Abstract
Most of our knowledge about pollutants and the way they are biodegraded in the environment has previously been shaped by laboratory studies using hydrocarbon-degrading bacterial strains isolated from polluted sites. In present study Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria, isolated from oily sludge, were found to be able to tolerate pure and mixture of saturated hydrocarbons, as well as pure and mixture of monoaromatic and polyaromatic hydrocarbons. Isolated Gram-negative bacteria were more tolerant to mixture of saturated (n-hexane, n-hexadecane, cyclohexane), monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons than Gram-positive bacteria. There were observed cellular and molecular modifications induced by mixture of saturated, monoaromatic and polyaromatic hydrocarbons to Gram-positive and Gram-negative bacteria. These modifications differ from one strain to another and even for the same bacterial strain, according to the nature of hydrophobic substrate.
Keywords: bacteria, multiple response, mixture of hydrocarbons
INTRODUCTION
Petroleum processing unavoidably generates considerable volumes of oily sludge. Common sources of these sludges are storage tank bottoms, oil-water separators, flotation and biological wastewater treatment units, cleaning of processing equipment and occasional minor spills on refinery grounds (15). Oily sludge contains several toxic hydrocarbon constituents, making the sites contaminated by them a major environmental concern, because many of the constituents of oily sludge are carcinogenic and potent immunotoxicants (47, 61). Oily sludge is a complex mixture of total petroleum hydrocarbon (TPH), water, and soil particles. The composition of these sludges varies according to their origin, storage, and treatment history. TPH is primarily composed of alkane, aromatics, NSO (nitrogen-, sulfur-, and oxygen-containing compounds), and asphaltene fractions. Among the many techniques employed to decontaminate the hydrocarbon-contaminated sites, in situ bioremediation using indigenous microorganisms is most widely used. These microorganisms can tolerate and also degrade a wide range of target constituents present in oily sludge (4, 7, 15, 16, 41, 47, 77).
The mechanisms of toxicity to microbial membranes caused by hydrocarbons have been well studied and comprehensively reviewed (28, 62, 69, 74, 78). The accumulation of toxic hydrocarbons in the membrane increases membrane fluidity (allowing the leakage of macromolecules such as RNA, phospholipids, proteins), increases membrane swelling, and reduces the normal functioning of membrane-associated proteins (52, 74, 84, 89). The accumulation of hydrocarbons results in the disruption of bilayer stability and membrane structure, causing a loss of membrane function and ultimately cell death. Despite this extreme toxicity, hydrocarbon-tolerant Gram-negative and Gram-positive bacteria that are capable to grow in a two-phase water-hydrocarbon system have been isolated (71, 89). Many of these tolerant bacterial species, including the first strain isolated, were Gram-negative bacteria such as Pseudomonas putida (12, 31, 37, 54, 63, 85) or closely related Pseudomonas sp. (49, 53, 71). Gram-positive bacteria such as Bacillus (34, 48, 68, 71, 89), Rhodococcus (58), Staphylococcus (89), Exiguobacterium (71) have also been found to be hydrocarbon-tolerant, although limited investigation has occurred towards understanding the mechanisms of their hydrocarbon tolerance. Because of the highly impermeable outer membrane of Gram-negative bacteria, it was generally accepted that this type of bacteria are more tolerant to hydrocarbons than Gram-positive bacteria (3, 25, 32, 34, 79). However, several Gram-positive bacteria seems to been more resistant (43, 48, 50, 58, 71, 89). Because of the different experimental set-ups used in the published literature, it has been difficult to compare the hydrocarbons tolerance of different strains, and extremely difficult to compare hydrocarbons tolerance between Gram-positive and Gram-negative strains (71). Only two recent studies have compared hydrocarbon tolerance properties in those two types of bacteria (71, 89).
The present study focuses on the isolation and characterization of several new Gram-positive and Gram-negative bacteria that are extremely tolerant to pure and mixture of saturated, monoaromatic and polyaromatic hydrocarbons. Although there are numerous studies on cellular and molecular modifications induced by hydrocarbons to different strains, still there are few studies on the modifications induced by hydrocarbons mixtures and there is no study to compare the effects of hydrocarbons mixtures on Gram-positive and Gram-negative bacteria. The modifications induced on cellular and molecular level on Gram-positive and Gram-negative bacteria grown in the presence of different mixture of saturated, monoaromatic and polyaromatic hydrocarbons are also presented in this study.
MATERIALS AND METHODS
Isolation and characterization of Gram-positive and Gram-negative bacterial strains
Cell suspensions in saline phosphate-buffered 0.8% (w/v) NaCl (PBS) were obtained according to Anderson et al. (2) and 10-fold dilution series in PBS were used for viable cell enumerations. The number of viable bacterial strains in Poeni oily sludge (Teleorman County, Romania) was estimated by a modified most probable number (MPN) procedure (22, 88). Aliquots of 20 μl were added into ten separate dilution series in 96-microwell plates (Iwaki, London, United Kingdom). The wells were pre-filled with 170 μl minimal medium (20) and 10 μl sterilized Poeni crude oil or hydrocarbons mixtures (saturated and aromatic). After 2 weeks incubation at 28°C, each well was added with 50 μl filter-sterilized solution of the respiration indicators 0.3% (w/v) INT [2-(4-iodophenyl)-3(4-nitrophenyl)-5-phenyltetrazolium chloride]. After over-night incubation in the dark at room temperature, red and pink wells were counted as positive for growth. A maximum-likelihood estimation of microbial numbers based on 10-fold dilution series was developed for the Microsoft Excel for Windows spreadsheet program (18, 22, 88).
Isolation of the bacterial strains (IBBPo1, IBBPo2, IBBPo3, IBBPo7, IBBPo10, IBBPo12) from Poeni oily sludge (Teleorman County, Romania) was carried out on minimal medium (20), using the enriched cultures method, with hydrocarbons mixtures (saturated and aromatic) as single carbon source. Bacterial strains were selected on the basis of different colony morphologies. For further characterization of bacterial strains several physiological and biochemical tests were performed: Gram reaction, morphology, endospore formation, mobility, respirator type, pigments production, growth on TTC (2, 3, 5-triphenyl tetrazolium chloride) medium, growth on CBB (Coomassie brilliant blue R250) agar, catalase and oxidase production. Gram-positive (IBBPo1, IBBPo2, IBBPo3) bacteria were investigated for: nitrates reduction, pyrazinamidase, pyrrolidonyl arylamidase, alkaline phosphatase, β-glucuronidase, β-galactosidase, α-glucosidase, N-acetyl-β-glucosaminidase, and urease production, esculin and gelatin hydrolysis, and fermentation of different carbohydrates such as glucose, ribose, xylose, mannitol, maltose, lactose, saccharose, glycogen. Gram-negative (IBBPo7, IBBPo10, IBBPo12) bacteria were investigated for: nitrates reduction, indole production, D-glucose fermentation, L-arginine dihydrolase and urease production, esculin and gelatin hydrolysis, β-galactosidase production, and assimilation of different substrates such as D-glucose, L-arabinose, D-mannose, D-mannitol, N-acetyl-glucosamine, D-maltose, potassium gluconate, capric acid, adipic acid, malic acid, trisodium citrate, phenylacetic acid. The taxonomic affiliation of Gram-positive and Gram-negative bacterial strains were determined based on their phenotypic characteristics and also based on the G+C content of the bacterial chromosome (13). MICs (minimum inhibitory concentration) of antibiotics (ampicillin, kanamycin) and toxic compounds (sodium dodecyl sulfate, rhodamine 6G) for Gram-positive and Gram-negative bacterial strains were also determined. Bacterial cells (105 CFU ml-1) were spotted on solid LB-Mg (30) medium (control) and on the same medium with antibiotics and toxic compounds at various concentrations (1-1000 μg ml-1). Petri dishes were incubated 24 hours at 28°C. MICs of antibiotics and toxic compounds were determined as the concentrations that severely inhibited bacterial cell growth. Rhodamine 6G accumulation in bacterial cells was observed under UV light after 24 hours of incubation at 28°C. Here, as elsewhere in this work, experiments were repeated at least three times.
