Abstract
The analysis of individual gene product should enable to clarify the role of a particular enzyme in a complex xylanase system of A. niger. The two genes encoding precursors of co-produced endo-1,4-β-D-xylanases, xynA1 and xynB, were isolated from Aspergillus niger SCTCC 400264 (SCTCC, China) by using RT-PCR technique and then successfully expressed in Escherichia coli BL21. The nucleotide sequences of the xynA1 and xynB genes revealed that they were only 52.5% homology to each other. Characterization of the recombinant enzymes revealed the different properties: the specific activity of recombinant XYNA1 was 16.58 U/mg compared to 1201.7 U/mg for recombinant XYNB; The optimum temperature and pH of the recombinant XYNA1 were 35 °C and 3.0, respectively, whereas the corresponding values for the recombinant XYNB were 55 °C and 5.0, respectively; The recombinant XYNB showed much more thermostability than recombinant XYNA1; The recombinant XYNB showed 94% of maximal activity after incubating in water for 60 min at 60 °C compared to no activity for recombinant XYNA1. Various metal ions had different effects on activity between the two recombinant xylanases.
Keywords: Aspergillus niger, xylanase, prokaryotic expression, enzymatic characterization
INTRODUCTION
Xylan, as the most abundant hemicellulose, accouts for more than 30% of the dry weight of terrestrial plants and represents the major renewable carbon resource in nature (6, 11). Xylanolytic enzymes are a group of enzymes that hydrolyze xylan and arabinoxylan polymers, of which the most important are the endo-1,4-β-xylanases (EC 3.2.1.8) (25). These enzymes degrade xylan to shortchain xylo-oligosaccharides varying lengths. As the applications of these enzymes are valuable in many biotechnological processes, they gain significant attention, and play important roles in the animal feed, pulp and paper, textile, and food industries (16). There are also other potential applications for the xylanases, such as the conversion of xylan that are from agriculture and food industries’ wastes to xylose, the production of fuel and the synthesis of chemical feedstocks (26). Therfore, a number of xylanolytic enzymes from various sources, especially from microorganisms, have been studied to understand their biochemical and physical characteristics (26).
Xylanases have been classified into two families, F/10 and G/11, based on hydrophobic cluster analysis and sequence homology (9). Family F/10 are endo-β-1,4-xylanses with higher molecular mass than family G/11 xylanses, and presenting (α/β)8 barrel folds in three-dimensional(3D) structure (7). Family G/11 are xylanases with lower molecular masses (<30kDa) (12) and are encoded as precursors composed of signal peptide and a mature xylanase. The 3D structures of family G/11 xylanases have the overall shape of a “right hand” as described by Torronen et al. (1994) (29). Fungi have been recognized as a target for screening and a sourse of new enzymes with useful and novel characteristics due to their diversity (24). Filamentous fungus Aspergillus niger produces a broad spectrum of xylan-degrading enzymes with different physicochemical properties (8, 20, 15), and most of which belong to family G/11. However, the inconsistency between the reported properties of homologous xylanases of family G/11 hampers comprehensive understanding of a xylanase system. Therefore, it would be of great importance to examine thoroughly the heterogonous xylanases co-produced by one strain of A. niger to determine their function. Nevertheless, detailed analysis of an enzyme was hampered by the presence of several enzyme activities in the fungal preparation and the inability to obtain the specific xylanolytic enzyme in a pure form, the recombinant DNA technology that enables to analyse the single gene products successfully.
Recently, several genes encoding Aspergillius niger xylanases have been cloned. Some of these genes were then either overexpressed in Aspergillus species (13) or heterologously expressed in the yeast Saccharomyces cerevisiae and Pichia pastoris (20, 14, 19), but in most cases the low secretion levels did not allow complete purification and characterization of the enzymes. Nevertheless, heterologous expression of A. niger xylanase was of interest for the production of large quantities of a single xylanolytic enzyme. The Escherichia coli BL21 due to its clear genetic background, simple operation, short growth period and high expression became an attractive expression system of foreign proteins, including these of eukaryotic origin.
