Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int. J. Syst. Evol. Microbiol. 2007;57:1055–1059. doi: 10.1099/ijs.0.64873-0. [DOI] [PubMed] [Google Scholar]
- 6.Ferri M.G. Contribuição ao conhecimentoda ecologia do cerrado e da caatinga. Estudo comparativo da economia d’água de sua vegetação. Boletim da Faculdade de Filosofia, Ciências e Letras da Universidade de São Paulo. 1955;195(Botânica 12):1–170. [Google Scholar]
- 7.Ildis (International Legume Database & Information Service). 2007. Acessado em 10 de junho 2007, Disponível em. http://www.ildis.org. [Google Scholar]
- 8.Jesus E.C., Moreira F.M.S., Florentino L.A., Rodrigues M.I.D., Oliveira M.S. Diversidade de bactérias que nodulam siratro em três sistemas de uso da terra da Amazônia Ocidental. Pesq. Agropec. Bras. 2005;40(8):769–776. [Google Scholar]
- 9.Lammel D.R., Brancalion P.H.S., Dias C.T.S., Cardoso E.J.B.N. Rhizobia and other legume nodule bacteria richness in brazilian Araucaria angustifolia forest. Sci. Agric. 2007;64:400–408. [Google Scholar]
- 10.Lewis G.P. Kew: Royal Botanic Gardens; 1987. Legumes of Bahia; p. 369. [Google Scholar]
- 11.Lima J.L.S. 1996. Plantas forrageiras das Caatingas—usos e potencialidades. EMBRAPA-CPATSA/ PNE/ RBG-KEW; p. 37. [Google Scholar]
- 12.Martins L.M.V., Neves M.C.P., Rumjanek N.G. Characteristics of cowpea rhizobia isolates from the northeast region of Brazil. Soil Biol. Biochem. 1997;5/6:1005–1010. [Google Scholar]
- 13.Martins L.M.V., Rangel F.W., Xavier G. R., Ribeiro J.R. A., Morgado L.B., Neves M.C.P., Rumjanek N. Contribution of biological nitrogen fixation to cowpea: a strategy for improving grain yield in the semi-arid region of Brazil. Biol. Fertil. Soils. 2003;38(6):333–339. [Google Scholar]
- 14.Moreira F.M.S., Cruz L., De Faria S.M., Marsh T., Martinez-Romero E., Pedrosa F.O., Pitard R.M., Young J.P.W. Azorhizobium doebereinerae sp. nov. microsymbiont of Sesbania virgata (Caz.) Pers.. System. Appli. Microbiol. 2006;29:197–206. doi: 10.1016/j.syapm.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 15.Moulin L., Munive A., Dreyfus B., Boivin-Masson C. Nodulation of legumes by members of the B-subclass of Proteobacteria. Nat. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 16.Neves M.C.P., Rumjanek N.G. Diversity and adaptability of soyben and cowpea rhizobia in tropical soils. Soil Biol. Biochem. 1997;29:889–895. [Google Scholar]
- 17.Pinto F.G.S., Hungria M., Mercante F.M. Polyphasic characterization of Brazilian Rhizobium tropici strains effective in fixing N2 with common bean (Phaseolus vulgaris L.). Soil Biol. Biochem. 2007;39:1851–1864. [Google Scholar]
- 18.Rohlf F.J. New YORK: Exeter SOFTWARE; 2000. NTSYS pc. Numerical Taxonomy and multivariate analysis system. Version 2.1. [Google Scholar]
- 19.Rumjanek N.G., Martins L.M.V., Xavier G.R., Neves M.C.P. Fixação Biológica de Nitrogênio. In: FREIRE FILHO, F. R.; LIMA, J. A. DE A.; SILVA, P. H. S. DA; VIANA, F. M. P. (Org.). Feijão caupi: avanços tecnológicos. 2005:279–335. [Google Scholar]
- 20.Santos C.E.R., Stamford N.P., Neves M.C.P., Runjanek N.G., Borges W.L., Bezerra R.V., Freitas A.D.S. Diversidade de rizóbios capazes de nodular leguminosas tropicais. Rev. Bras. Ciênc. Agrár. 2007;2(4):249–256. [Google Scholar]