Tolerance of Gram-positive and Gram-negative bacterial strains to pure and mixture of saturated (n-hexane, n-hexadecane, cyclohexane, mixture of them), monoaromatic (benzene, toluene, ethylbenzene, mixture of them) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene, mixture of them) hydrocarbons. Bacterial cells (105 CFU ml-1) were spotted on solid LB-Mg (30) medium (control) or solid minimal (20) medium, added with 1% (w/v) yeast extract (control) and on the same media with hydrocarbons, supplied in the vapor phase. Petri dishes were sealed and the formation of hydrocarbon-resistant bacterial colonies on the agar was determined after 24 hours incubation at 28°C.
Cellular and molecular modifications induced by mixture of saturated (n-hexane, n-hexadecane, cyclohexane), monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons on Gram-positive and Gram-negative bacterial strains. Bacterial cells were cultivated in liquid LB-Mg (30) medium (control) and incubated at 28°C on a rotary shaker (150-200 rpm) until 105 CFU ml-1. 0.5-5% (v/v) mixtures of saturated, monoaromatic or polyaromatic hydrocarbons were then supplied in culture liquid. Flasks were sealed and incubated 24 hours at 28°C on a rotary shaker.
Modifications induced by mixture of hydrocarbons to cells viability
The growth of the bacterial strains was determined by spetrophotometric measurement of the optical density (OD660nm). Serial dilutions of culture liquid were also spread on agar LB-Mg medium (30) using the method of Ramos et al. (64) and the number of viable cells (CFU ml-1) was determined.
Effects of mixture of hydrocarbons to β-galactosidase activity
Bacterial cells were permeabilized by using chloroform and sodium dodecyl sulfate according to the Miller assay (46). β-galactosidase activity was determined in triplicate by measuring the hydrolysis of the chromogenic substrate, o-nitrophenyl-β-D-galactoside (46).
Modifications induced by mixture of hydrocarbons to lipids
Lipids were extracted with chloroform-methanol (2:1) mixture using the method of Benning and Somerville (5). The samples were spotted onto 20×20cm Silica gel 60 TLC aluminium sheets (Merck), and the separation was performed using chloroform-methanol-acetic acid-water (85:22.5:10:4 v/v/v/v) mixture as mobile phase, in saturated athmosphere (simultaneous separating chamber of 21×9×21 cm, Desaga type). The plates were treated with 10% (w/v) molybdatophosphoric acid hydrate in ethanol, and after drying the spots appeared on green background. The identification of the phospholipids was done based on their motilities (Rf) and their comparison with those of phospholipids standards (Sigma-Aldrich, Supelco).
Modifications induced by mixture of hydrocarbons to proteins
Membrane and periplasmic protein fractions were extracted with HE buffer (10 mM HEPES-NaOH, pH 7.6, 10 mM EDTA, 10 mM MgCl2), solved in Laemmli buffer and denaturated at 95°C, for 5 min. 50 μg of protein per lane were loaded onto a 12% (w/v) polyacrylamide gel (66). Gels were stained with Bio-Safe colloidal Coomassie Blue G-250 (Bio-Rad). Protein content was measured by the method of Bradford (9).
Modifications induced by mixture of hydrocarbons to DNA
The bacterial cells were lysed with Tri-Reagent (mixture of guanidine thiocyanate and phenol). The extraction of DNA was performed as recommended by the manufacturer (Sigma-Aldrich) and the DNA was precipitated with ethanol and resuspended in TE (10 mM Tris-HCl, 1 mM EDTANa2) buffer. After separation on 0.8% (w/v) TBE (Tris-Borate-EDTA) agarose gel and ethidium bromide staining the DNA was visualized with ultraviolet light. DNA content and purity were measured by the method of Sambrook et al. (66). DNA concentration of each variant was adjusted to 100 μg ml-1 for spectral comparison. Template DNA for PCR was obtained using the method of Whyte et al. (87). For PCR amplification, 5 μl of DNA extract was added to a final volume of 50 μl reaction mixture, containing: 5×GoTaq flexi buffer, MgCl2, dNTP mix, primers (A24f2 5’-CCSRTITTYGCITGGGT-3’, A577r2 5’-SAICCARAIRCGCATSGC-3’), GoTaq DNA polymerase (Promega). PCR was performed with a C1000 thermal cycler (Bio-Rad). PCR program consisted in initial denaturation for 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 50 or 52°C, 1 min at 72°C, and a final extension of 5 min at 72°C. PCR were performed in duplicate. After separation on 1.6% (w/v) TBE agarose gel and staining with fast blast DNA stain the amplified fragments were analyzed.
Reagents
n-hexane (96% pure), n-hexadecane (99% pure), cyclohexane (99.7% pure), benzene (99% pure), toluene (99% pure) were obtained from Merck (E. Merck, Darmstadt, Germany), ethylbenzene (98% pure), naphthalene (99% pure), 2-methylnaphthalene (97% pure), fluorene (90% pure) were obtained from Sigma-Aldrich (Saint-Quentin-Fallavier, France). Other reagents used were procured from Merck, Sigma-Aldrich, Difco Laboratories (Detroit, Michigan, USA), Promega (Promega GmbH, Mannheim, Germany), bioMérieux (Marcy-l’Etoile, France) or Bio-Rad Laboratories (Alfred Nobel Dr., Hercules, CA, USA).
RESULTS AND DISCUSSION
In the latest decades the researches regarding microorganisms involved in environments polluted with oil and oily sludge bioremediation have been intensified, the use of the bacteria for this purpose becoming an advantageous and cheap alternative compared with other traditional methods. Microbial remediation of a hydrocarbon-contaminated site is accomplished with the help of a diverse group of microorganisms, particularly the indigenous bacteria, but their population and efficiency are affected when any toxic contaminant is present at high concentrations (4, 16, 56).