Since many industrial processes need high temperature, thermostability is one of the most desirable enzyme characteristics. Moreover, it has long been recognized that thermophiles represent a source of novel thermostable enzymes (4). Previous study conducted in our laboratory showed the crude proteins from a thermoresistant fungus Aspergillus niger SCTCC 400264 retained high xylanolytic activity after heat treatment, however, there was little information about the genes encoded the xylanases from the strain. In this study, we presented genes retrieval of two highly expressed endoxylanases of family 11 from A. niger SCTCC 400264 that were later named: xynA1 and xynB, and contributed greatly to understand the complexity of the xylanase system of A. niger through characterization of the two recombinant xylanases.
MATERIALS AND METHODS
Microbial strains and plamids
A. niger SCTCC 400264 from our Institute collection (SCTCC, China) was grown in a complex medium (22) containing 0.5% oat spelt xylan as the sole carbon source for induction of the xylanolytic enzymes. E. coli JM109 was used as a cloning host and DNA propagation and was grown in LB medium. E. coli BL21 was used to express recombinant protein using the expression vector pET32a (Novagen/Merck, America).
RNA isolation, cDNA synthesis and subcloning of genes encoding xylanases from A. niger SCTCC 400264
Total RNA from A. niger SCTCC 400264 was isolated using trizol reagent (Invitrogen, America). Copy DNA synthesis was initiated by cDNA kit [TaKaRa RNA PCR Kit (AMV) ver. 2.1, Japan] following the instruction. Total RNA concentration was determined spectrophotometrically. The samples were reverse transcribed into cDNA immediately or stored at -20 °C until used. We designed the primers of xynA1 and xynB encoding xylanases from A.niger SCTCC 400264 based on the 5’terminal and 3’ terminal acid sequences of xylanases from A.niger (U39784, DQ147775, AY126481, AF490982) reported in GenBank. The genes of xylanases were amplified by the PCR technique (23), using the following primers for xynA1: 5’- GGAATTCATGAAGGTCACTGCGG CTT-3’ (forward), 5’- CCGCTCGAGTTAAGAGGAGATCG TGACACTGG-3’ (reverse) and for xynB: 5' - GGAATTCATG CTCACCAAGAACCTTCTC -3' (forward), 5'- CCGCTCGAG TTACTGAACAGTGATGGAGGAAGA-3' (reverse), containing the EcoRI site and XholI site (shown by underlining), respectively. DNA fragments were amplified in 50 ul reaction mixtures using Mastercycler (Eppendorf, Germany). The reaction mixture contained 0.5μl DNA template, 5μl Ex Taq buffer(10×) (TaKaRa , Japan) , 4μl MgCl2 (25 mM), 4μl dNTP mixture (each 2.5 mM ), 1μl each primers(20μM) , 0.5μl Ex Taq polymerase(5U/μl ) (TaKaRa , Japan) in a total volume of 50μl. The reactions conditions were optimized until the highly specific products were formed. Finally, denaturation, annealing, and polymerization were carried out for 1 min at 94 °C, 30 s at 50 °C, and 60 s at 72 °C, respectively, for 30 cycles followed by the final extension for 10 min at 72 °C. The resulting PCR products from two parallel experiments were analysed by gel electrophoresis in 0.9% agarose. The amplified segments were ligated into the cloning vector pMD18-T, sequencing was performed by Invitrogen Inc., Shanghai, China.
Expression and purification of recombinant xylanases
Based on the nucleotide sequenses of xynA1 and xynB, the entire coding regions of the two genes without their signal peptides, were amplified by PCR, both the forward primers incorporated EcoRI endonuclease site (shown by underlining), : 5’-GGAATTCAGTGCCGGTATCAACTACGTG-3’ and 5’-GGAATTCTCGACCCCGAGCTCGACCGGCGAGAA-3’ for xynA1 and xynB, respectively. The reverse primers were exactly the same as used for amplification full-length DNA. The amplified fragments were ligated into pMD18-T vector and the inserts were subjected to DNA sequencing. The resulting vectors were digested with EcoRI and XhoI and ligated at EcoRI -XhoI sites of pET32a vector (Novagen/Merck, America) previously treated with the same enzymes. The constructed plasmids were named pET32a-xynA1 and pET32a-xynB, and the recombinant enzymes named XYNA1 and XYNB, respectively.