- 21.Soares A.L.L., Pereira J.P.A.R., Ferreira P.A.A., Do Vale H.M.M., Lima A.S., Andrade M.J.B., Moreira F.M.S. Eficiência agronômica de rizóbios selecionados e diversidade de populações nativas nodulíferas em Perdões (MG). I—Caupi (1). Rev. Bras. Ciênc. Solo. 2006a;30(5):795–802. [Google Scholar]
- 22.Soares A.L.L., Ferreira P.A.A., Pereira J.P.A.R., Do Vale H.M.M., Lima A.S., Andrade M.J.B., Moreira F.M.S. Eficiência agronômica de rizóbios selecionados e diversidade de populações nativas nodulíferas em Perdões (MG). II—Feijoeiro. Rev. Bras. Ciênc. Solo. 2006b;30(5):803–811. [Google Scholar]
- 23.Sprent J.I. Evolution and diversity in the legume-rhizobium symbiosis: chaos theory? Plant Soil. 1994;161:1–10. [Google Scholar]
- 24.Starin S.R., Leung K., Whittam T.S., De Bruijin F. S., Bottomley P.J. Genetic structure of Rhizobium leguminosarum biovar trifoli and viviae populations found in two Oregon soils under different plant communities. Appl. Environ. Microbiol. 1994;60:2772–2778. doi: 10.1128/aem.60.8.2772-2778.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taurian T., Ibañez F., Fabra A., Mario Aguilar O. Genetic diversity of rhizobial nodulating Arachis hypogaea L. in Central Argentinean Soils. Plant Soil. 2006;282:41–52. [Google Scholar]
- 26.Teixeira F.C.P., Reinert F., Rumjanek N.G., Boddey R.M. Quantification of the contribution of biological nitrogen fixation to Cratylia mollis using the 15N natural abundance technique in the semi-arid Caatinga region of Brazil. Soil Biol. Biochem. 2006;38:1989–1993. [Google Scholar]
- 27.Vincent J.M. Oxford: Blackwell Scientific Publications; 1970. A manual for the practical study of root nodulate bacteria; p. 164. IPB Handbook, 15. [Google Scholar]
- 28.Xavier D.F., Carvalho M.M., Botrel M.A. Cratylia argentea: Informações Preliminares para sua Utilização como Forrageira. Coronel Pacheco: EMBRAPA-CNPGL (EMBRAPA-CNPGL) Circular Técnica. 1995;40:18. [Google Scholar]
- 29.Xavier G.R., Martins L.M.V., Neves M.C.P., Rumjanek N.G. Edaphic factors as determinants for the distribution of intrinsic antibiotic resistance in a cowpea rhizobia population. Biol. Fertil. Soils (1998) 1998;27:386–392. [Google Scholar]
- 30.Yang K.J., Xie F.L., Zou J., Zhou Q., Zhou J.C. Polyphasic characteristics of bradyrhizobia isolated from nodules of peanut (Arachis hypogaea) in China. Soil Biol. Biochem. 37:141–153. [Google Scholar]
- 31.Young J.P.W., Downer H.L., Eardly B.D. Phylogeny of the phototrophic Rhizobium strain BTAi1 by polymerase chain reaction sequencing of a 16S rRNA gene segment. J. Bacteriol. 1991;173:2271–2277. doi: 10.1128/jb.173.7.2271-2277.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Willems A. The taxonomy of rhizobia: an overview. Plant Soil. 2006;287(1–2):3–14. [Google Scholar]
- 33.Zilli J.É., Valisheski R.R., Freire Filho F.R., Neves M.C.P., Rumjanek N.G. Assessment of cowpea rhizobium diversity in Cerrado areas of northeastern Brazil. Braz. J. Microbiol. 2004;35:281–287. [Google Scholar]
- 34.Zilli J.É., Valicheski R.R., Rumjanek N.G., Simões-Araújo J.L., Freire Filho F.R.E., Neves M.C.P. Eficiência simbiótica de estirpes de Bradyrhizobium isoladas de solo do Cerrado em caupi. Pesq. Agropec. Bras. 2006;41(5):811–818. [Google Scholar]