Isolation and characterization of Gram-positive and Gram-negative bacterial strains. Bacteria with specific metabolic capabilities, such as oil degradation, can be enumerated based on their ability to grow on selective media. The most probable number (MPN) procedure is particularly well suited for bacteria that grow on insoluble substrates, due to the difficulties in preparation of solid media containing a homogenous distribution of the appropriate carbon source. Also, many agar based solid media contain impurities that allow growth of organisms that cannot degrade the target substrate, leading to overestimation of the size of the population of interest. Haines et al. (22) developed a 96-well microtiter plate MPN procedure to enumerate hydrocarbon-degrading bacteria. Selective minimal medium with Poeni crude oil or hydrocarbons mixtures (saturated and aromatic), as sole carbon source, was used to enumerate the hydrocarbon-degrading bacteria in Poeni oily sludge (Teleorman County, Romania). Growth in 96-microwell plates was identified using INT, which forms an insoluble red precipitate in wells containing bacteria that can use crude oil or hydrocarbons mixtures (saturated and aromatic), as sole carbon source. After each plate from the 96-microwell plate had been scored, the number of positive wells at each serial dilution was entered into a computer program to determine the most probable number of bacteria per g of oily sludge (MPN g-1 were between 102 and 107) (data not shown).
Isolation of some new bacterial strains from Poeni oily sludge was carried out on minimal medium, using the enriched cultures method, with hydrocarbons mixtures (saturated, monoaromatic, polyaromatic), as sole carbon source. The use of minimal medium, with hydrocarbons mixtures, as sole carbon source, allowed the selective development of Gram-positive (IBBPo1, IBBPo2, IBBPo3) and Gram-negative (IBBPo7, IBBPo10, IBBPo12) hydrocarbon-degrading bacterial strains. The taxonomic affiliation of isolated bacterial strains was determined based on their phenotypic characteristics and also based on the G+C content of the bacterial chromosome (Table 1). Isolated bacterial strains could be included in phylum Actinobacteria (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Proteobacteria (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10 - γ-subclass of Proteobacteria and Burkholderia sp. IBBPo12 - β-subclass of Proteobacteria). Although many bacteria are capable to degrade petroleum hydrocarbons, only a few of them seem to be important for petroleum biodegradation in natural environments. According with literature, these bacteria belong to a new taxonomic group of phylogenetically related oil-degrading Proteobacteria (17, 23, 24, 35, 78, 80) which has been isolated during the last decade from different sites all over the world.
Table 1.
Phenotypic characteristics of Gram-positive and Gram-negative bacterial strains
| Characteristics | Bacterial strains: | Characteristics | Bacterial strains: | ||||||
|---|---|---|---|---|---|---|---|---|---|
| IBBPo1 | IBBPo2 | IBBPo3 | IBBPo7 | IBBPo10 | IBBPo12 | ||||
| Gram reaction | + | + | + | Gram reaction | - | - | - | ||
| Optimal growth temperature | 28°C | 28°C | 28°C | Optimal growth temperature | 28°C | 28°C | 28°C | ||
| Morphology | rods | rods | rods | Morphology | rods | rods | rods | ||
| Endospore formation | - | - | - | Endospore formation | - | - | - | ||
| Mobility | - | - | - | Mobility | + | + | + | ||
| Respirator type | A | A | A | Respirator type | A | A | A | ||
| Pigment formation on: | LB | yellow | - | - | Pigment formation on: | LB | - | green | orange |
| King A | - | - | - | King A | - | + | - | ||
| King B | - | - | - | King B | - | + | - | ||
| Growth on TTC medium | - | - | - | Growth on TTC medium | - | + | - | ||
| Growth on CBB R250 | - | + | - | Growth on CBB R250 | + | - | - | ||
| Catalase | + | + | + | Catalase | + | + | - | ||
| Oxidase | - | - | + | Oxidase | - | + | + | ||
| Nitrates reduction | - | + | - | Nitrates reduction | + | + | + | ||
| Pyrazinamidase | + | + | + | Indole production | - | - | - | ||
| Pyrolidonyl arylamidase | + | + | + | D-glucose fermentation | - | - | + | ||
| Alkaline phosphatase | + | + | + | L-arginine dihydrolase | - | + | - | ||
| β-glucuronidase | - | - | - | Urease | - | + | + | ||
| β-galactosidase | + | - | - | Esculin hydrolysis | + | - | + | ||
| α-glucosidase | + | + | - | Gelatin hydrolysis | + | + | + | ||
| N-acetyl-β-glucosaminidase | - | + | - | β-galactosidase | - | + | + | ||
| β-glucosidase/esculin | + | + | - | D-glucose assimilation | + | + | + | ||
| Urease | - | - | - | L-arabinose assimilation | + | - | + | ||
| Gelatin hydrolysis | + | + | - | D-mannose assimilation | + | - | + | ||
| Fermentation (negative control) | - | - | - | D-mannitol assimilation | + | + | + | ||
| Glucose fermentation | + | + | - | N-acetyl-glucosamine assimilation | + | + | + | ||
| Ribose fermentation | + | + | - | D-maltose assimilation | + | - | + | ||
| Xylose fermentation | + | - | - | Potassium gluconate assimilation | + | + | + | ||
| Mannitol fermentation | + | - | - | Capric acid assimilation | + | + | + | ||
| Maltose fermentation | + | + | - | Adipic acid assimilation | - | + | + | ||
| Lactose fermentation | + | - | - | Malic acid assimilation | + | + | + | ||
| Saccharose fermentation | + | - | - | Trisodium citrate assimilation | - | + | + | ||
| Glycogen fermentation | - | + | - | Phenylacetic acid assimilation | - | - | + | ||
| G+C content (%) | 71.2 | 70.5 | 68.0 | G+C content (%) | 51.1 | 66.6 | 66.9 | ||
| MIC90 (μg ml-1) | Ampicillin | 500 | 800 | 250 | MIC90 (μg ml-1) | Ampicillin | 1000 | 1000 | 1000 |
| Kanamycin | 50 | 50 | 75 | Kanamycin | 45 | 550 | 250 | ||
| SDS | 40 | 75 | 50 | SDS | 1000 | 1000 | 1000 | ||
| Rhodamine 6G | 100 | 100 | 100 | Rhodamine 6G | 1000 | 1000 | 1000 | ||
| PCRHAE1 | A24f2-A577r2 | - | - | - | PCRHAE | A24f2-A577r2 | - | + | + |
Legend: + = positive, - = negative, A = aerobic, MIC90 = Minimum Inhibitory Concentration required to inhibit the growth of 90% of the bacteria; PCRHAE1 = PCR amplification of HAE1 (hydrophobe/amphiphile efflux 1) fragment using as template the DNA extracted from Gram-positive and Gram-negative bacterial strains, + = expected size (550 bp) fragment was amplified, - = expected size fragment was not amplified.
The resistance of Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacterial strains to antibiotics (ampicillin, kanamycin) and toxic compounds (sodium dodecyl sulfate, rhodamine 6G) differs from one strain to another (Table 1). Antimicrobial effect of all these compounds was more pronounced for Gram-positive (MIC90 = 40 - 800 μg ml-1) bacteria, probably due to the lack of additional permeability barriers, particularly the outer membrane of Gram-negative (MIC90 = 45 - 1000 μg ml-1) bacteria. Nevertheless, the outer membrane itself does not provide resistance to antimicrobial agents as it only decreases permeability. The resistance to antimicrobial agents is dependent on the other resistance mechanisms such as efflux but these mechanisms have enhanced effectiveness in the presence of the outer membrane in Gram-negative bacteria. The synergy between the efflux pumps and the outer membrane probably explains the variable effectiveness of related multidrug efflux systems in providing resistance in organisms with differences in intrinsic outer membrane permeability properties (60, 81).