E. coli BL21 strains containing expression vector pET32a-xynA1 and pET32a-xynB was used to express recombinant xylanases of A. niger. The two recombinant proteins fused six histidine residues and S•Tag with xylanases. E. coli BL21 grew in LB media at 37 °C in the presence of 50μg/ml ampicillin. Expression was induced at OD600 of 0.6-0.8 by the addition of 0.5 mM isopropylthion-β-D-galactoside (IPTG) and culture for another 4h at 30 °C for recombinant XYNA1, and for recombinant XYNB, 0.2 mM IPTG and culture for another 12 h at 20 °C. Bacteria were pelleted by centrifugation at 5,000 r/min for 10 min and resuspended in 1×Phosphate Buffer (PB). Cells were ruptured 60 s at 10 s per cycle (150 w, 5 s pulse on, 5 s pulse off) by ultrasonic. The lysate was clarified by centrifugation at 12,000 r/min for 10 min. The supernatant containing His-xylanase was purified by Ni(2+)-NTA affinity chromatography. Recombinant XYNA1 and recombinant XYNB were both eluted with 100mM imidazole. The solution was dialyzed by 1×PB.
Enzyme and protein assays, SDS-PAGE analysis
The xylanase activity was assayed by measuring the reducing groups liberated from 0.8% (w/v) oat spelt xylan (Sigma, America) by the DNS method using xylose as standard (21). One unit of enzyme activity was defined as the amount of enzyme that produced reducing sugars equivalent to 1μmol of xylose per minute. Kinetic parameters of the enzyme were determined by measuring the enzymatic activity toward birchwood xylan at various concentrations (2.5–20 mg ml−1 in 100mM citrate buffer pH 5) (22). The activity determination of the enzyme was carried out at each optimal temperature of enzymes for 10 min by the DNS method. The kinetic parameters were then determined by fitting the initial velocity data to the Michaelis–Menten equation using the Kaleida Graph software (Synergy Software, Reading, PA) (5).
Protein was estimated using the method of Bradford (3).
SDS-PAGE was done according to the method of Laemmli (17). Gels were stained for protein with Coomassie Brilliant Blue R-250.
To investigate the effects of different metal ions on the recombinant xylanases activities, 10mM CuSO4, 10mM ZnSO4, 10mM CaCl2, 10mM Fe2(SO4)3, 10mM FeSO4, 10mM MgSO4, 10mM MnSO4, 10mM KCl were added separately to the reaction solution, and xylanase activities were measured under the standard assay.
Sequence analysis and Nucleotide sequences accession numbers
Sequence alignments were performed using a BLAST program. The xynA1 and xynB sequences were deposited at GenBank under the accession numbers FJ 785738 and FJ 772090, respectively.
RESULTS AND DISCUSSION
Isolation and analysis of genes encoding xynA1 and xynB
The genes encoding two co-produced endo-1,4-β-xylanases, named: xynA1 and xynB, were isolated from A. niger SCTCC 400264 by RT-PCR technique. Sequence analysis by NCBI BLAST (http:// www.ncbi.nlm.nih.gov/blast/) showed that the sequence has high levels of homology with the other xylanase sequences available in the GenBank database.
A BLAST search showed that nucleotide sequence of xynB (AC: FJ772090) coding sequence was 99% identical to the endoxylanases from A.niger (AC: DQ302412), A.niger (AC: AM269952), A.niger (AC: D38071), the deduced amino acid sequence was identical to A.niger (AC: XP 001388522), A.niger (AC: P55330), A.niger (AC: BAA07265), however, their properties have not been reported. Besides, xynB was 99% identical to xylanases produced by other strains of A. niger (AC:AY126481, AF490982, AY551187, AY 536639) at the amino acid level. The alignment of xynA1 (AC: FJ785738) revealed that the gene encodes xylanase that differed from other known A. niger xylanases at the nucleotide as well as the protein level. xynA1 was the most homology (99%) to A.niger (AC: U39784), and in 95.3% homology to A.niger (AC: P55329), 94% to A.niger (AC: AY536688),and 93% to A.niger (AC: DQ147775) at the amino acid level.
Most of the detailed informations on enzyme properties, functions, and structure available were derived from the study of endoxylanase type A (10, 2, 27). Based on structural analysis, both endoxylanase A and B belong to family 11. However, they are classified as type A and B because their amino acid sequences share only 41% identity (1). The xynA1 gene (AC: FJ785738) showed merely 51.5% homology to the xynB gene (AC: FJ772090) encoding endoxylanases at the amino acid level.
The DNA sequences comprising the pre(pro)xylanases revealed the open reading frames (ORF) coding for 211 amino acid residues in case of xynA1 and 225 amino acids when it came to xynB. Analysis of the deduced xynA1 and xynB proteins sequence using the program SignalP 3.0 Server (http://www.cbs.dtu.dk/services/SignalP ) predicted a 27 aa signal sequence for xynA1 and a 37 aa signal sequence for xynB respectively, which were exactly the same as the reported by Korona, et al. (14). Analysis using ProtParam Tool (http://www.expasy.org/tools/protparam.html) revealed that the calculated molecular weight of the mature protein (without peptide) containing 184 amino acids was 19.837 kDa and the deduced isoelectric point was 4.19 for xynA1, compared to 188 amid acids, 20.08 kDa, the deduced isoelectric point 4.57 for xynB. From the sequence analysis, it would appear the two recombinant enzymes belonged to the family 11 of glycoside hydrolases. Moreover, two glutamic acid residues conserved in all the xylanases can be identified as the active site of xynA1: E79, E170 (15) and xynB: E121, E212 (13). Taken together, our studies have confirmed that genes of equivalent xylanases from different strains of A. niger were diversified.
Construction of expression plasmid and expression in E.coli
The entire coding regin of xynA1 and xynB (without peptide), were amplified by PCR from the cloning plasmids pMD-xynA1and pMD-xynB respectively, and then cloned into pET32a vector. They expressed in different condition in E.coli BL21. The enzymes fractions appeared homogeneous and visible as protein bands of about 41 kDa for recombinant XYNA1 and recombinant XYNB (Fig.1). They were both in an excellent agreement with the calculated molecular mass.
Figure 1.
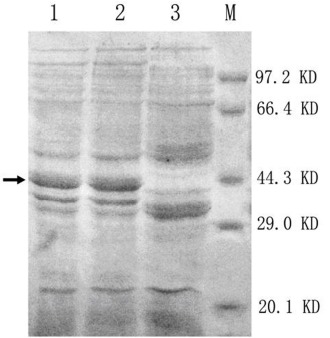
SDS-PAGE analysis of the recombinant xylanases produced by E.coli BL21 .lanes 1 and 2: recombined proteins: XYNA1 and XYNB, respectively; Lane3: pET32a host; M: Standard protein molecular weight. The protein gels were stained with Coomasie Brilliant Blue R250; →: interest proteins
Purification of the recombinant xylanase from E.coli
According to SDS-PAGE, there were both only one clear band of the two recombinant xylanases with molecular weights 41 kDa (Fig. 2). The specific activity of the purified enzymes showed 13.8-fold (XYNA1) and 14.5-fold (XYNB) increase compared with the crude proteins in culture supernatant of E.coli BL21 (DE3). However, the highest specific xylanase activity for recombinant XYNA1 and XYNB were 16.58 U/mg protein and 1201.7 U/mg protein respectively. The latter was also higher than that of the other recombinant xylanases from A. niger reported (14, 19).
Figure 2.