Accumulation of rhodamine 6G (1-1000 μg ml-1) in Gram-positive and Gram-negative bacterial cells was observed by the fluorescence of rhodamine 6G under UV light. Rhodamine 6G is P-glycoprotein substrates, which mediate the energy-dependent efflux of certain toxic compounds, from the bacterial cells (51). P-glycoprotein has been shown to bind a large number of lipophilic compounds (antibiotics, dyes, polyaromatic hydrocarbons, polycholorinated biphenyls, heavy metals, pesticides and other toxic compounds) with no apparent structural or functional similarities (8, 74). Both Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria tolerate the presence of rhodamine 6G in culture medium and also they accumulate this toxic compound in bacterial cells (Figure 1a). Gram-negative bacteria were more tolerant (1000 μg ml-1) to rhodamine 6G than Gram-positive bacteria (100 μg ml-1). Rhodamine 6G accumulation in Gram-negative bacteria was higher (75-100%) than rhodamine 6G accumulation in Gram-positive bacteria (50%) (data not shown).
Figure 1.
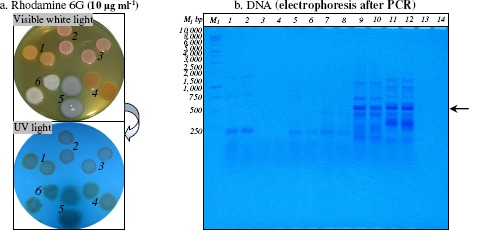
Rhodamine 6G accumulation in Gram-positive and Gram-negative bacterial cells (panel a.) and PCR amplification of HAE1 fragment using as template the DNA extracted from Gram-positive and Gram-negative bacteria (panel b.)
Panel a. After incubation at 28°C for 24 h, bacterial colonies was observed under visible white light and UV light; Mycobacterium sp. IBBPo1 (spots 1), Oerskovia sp. IBBPo2 (spots 2), Corynebacterium sp. IBBPo3 (spots 3), Chryseomonas sp. IBBPo7 (spots 4), Pseudomonas sp. IBBPo10 (spots 5), Burkholderia sp. IBBPo12 (spots 6).
Panel b. 1 kb DNA ladder, Promega (lane M1); Mycobacterium sp. IBBPo1 (lanes 1, 2), Oerskovia sp. IBBPo2 (lanes 3, 4), Corynebacterium sp. IBBPo3 (lanes 5, 6), Chryseomonas sp. IBBPo7 (lanes 7, 8), Pseudomonas sp. IBBPo10 (lanes 9, 10), Burkholderia sp. IBBPo12 (lanes 11, 12), negative control DNA (lanes 13, 14); anneling at 50.0°C (lanes 1, 3, 5, 7, 9, 11, 13), anneling at 52.0°C (lanes 2, 4, 6, 8, 10, 12, 14); the positions of the expected HAE1 fragment (550 bp) are indicated with arrow.
An RND (resistance-nodulation-cell division) efflux pump comprises of three subunits: an RND transporter, a membrane fusion protein and an outer membrane protein. RND pumps have been found in all major domains and constitute a superfamily of transporter proteins with a variety of substrates, including hydrocarbons, antibiotic drugs, and heavy metals. Phylogenetic analysis of RND transporters have shown that they could be separated into seven distinct families, including HAE1 (hydrophobe/amphiphile efflux pumps of Gram-negative bacteria), HME (heavy metal efflux pumps), SecDF (SecDF protein secretion accessory proteins), and NFE (nodulation factor exporters) families (45). To investigate wheather efflux pumps of the RND family were present in new isolated bacterial strains (Figure 1b), primer set A24f2-A577r2 was used to amplify members of the HAE1 family of transporters (45). In order to reduce biases in template/product ratios occurring due to the use of degenerate primers, inosine residues were incorporated into the primers at some degenerate positions. Other studies have suggested that interactions of inosine with the four natural bases exhibit different binding energies, although the differences were much less than the differences between the proper Watson-Crick base pairs and mismatch pairs (45). As anticipated, Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) strains did not amplified bands of the predicted size of 550 bp. Despite the fact that these transporters are widespread among Gram-negative bacterial strains, only Pseudomonas sp. IBBPo10 and Burkholderia sp. IBBPo12 amplified the expected 550 bp fragment, while Chryseomonas sp. IBBPo7 did not amplified bands of the predicted size. It was also obtained the unspecific amplification of other fragments when it was used as template the DNA extracted from Pseudomonas sp. IBBPo10 and Burkholderia sp. IBBPo12. The optimum temperatures for primer annealing were 50.0°C for both Pseudomonas sp. IBBPo10 and Burkholderia sp. IBBPo12.
Tolerance of Gram-positive and Gram-negative bacterial strains to pure and mixture of saturated, monoaromatic and polyaromatic hydrocarbons
The key factor for hydrocarbons degradation is represented by the tolerance that some microorganisms exhibit toward these compounds. Toxicity of hydrocarbons, when they are present in the medium at supersaturating concentrations (second phase), has been correlated with the logarithm of the partition coefficient of the hydrocarbons in a mixture of octanol and water (log POW) (73). Compounds with log POW value between 1.5 and 4 are extremely toxic for microorganisms (14, 57), and within this range, those with lower log POW values are considered more toxic than those with higher log POW values (32). Compounds with log POW below 1 is too low to allow compounds to enter the membrane and above 4 is extremely poorly water soluble and, as it is not bioavailable, it cannot cause any toxic effect (28). Hydrocarbons tolerance does not appear to be related to degradation, as most hydrocarbon-tolerant organisms tolerate a wide variety of aromatic hydrocarbons, alkanes, and alcohols but they are incapable of degrading or transforming them (26, 33, 34, 59).
The tolerance of Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria to pure and mixture of saturated (n-hexane, n-hexadecane, cyclohexane), monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons differ from one strain to another and even for the same bacterial strain according to the nature of the hydrophobic substrate (Table 2). There were observed that aromatic hydrocarbons (benzene, toluene, ethylbenzene, naphthalene, 2-methylnaphthalene, fluorene), with log POW value between 2.14 and 4.49, were more toxic for tested bacterial strains, compared with saturated hydrocarbons (n-hexane, n-hexadecane, cyclohexane), with log POW value between 3.35 and 9.15. The growth of Gram-positive and Gram-negative bacteria on agar LB-Mg medium supplied with hydrocarbons in the vapor phase was 50-100% in the presence of pure and mixture of saturated hydrocarbons, and 25-75% in the presence of pure and mixture of monoaromatic and polyaromatic hydrocarbons. The growth of Gram-positive and Gram-negative bacteria on agar minimal medium supplied with hydrocarbons in the vapor phase was 10-100% in the presence of pure and mixture of saturated hydrocarbons, and 0.01-75% in the presence of pure and mixture of monoaromatic and polyaromatic hydrocarbons. The high sensitivity of Mycobacterium sp. IBBPo1 strain can be observed towards the presence in the vapor phase of pure and mixture of monoaromatic hydrocarbons (0.01%). According with literature (62, 71) hydrocarbon toxicity depends not only on its physicochemical properties but also on the specific response of the cells, and that this cellular response is not the same in all strains.