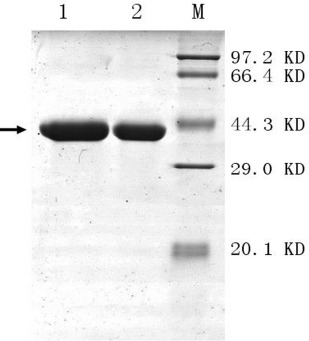
SDS-PAGE analysis of the purified xylanases obtained from E.coli BL21. Lane 1: 100 mM imidazole wash of the recombined protein XYNB to Ni-nitrilotriacetic acid resin; Lane2: 100 mM imidazole wash of the recombined protein XYNA1 to Ni-nitrilotriacetic acid resin; M: Standard protein molecular weight. The protein gels were stained with Coomasie Brilliant Blue R250. →:interest proteins
Properties of the recombinant xylanase
The kinetic parameters of recombinant XYNA1 and XYNB were determined. The Km and Vmax values of the XYNA1 enzyme for oat spelt xylan were 13.8 mg ml−1, and 43.66 U/mg protein, respectively. The XYNB enzyme showed slightly lower affinity for substrate with Km of 18.7 mg ml−1 and a decrease in Vmax with the value of 1666.7 U/mg protein compared to XYNA1.
Characterization of the recombinant enzymes revealed the great difference between the recombinant XYNA1 and XYNB: The optimum reaction temperature and pH of the recombinant XYNA1 were 35 °C and 3.0, respectively, but the recombinant XYNB were 55 °C and 5.0(Fig.3, Fig.4). The properties for the optimal enzyme activity of the recombinant XYNB were in the range of that reported for other fungi and bacteria (30). The XynB from A. niger IBT-90 and XynNB described by Kinoshita et al. (13) showed the same pH optima of about 5.0. The pH determined for XYNA1 was exactly the same as the pH optimum reported for XynI derived from A. niger (15). As anticipated from the resolved crystal structure of A. niger XynI (15) and further determined by means of mutagenesis by Tahir et al. (28), the Asp37 presented in the active site of acidic β-xylanases was the key residue responsible for low pH optimum (18), so the lower pH optimum of XYNA1 would be attributed to the presence of Asp at the mentioned position.
Figure 3.
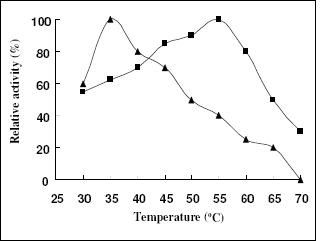
Effects of temperature on the activity of recombinant xylanase. The optimal temperature was determined by incubating the enzyme in 0.1M citrate buffer, pH 5, for 10 min at different temperatures as indicated in the graph. The maximal enzyme activity was defined as 100% relative activity. Symbols: recombinant XYNA1 (▲); recombinant XYNB (■).
Figure 4.
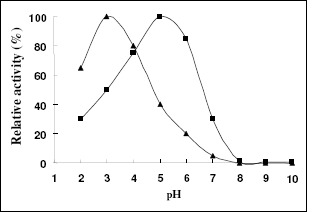
Effect of pH on the activity of the recombinant xylanase. The optimal pH was determined by incubating the enzyme at 50 °C for 10 min at different pH. To determine the optimal pH, pH range from 2 to 10 was used with the following (100mM) buffers: KCl-HCl (pH 2), sodium citrate (pH 3–7), Na2HPO4- NaH2PO4 (pH 8.0), glycine -NaOH (pH 9-10). The maximal enzyme activity was defined as 100% relative activity. Symbols: recombinant XYNA1 (▲); recombinant XYNB (■).
The results of the effects of pH stability on the two recombinant xylanase revealed that, the recombinant XYNA1 was more stable than recombinant XYNB after being incubated at pH 2.0-5.0 for 30 min at 37 °C, however, there had little difference after being incubated at pH 6.0-10.0 between them , and both retained 75%~85% of maximal activity (Fig.5).
Figure 5.