Table 2.
Tolerance of Gram-positive and Gram-negative bacterial strains to pure and mixture of saturated, monoaromatic and polyaromatic hydrocarbons
| Variant: | log POW1 | Bacterial strains growth on solid medium2with hydrocarbons in vapor phase | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mycobacterium sp. IBBPo1 | Oerskovia sp. IBBPo2 | Corynebacterium sp. IBBPo3 | Chryseomonas sp. IBBPo7 | Pseudomonas sp. IBBPo10 | Burkholderia sp. IBBPo12 | ||||||||
| LBMg | MM | LBMg | MM | LBMg | MM | LBMg | MM | LBMg | MM | LBMg | MM | ||
| Control | - | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% | 100% |
| Cyclohexane | 3.35 | 50% | 25% | 75% | 50% | 50% | 25% | 50% | 10% | 75% | 25% | 50% | 25% |
| n-Hexane | 3.86 | 100% | 75% | 100% | 100% | 100% | 100% | 100% | 50% | 75% | 25% | 100% | 75% |
| n-Hexadecane | 9.15 | 100% | 75% | 100% | 100% | 100% | 100% | 100% | 75% | 100% | 75% | 100% | 75% |
| Mixture of saturated hydrocarbon | - | 100% | 75% | 100% | 100% | 100% | 100% | 100% | 75% | 100% | 100% | 100% | 100% |
| Benzene | 2.14 | 25% | 0.01% | 75% | 50% | 25% | 25% | 25% | 10% | 50% | 10% | 25% | 10% |
| Toluene | 2.64 | 25% | 0.01% | 75% | 75% | 50% | 25% | 25% | 10% | 50% | 10% | 50% | 10% |
| Ethylbenzene | 3.17 | 25% | 0.01% | 75% | 75% | 50% | 50% | 25% | 25% | 75% | 50% | 75% | 50% |
| Mixture of monoaromatic hydrocarbons | - | 25% | 0.01% | 75% | 75% | 75% | 50% | 25% | 10% | 50% | 10% | 50% | 25% |
| Naphthalene | 3.31 | 50% | 50% | 75% | 75% | 50% | 50% | 50% | 25% | 50% | 25% | 75% | 50% |
| 2-Methylnaphthalene | 3.81 | 25% | 10% | 50% | 25% | 75% | 75% | 75% | 25% | 75% | 50% | 75% | 75% |
| Fluorene | 4.49 | 75% | 50% | 75% | 75% | 75% | 75% | 75% | 50% | 75% | 25% | 75% | 50% |
| Mixture of polyaromatic hydrocarbons | - | 50% | 50% | 75% | 50% | 75% | 50% | 75% | 75% | 75% | 25% | 50% | 25% |
Legend: 1 = logarithm of the partition coefficient of the hydrocarbons in octanol-water mixture; 2 = the growth on agar medium (LBMg = LB-Mg medium, MM = minimal medium) was estimated by determining the formation of resistant bacterial colonies and the hydrocarbons resistance is represented by the frequency of colony formation, with that observed in the absence of any hydrocarbon taken as 100%.
Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria isolated from oily sludge were able to tolerate pure and mixture of saturated (n-hexane, n-hexadecane, cyclohexane) hydrocarbons, as well as pure and mixture of monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons (except Mycobacterium sp. IBBPo1). These capabilities of isolated bacterial strains makes them interesting candidates for studying the cellular and molecular modifications induced by mixture of saturated, monoaromatic and polyaromatic hydrocarbons.
Cellular and molecular modifications induced by mixture of saturated, monoaromatic and polyaromatic hydrocarbons on Gram-positive and Gram-negative bacterial strains. Research has begun to uncover the mechanisms responsible for the unique property of hydrocarbons tolerance since the first hydrocarbons-tolerant strain was isolated. Because the membrane is the main target of the toxic action of hydrocarbons, it is not surprising that, along with the discovery of hydrocarbon-tolerant bacteria, changes in the membrane composition were considered to play a pivotal role in the mechanisms contributing to hydrocarbons tolerance (31). When hydrocarbons accumulate in the membrane, its integrity is affected, eventually resulting in loss of function as a permeability barrier, as a protein and reaction matrix and as an energy transducer concomitantly leading to damages of the cellular metabolism, growth inhibition, and, finally, cell death (34). Hydrocarbons accumulating within the hydrophobic layer of the membrane cause an increase in fluidity (29, 30, 74, 85), which leads to inactivation and denaturation of membrane embedded proteins, such as ion pumps and ATPases, it provokes leakage of ions and intracellular macromolecules, such as RNA, phospholipids, and proteins (34). Increase in membrane permeability is considered the main reason for cell death (28, 29, 74).
Modifications induced by mixture of hydrocarbons to cells viability
To further characterize the hydrocarbons tolerance of Gram-positive and Gram-negative bacterial strains, the cell viability was determined in the presence of 5% (v/v) mixture of saturated (n-hexane, n-hexadecane, cyclohexane) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons and 0.5% (v/v) mixture of monoaromatic (benzene, toluene, ethylbenzene) hydrocarbons (Table 3). The maintenance or loss of viability of the cells grown in the presence of mixture of hydrocarbons, estimated by determining the most probable number of bacterial cells on solid medium (with values between 10 and 109 CFU ml-1), were correlated with growth (OD660 with values between 0.150 and 1.141). The survival rate of Gram-positive (Mycobacterium sp. IBBPo1, Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) bacteria was 104-107 CFU ml-1 in the presence of mixture of saturated hydrocarbons, whereas in the presence of mixture of monoaromatic and polyaromatic hydrocarbons the survival rate was 10-104 CFU ml-1. The survival rate of Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria was 107-109 CFU ml-1 in the presence of mixture of saturated hydrocarbons, whereas in the presence of mixture of monoaromatic and polyaromatic hydrocarbons the survival rate was 103-107 CFU ml-1. As mentioned, Gram-negative bacteria are less sensitive to hydrocarbons than Gram-positive bacteria. However, no differences between Gram-positive and Gram-negative bacteria were observed with regard to the critical concentration of molecules dissolved in the cytoplasmic membrane (28, 34, 79). Hence, the differences in hydrocarbons tolerance must be based on other alterations.
Table 3.
The cell viability modifications of Gram-positive and Gram-negative bacteria in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons
| Bacterial strain | Variant: | |||||||
|---|---|---|---|---|---|---|---|---|
| Control | Mixture of saturated hydrocarbon1 (5% v/v) | Mixture of monoaromatic hydrocarbons2 (0.5% v/v) | Mixture of polyaromatic hydrocarbons3 (5% v/v) | |||||
| OD6604 | CFU ml-1* | OD6604 | CFU ml-1* | OD6604 | CFU ml-1* | OD6604 | CFU ml-1* | |
| Mycobacterium sp. IBBPo1 | 0.893 | 2.1×109 | 0.369 | 2.1×104 | 0.150 | 1.4×10 | 0.250 | 1.2×102 |
| Oerskovia sp.IBBPo2 | 0.984 | 2.3×109 | 0.650 | 2.1×106 | 0.395 | 3.3×104 | 0.177 | 2.6×102 |
| Corynebacterium sp. IBBPo3 | 1.010 | 2.6×109 | 0.745 | 3.2×107 | 0.185 | 1.2×102 | 0.235 | 1.6×103 |
| Chryseomonas sp. IBBPo7 | 0.996 | 2.5×109 | 0.735 | 2.0×107 | 0.786 | 2.4×107 | 0.311 | 1.1×103 |
| Pseudomonas sp. IBBPo10 | 1.498 | 3.6×109 | 1.141 | 1.3×109 | 0.809 | 3.4×107 | 0.511 | 2.5×105 |
| Burkholderia sp. IBBPo12 | 0.938 | 2.4×109 | 0.712 | 3.2×108 | 0.622 | 1.2×106 | 0.705 | 1.5×107 |
Legend: 1 = mixture of n-hexane, n-hexadecane, cyclohexane; 2= mixture of benzene, toluene, ethylbenzene; 3 = mixture of naphthalene, 2-methylnaphthalene, fluorene; 4 = the intensity of bacterial growth on liquid LB-Mg medium, were determined by spetrophotometric measurement of the optical density (OD660) after 24 hours incubation at 28°C on a rotary shaker (150-200 rpm); * = serial dilutions of culture liquid were spread on agar LB-Mg medium and the number of viable cells (CFU ml-1) was determined after 24 hours incubation at 28°C.
Effects of mixture of hydrocarbons to β-galactosidase activity
After Gram-positive and Gram-negative cells permeabilization with chloroform and sodium dodecyl sulfate, o-nitrophenyl-β-D-galactoside diffused into the cells and it was hydrolyzed by the β-galactosidase enzyme entrapped within the cell. Mycobacterium sp. IBBPo1, Pseudomonas sp. IBBPo10 and Burkholderia sp. IBBPo12 were β-galactosidase-positive, while Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3 and Chryseomonas sp. IBBPo7 were β-galactosidase-negative. The level of β-galactosidase activity was measured in the late exponential phase of cultures exposed to mixture of hydrocarbons. β-galactosidase activity measurements revealed that the lacZ gene was induced in Mycobacterium sp. IBBPo1, Pseudomonas sp. IBBPo10 and Burkholderia sp. IBBPo12 grown in the presence of 5% (v/v) mixture of saturated (n-hexane, n-hexadecane, cyclohexane) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons and 0.5% (v/v) mixture of monoaromatic (benzene, toluene, ethylbenzene) hydrocarbons (Table 4). Mixture of hydrocarbons induced β-galactosidase activity in Mycobacterium sp. IBBPo1 to a level between 1.6 and 3.9 times greater than basal level; in Pseudomonas sp. IBBPo10 to a level between 1.5 and 1.8 times greater than basal level; and in Burkholderia sp. IBBPo12 to a level between 1.3 and 1.9 times greater than basal level.
Table 4.
β-galactosidase activity of Gram-positive and Gram-negative bacteria in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons
| Bacterial strain | Variant: | |||||||
|---|---|---|---|---|---|---|---|---|
| Control | Mixture of saturated hydrocarbon1 (5% v/v) | Mixture of monoaromatic hydrocarbons2 (0.5% v/v) | Mixture of polyaromatic hydrocarbons3 (5% v/v) | |||||
| β-Gal4 | Ind. fold | β-Gal4 | Ind. fold | β-Gal4 | Ind. fold | β-Gal4 | Ind. fold | |
| Mycobacterium sp. IBBPo1 | 10.7±0.5 | 1.0 | 39.3±0.7 | 3.7 | 42.2±0.6 | 3.9 | 17.1±0.8 | 1.6 |
| Oerskovia sp.IBBPo2 | - | - | - | - | - | - | - | - |
| Corynebacterium sp. IBBPo3 | - | - | - | - | - | - | - | - |
| Chryseomonas sp. IBBPo7 | - | - | - | - | - | - | - | - |
| Pseudomonas sp. IBBPo10 | 8.6±0.7 | 1.0 | 13.2±0.5 | 1.5 | 15.9±0.9 | 1.8 | 13.6±0.8 | 1.6 |
| Burkholderia sp. IBBPo12 | 9.7±0.6 | 1.0 | 12.2±0.7 | 1.3 | 17.6±0.6 | 1.8 | 18.7±0.9 | 1.9 |
Legend: 1= mixture of n-hexane, n-hexadecane, cyclohexane; 2= mixture of benzene, toluene, ethylbenzene; 3 = mixture of naphthalene, 2-methylnaphthalene, fluorene; 4= β-galactosidase (β-Gal) activity, expressed as the change in absorbance at 420 nm minute-1 milliliter of cells-1 optical density units at 650 nm-1, was measured in permeabilized cells and was calculate the induction fold (Ind. fold).
Several hydrophobic organic solvents, including aromatic compounds, aliphatic compounds, and aliphatic alcohols, were able to induce the srp-lacZ gene in Pseudomonas putida S12. The level of induction seems to be correlated with increasing side chain length in the case of the alkyl-substituted aromatics and with chain length in the case of the aliphatic alcohols (36). Biphenyl induced β-galactosidase activity in Pseudomonas strain Cam-10 to a level approximately six times greater than the basal level in cells incubated with pyruvate. In contrast, the β-galactosidase activities in Burkholderia strain LB400-1 incubated with biphenyl or pyruvate were indistinguishable. At a concentration of 1 mM, most of the 40 potential inducers tested were inhibitory to induction by biphenyl of β-galactosidase activity in Pseudomonas strain Cam-10. The exceptions were naphthalene, salicylate, 2-chlorobiphenyl, and 4-chlorobiphenyl, which induced β-galactosidase activity in Pseudomonas strain Cam-10, although at levels that were no more than 30% of the levels induced by biphenyl (42). Furthermore, in Escherichia coli (pRD579, pTS174), the upper pathway substrates (e.g., toluene and m- and p-xylene) and the pathway intermediates (e.g., benzyl alcohol and m- and p-methylbenzyl alcohol) induced high levels of β-galactosidase synthesis (1).
Modifications induced by mixture of hydrocarbons to lipids. The thin-layer chromatography (TLC) studies revealed the existence of some differences (motilities and phospholipid headgroups composition) between phospholipids extracted from Gram-positive and Gram-negative bacterial cells incubated without hydrocarbons (control) and those extracted from cells incubated in the presence of 5% (v/v) mixture of saturated and polyaromatic hydrocarbons and 0.5% (v/v) mixture of monoaromatic hydrocarbons (Figure 2a). The phospholipids found, based on their motilities (Rf), in Gram-positive and Gram-negative bacterial cells were phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI) and cardiolipin (CL).
Figure 2.