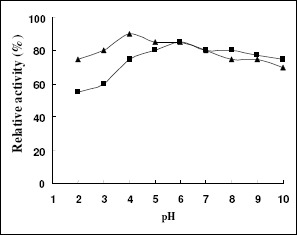
Acid-alkali stability of recombinant xylanases. To estimate pH stability, the recombinant enzymes were incubated in the different pH buffers (pH range from 2 to 10) at 37°C for 30 min, and then the residual xylanase activity was determined at optimal pH. The activity determined under the standard conditions of optimal pH without incubation was defined as 100% relative activity. Symbols: recombinant XYNA1 (▲); recombinant XYNB (■).
Moreover, it was worth stressing that the effects of temperature on xylanases activity and the thermostability of the recombinant enzymes revealed much more difference which were presented in Fig.6. The recombinant XYNB showed much more thermostability than recombinant XYNA1; the recombinant XYNB showed 94% of maximal activity after incubating in water for 60 min at 60 °C compared to no activity for recombinant XYNA1. In order to futher study their thermostability, the incubatied temperature was risen to 85 °C. The recombinant XYNB remained 94% of maximal activity after being incubated in water for 10 min at 85 °C compared to no activity for recombinant XYNA1. Hence, the discrepancies between these two low molecular weight endo-1,4-β-D-xylanases co-expressed by A. niger are not only in the pH optima, but also in temperature optima and thermostability. It can be implied that the co-existence of these two similar enzymes may be due to their different catalytic activity. Moreover, the recombinant XYNB showed higher thermostability than crude enzyme from A. niger SCTCC 400264 preparation. Hence, we can deduced that the xynB coded protein played a key role in the thermostability of xylanase system of A. niger SCTCC 400264. In addition, comparing to the other xylanases from A.niger (19, 30), the recombinant XYNB in our research also showeded higher thermostability.
Figure 6.
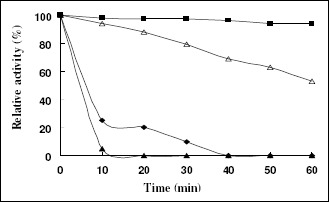
Thermostability of the recombinant xylanases.The enzyme solutions were incubated at 60°C and 85°C without the presence of substrate for various times, respectively. The residual xylanases activity were measured by 10 min incubation at optimal temperature for recombinant XYNA1 and XYNB, respectively. The activity determined under the standard conditions of optimal temperature without incubation was defined as 100% relative activity. Symbols: recombinant XYNB treated by 60°C (■), recombinant XYNB treated by 85°C (∆), recombinant XYNA1 treated by 60°C (♦), recombinant XYNA1 treated by 85°C (▲).
Various metal ions have different effects on activity between the two recombinant xylanases (Table 1). The potassium, calcium, zinc, iron had defferent stimulation effects on recombinant XYNB activity, copper inhibited its activity. However, manganese, copper and iron had an inhibitory effect on recombinant XYNA1 activity, only potassium stimulated its activity by 41%.
Table 1.
The effect of metal ions on recombinant xylanases activity
| Metal iron | Relative activity of recombinant XYNA1 (%) | Relative activity of recombinant XYNB (%) |
|---|---|---|
| control | 100 | 100 |
| Zn2+ | 122 | 101.64 |
| Ca2+ | 122.77 | 96.85 |
| Mg2+ | 91.08 | 91.56 |
| Cu2+ | 86.15 | 73.68 |
| Fe2+ | 93.19 | 91.44 |
| Fe3+ | 104.46 | 64.86 |
| K+ | 119.01 | 141.06 |
| Mn2+ | 104.69 | 80.1 |
In conclusion, this was the first report on isolation of two genes encoding: xynA1 and xynB from different groups of glycosyl hydrolase family 11 co-produced by one strain of A. niger and their subsequent expression in Escherichia coli BL21. The fact that two xylanases come from the common origin, the same fungal organism, will make the comparative studies of both enzymes more accurate. Characterization of the recombinant enzymes revealed the great differences between the recombinant XYNA1 and XYNB. The protein encoded by xynB played a key role to the thermostability of xylanase system of A. niger SCTCC 400264. Since high temperatures were used in many industrial processes, and always made the xylanase activity destroyed, thermostability was one of the most desirable enzyme characteristics. The excellent thermostability of recombibant XYNB could establish a foundation for its industry application.