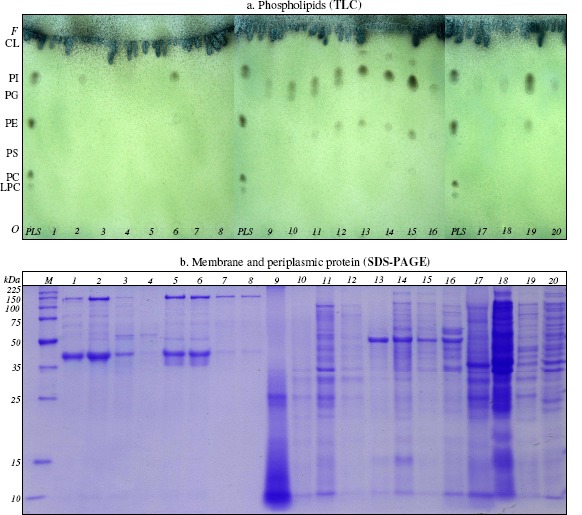
The lipid profile (a.) and protein profile (b.) modifications of Gram-positive and Gram-negative bacteria in the presence of mixture of saturated (n-hexane, n-hexadecane, cyclohexane), monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons Oerskovia sp. IBBPo2 (lanes 1-4), Corynebacterium sp. IBBPo3 (lanes 5-8), Chryseomonas sp. IBBPo7 (lanes 9-12), Pseudomonas sp. IBBPo10 (lanes 13-16), Burkholderia sp. IBBPo12 (lanes 17-20); bacterial strains cultivated onto liquid LB-Mg medium without hydrocarbons (lanes 1, 5, 9, 13, 17); bacterial strains cultivated onto liquid LB-Mg medium with: mixture of saturated hydrocarbons (lanes 2, 6, 10, 14, 18), mixture of monoaromatic (lanes 3, 7, 11, 15, 19), and mixture of polyaromatic hydrocarbons (lanes 4, 8, 12, 16, 20). Panel a. Phospholipids standards, Sigma-Aldrich, Supelco (lane PLS); origin (O), solvent front (F), lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), phosphatidylglycerol (PG), phosphatidylinositol (PI), cardiolipin (CL). Panel b. Broad range protein molecular weight marker, Promega (lane M).
The phospholipid found, based on their motilities (Rf), in Oerskovia sp. IBBPo2 cells (control) was CL (Rf = 0.97). The phospholipids found in bacterial cells incubated in the presence of mixture of saturated hydrocarbons were PI (Rf= 0.79) and CL (Rf= 0.97), while the phospholipid found in bacterial cells incubated in the presence of mixture of monoaromatic and polyaromatic hydrocarbons was CL (Rf= 0.93).
The phospholipid found in Corynebacterium sp. IBBPo3 cells (control) was CL (Rf = 0.97). The phospholipids found in bacterial cells incubated in the presence of mixture of saturated hydrocarbons were PE (Rf= 0.59), PI (Rf= 0.81), CL (Rf= 0.97), while the phospholipid found in bacterial cells incubated in the presence of mixture of monoaromatic and polyaromatic hydrocarbons was CL (Rf = 0.96-0.97).
The phospholipids found in Chryseomonas sp. IBBPo7 cells (control) were PE (Rf= 0.48), PG (Rf= 0.71), CL (Rf= 0.96). The phospholipids found in bacterial cells incubated in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons were the same, but with modified motility: PE (Rf= 0.44-0.52), PG (Rf= 0.70-0.76), CL (Rf= 0.96-0.97).
The phospholipids found in Pseudomonas sp. IBBPo10 cells (control) were PS (Rf = 0.36), PE (Rf = 0.53), PG (Rf = 0.78), PI (Rf = 0.86) and CL (Rf = 0.95). The phospholipids found in bacterial cells incubated in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons were PS (Rf = 0.30-0.33), PE (Rf = 0.48-0.51), PG (Rf = 0.73-0.77), PI (Rf = 0.83-0.86), CL (Rf = 0.93-0.96). There were observed the absence of PS and PI in the bacterial cells incubated in the presence of mixture of plyaromatic hydrocarbons.
The phospholipids found in Burkholderia sp. IBBPo12 cells (control) were PG (Rf = 0.72) and CL (Rf = 0.97). The phospholipids found in bacterial cells incubated in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons were PE (Rf = 0.48-0.52), PG (Rf = 0.71-0.73), CL (Rf = 0.96-0.99).
Cell wall lipids play a crucial role in the uptake of hydrophobic carbon sources. In addition to membrane abnormalities caused by specific inhibitors of lipid synthesis, structural changes in cell wall lipids occur in response to various stress conditions (75). In the cytoplasmic membrane, changes at the level of the lipids and proteins have been observed. These adaptations reestablish the stability and fluidity of the membrane once it is disturbed by the hydrocarbons after hydrocarbon exposure (84). In principle, several mechanisms are possible and may vary from strain to strain. Four main mechanisms were discovered: degree of saturation of the fatty acids, cis-trans isomerisation of unsaturated fatty acids, composition of phospholipid headgroups, and dynamics of phospholipid turnover. Generally spoken, such adaptation mechanisms prevent the influx of hydrocarbons by decreasing the membrane’s permeability and fluidity (21, 28, 34, 59, 62, 83).
Modifications induced by mixture of hydrocarbons to proteins
To investigate the modifications induced by hydrocarbons to membrane (for Gram-positive and Gram-negative bacteria) and periplasmic (for Gram-negative bacteria) protein profile, one-dimensional sodium dodecyl sulfate polyacrylamide gel electrophoresis (1D SDS-PAGE) was used. Many more types of variation in protein sequences can be distinguished on one-dimensional gels in the absence of denaturants such as urea used in two-dimensional electrophoresis (44). The electrophoresis studies showed the existence of some differences between protein profiles extracted from Gram-positive and Gram-negative bacterial cells incubated without hydrocarbons (control) and those extracted from bacterial cells incubated 24 hours in the presence in the presence of 5% (v/v) mixture of saturated and polyaromatic hydrocarbons and 0.5% (v/v) mixture of monoaromatic hydrocarbons. Modifications induced were different from one strain to another and even for the same bacterial strain according to the nature of the hydrophobic substrate (Figure 2b).
Strong induction of the synthesis of some proteins was observed in the membrane profile of the Oerskovia sp. IBBPo2 cells grown in the presence of mixture of saturated hydrocarbons, while repression of the synthesis of some proteins was observed in the presence of mixture of monoaromatic and polyaromatic hydrocarbons.
No modifications were observed in the membrane protein profile of the Corynebacterium sp. IBBPo3 cells grown in the presence of mixture of saturated hydrocarbons, compared with the control. Strong repression of the synthesis of some proteins was observed in the membrane profile of the cells grown in the presence of mixture of monoaromatic and polyaromatic hydrocarbons.
No modifications were observed in the membrane and periplasmic protein profile of the Chryseomonas sp. IBBPo7 cells grown in the presence of mixture of saturated hydrocarbons, compared with the control. Strong induction of the synthesis of some proteins was observed in the membrane and periplasmic protein profile of the cells grown in the presence of mixture of monoaromatic and polyaromatic hydrocarbons.