ACKNOWLEDGEMENTS
This study was supported by the National Natural Science Foundation of China (NSFC 30871321, 30740055, 30771312), and Program for New Century Excellent Talents in University (NCET-05-0785).
REFERENCES
- 1.Asano, K.; Sriprang, R.; Gobsuk, J.; Eurwilaichitr, L.; Tanapongpipat, S.; Kirtikara, K. (2005). Endo-1,4-β-xylanase B from Aspergillus niger BCC14405 Isolated in Thailand: Purification, Characterization and Gene Isolation. J. Biochem. Mol. Biol, 38, 17-23. [DOI] [PubMed] [Google Scholar]
- 2.Berrin, J.G.; Williamson, G.; Puigserver, A.; Chaix, J.C.; McLauchlan, W.R.; Juge, N. (2000). High-level production of recombinant fungal endo-beta-1,4-xylanase in the methylotrophic yeast Pichia pastoris Protein Expr. Purif., 19, 179-187. [DOI] [PubMed] [Google Scholar]
- 3.Bradford, M.M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 4.Bruins, M.E.; Jasen, A.E.; Boom, R.M. (2001). Thermozymes and their applications: a review of recent literature and patents. Appl. Biochem. Biotechnol, 90, 155-186. [DOI] [PubMed] [Google Scholar]
- 5.Chen, A.P.C.; Chang, S.; Lin, Y.; Sun, Y.; Chen, C.; Wang, A.H.J.; Liang, P. (2005). Substrate and product specificities of cistype undecaprenyl pyrophosphate synthase. Biochem. J, 386, 169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coughlan, M.P.; Hazlewood, G.P. (1993). Β-1,4-d-Xylan-degrading enzyme systems:biochemistry, molecular biology and applications. Biotechnol. Appl. Biochem, 17, 259–289. [PubMed] [Google Scholar]
- 7.Dominguez, R.; Souchon, H.; Spinelli, S.; Dauter, Z.; Wilson, K.S.; Chauvaux, S.; Beguin, P.; Alzari, P.M. (1995). A common protein fold and similar active site in two distinct families of β-glycanases. Nat. Struct. Biol, 2, 569–576. [DOI] [PubMed] [Google Scholar]
- 8.Frederick, M.M,; Kiang, C.; Frederick, J.R.; Reilly, P.J. (1985). Purification and characterization of endo-xylanases from Aspergillus niger 1. Two isozymes active on xylan backbones near branch points. Biotechnol. Bioeng, 27, 525–532. [DOI] [PubMed] [Google Scholar]
- 9.Gilkes, N.R.; Henrissat, B.; Kilburn, D.G.; Miller, R.C.; Warren, R.A.J. (1991). Domains in microbial β-1,4-glycanases: sequence conservation, function,and enzyme families. Microbiol. Rev, 55, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbacheva, I.V.; Rodionova, N.A. (1977). Studies on xylan degrading enzymes. I. Purification and characterization of endo-1,4-beta-xylanase from Aspergillus niger str. 14. Biochim.Biophys. Acta, 484, 79-93. [DOI] [PubMed] [Google Scholar]
- 11.Haki, G.D; Rakshit, S.K. (2003). Developments in industrially important thermostable enzymes: a review. Bioresour. Technol, 89, 17–34 [DOI] [PubMed] [Google Scholar]
- 12.Henrissat, B.; Bairoch, A. (1993). New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J, 293, 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinoshita, K.; Takano, M.; Koseki, T.; Ito, K.; Iwano, K. (1995). Cloning of the xynNB gene encoding xylanase B from Aspergillus niger and its expression in Aspergillus kawachii J.Ferment. Bioeng, 79, 422–428. [Google Scholar]
- 14.Korona, B.; Korona, D.; Bielecki, S. (2006). Efficient expression and secretion of two co-produced xylanases from Aspergillus niger in Pichia pastoris directed by their native signal peptides and the Saccharomyces cerevisiae α-mating factor. Enzyme Microb. Technol., 39, 683-689. [Google Scholar]
- 15.Krengel, U.; Dijkstra, B.W. (1996). Three-dimensional structure of endo-1, 4-xylanase I from Aspergillus niger: molecular basis for its low pH optimum. J. Mol. Biol., 263, 70–78. [DOI] [PubMed] [Google Scholar]
- 16.Kulkarni, N.; Shendye, A.; Rao, M. (1999). Molecular and biotechnological aspects of xylanases. FEMS. Microbiol. Rev., 23, 411–456. [DOI] [PubMed] [Google Scholar]
- 17.Laemmli, U.K. (1970). Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- 18.Liu, L.; Li, X.; Li, X.; Shao, W. (2004). Computational analysis of responsible dipeptides for optimum pH in G/11 xylanase. Biochem. Biophys. Res. Commun., 321, 391–396. [DOI] [PubMed] [Google Scholar]
- 19.Liu, M.Q.; Weng, X.Y.; Sun, J.Y. (2006). Expression of recombinant Aspergillus niger xylanase A in Pichia pastoris and its action on xylan. Protein Expr. Purif., 48, 292-299. [DOI] [PubMed] [Google Scholar]
- 20.Luttig, M.; Pretorius, I.S.; Van-Zyl, W.H. (1997). Cloning of two β-xylanase encoding genes from Aspergillus niger and their expression in Saccharomyces cerevisiae Biotechnol. Lett., 19, 411–415. [Google Scholar]
- 21.Miller, G.L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem., 31, 426–428. [Google Scholar]
- 22.Rogalski, J.; Oleszek, M.; Tokarzewska-Zadora, J. (2001). Purification and characterization of two endo-1,4 beta-xylanase and a 3-xylosidase from Phlebia adiate Acta. Microbiol. Pol., 50, 117–128. [PubMed] [Google Scholar]
- 23.Saiki, R.K.; Gelvand, D.H.; Stoffel, S.; Scharf, S.J.; Higuchi, R.; Horn, G.T.; Mullis, K.B.; Erlich, H.A. (1988). Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science, 239, 487–491. [DOI] [PubMed] [Google Scholar]
- 24.Singh, S.; Madlala, A.M.; Prior, B.A. (2003). Thermomyces lanuginosus: properties of strains and their hemicellulases. FEMS. Microbiol. Rev., 27, 3-16. [DOI] [PubMed] [Google Scholar]
- 25.Subramaniyan, S.; Prema, P. (2002). Biotechnology of microbial xylanases: enzymology. Crit. Rev. Biotechnol., 22, 33–64 [DOI] [PubMed] [Google Scholar]
- 26.Sunna, A.; Antranikian, G. (1997). Xylanolytic enzymes from fungi and bacteria. Crit. Rev. Biotechnol., 17, 39-67. [DOI] [PubMed] [Google Scholar]
- 27.Tahir, T.A.; Berrin, J.G.; Flatman, R.; Roussel, A.; Roepstorff, P.; Williamson, G.; Juge, N. (2002). Specific characterization of substrate and inhibitor binding sites of a glycosyl hydrolase family 11 xylanase from Aspergillus niger J. Biol. Chem., 277, 44035-44043. [DOI] [PubMed] [Google Scholar]
- 28.Tahir, T.A.; Durand, A.; Gebruers, K.; Roussel, A.; Williamson, G.; Juge, N. (2004). Functional importance of Asp37 from a family 11 xylanase in thebinding to two proteinaceous xylanase inhibitors from wheat. FEMS. Microbiol. Lett, 239, 9-15. [DOI] [PubMed] [Google Scholar]
- 29.Torronen, A.; Harkki, A.; Rouvinen, J. (1994). Three-dimensional structure of endo-1,4-β-xylanase II from Trichoderma reesei: two conformational states in the active site. EMBO. J., 13, 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, K.K.Y.; Tan, L.U.L.; Saddler, J.N. (1988). Multiplicity of β-1,4-xylanase in microorganisms: function and applicants. Microbiol. Rev., 52, 305–317. [DOI] [PMC free article] [PubMed] [Google Scholar]