Modifications induced by mixture of hydrocarbons to DNA
Exposures of a microbial community to hydrocarbons have been shown to result in an increase in the number of bacterial plasmids types. Indirect evidence for the role of plasmids in adaptation to hydrocarbons has been provided by some studies in which a greater frequency and/or multiplicity of plasmids has been observed among bacterial isolates from hydrocarbon-contaminated environments than among isolates from uncontaminated sites (40). Catabolic plasmids are non-essential genetic elements in so far as viability and reproduction of an organism is concerned, but they do provide a metabolic versatility not normally present in the cell. Such genetic potential allows for the evolution of integrated and regulated pathways for the degradation of hydrocarbons (55).
There were observed, by agarose gel electrophoresis studies, the existence of some plasmids in Gram-positive (Corynebacterium sp. IBBPo3) and Gram-negative bacteria (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10). Despite in a high tolerance of Oerskovia sp. IBBPo2 and Burkholderia sp. IBBPo12 to mixture of saturated, monoaromatic and polyaromatic hydrocarbons, these bacterial strains do not harbor plasmid. According with the literature, most of hydrocarbon-tolerant bacteria do not harbor plasmids, indicating that the resistance factors reside on the chromosome (12, 59). Carney and Leary (10) observed that maintenance of phenotype in Pseudomonas putida R5-3 is achieved through a complex system involving an interaction between plasmid and chromosomal DNA. The particular aromatic hydrocarbon substrate utilized as the sole carbon source apparently induced the alterations in the biodegradative plasmids. Restriction enzyme analysis and Southern blot hybridizations of total plasmid DNA indicated deletions and rearrangements of DNA restriction fragments in the derivatives maintained on m-xylene and toluene, when compared with the original R5-3A. Southern blot hybridizations revealed that part of the plasmid DNA lost from the original plasmid profile was integrated into the chromosomal DNA of xylene grown R5-3B and that these plasmid fragments were associated with aromatic hydrocarbon metabolism (10).
The spectral studies revealed the existence of some differences between DNA extracted from Gram-positive and Gram-negative bacterial cells incubated without hydrocarbons (control) and those extracted from cells incubated in the presence of 5% (v/v) mixture of saturated and polyaromatic hydrocarbons and 0.5% (v/v) mixture of monoaromatic hydrocarbons. Modifications induced were different from one strain to another and even for the same bacterial strain, according to the nature of the hydrophobic substrate (Figure 3).
Figure 3.
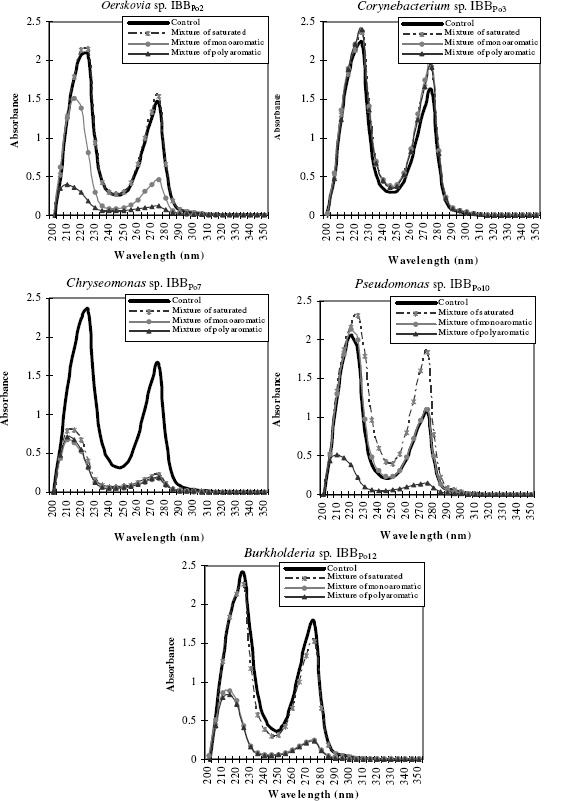
The DNA modifications of Gram-positive (Oerskovia sp. IBBPo2, Corynebacterium sp. IBBPo3) and Gram-negative (Chryseomonas sp. IBBPo7, Pseudomonas sp. IBBPo10, Burkholderia sp. IBBPo12) bacteria in the presence of mixture of saturated (n-hexane, n-hexadecane, cyclohexane), monoaromatic (benzene, toluene, ethylbenzene) and polyaromatic (naphthalene, 2-methylnaphthalene, fluorene) hydrocarbons. DNA concentration of each variant was adjusted to 100 μg ml-1 for spectral comparison (OD200nm-OD350nm).
There are no modifications of the spectrum of the DNA extracted from Oerskovia sp. IBBPo2 and Burkholderia sp. IBBPo12 cells cultivated in the absence of mixture of hydrocarbons (control) and the spectrum of the DNA extracted from bacterial cells cultivated in the presence of mixture of saturated hydrocarbons. It was observed the existence of some modifications of the spectrum of the DNA extracted from the bacterial cells grown in the presence of mixture of monoaromatic and polyaromatic hydrocarbons.
Only few modifications were observed in the spectrum of the DNA extracted from Corynebacterium sp. IBBPo3 cells cultivated in the absence of mixture of hydrocarbons (control) and those extracted from bacterial cells cultivated in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons.
There were observed significant modifications of the spectrum of the DNA extracted from Chryseomonas sp. IBBPo7 cells cultivated in the absence of mixture of hydrocarbons (control) and the spectrum of the DNA extracted from bacterial cells cultivated in the presence of mixture of saturated, monoaromatic and polyaromatic hydrocarbons.
There were no significant modifications of the spectrum of the DNA extracted from Pseudomonas sp. IBBPo10 cells cultivated in the absence of mixture of hydrocarbons (control) and the spectrum of the DNA extracted from bacterial cells cultivated in the presence of mixture of monoaromatic hydrocarbons. It was observed the existence of some modifications of the spectrum of the DNA extracted from the bacterial cells grown in the presence of mixture of saturated and polyaromatic hydrocarbons.
There were no significant modifications of the spectrum of the DNA extracted from Pseudomonas sp. IBBPo10 cells cultivated in the absence of mixture of hydrocarbons (control) and the spectrum of the DNA extracted from bacterial cells cultivated in the presence of mixture of monoaromatic hydrocarbons. It was observed the existence of some modifications of the spectrum of the DNA extracted from the bacterial cells grown in the presence of mixture of saturated and polyaromatic hydrocarbons.
According with literature (19, 39, 86) the hydrocarbons often produce lesions on DNA level (in purine base). By DNA sequence analysis, activated polyaromatic hydrocarbons have been found to induce G-C and T-A base pair insertions and deletions (86). Aromatic hydrocarbons are metabolically activated in cells to yield highly reactive bay region dihydrodiol epoxide derivatives. Dihydrodiol epoxides are electrophilic and can effectively attack DNA, forming covalently linked bulky adducts on DNA bases. This adducts cause structural changes in DNA, thus leading to disruption of normal cellular functions, such as transcription and replication. Furthermore, if not repaired, damaged nucleotides can result in mutations during replication (19, 86).
Obviously, the variety of presented hydrocarbons adaptation mechanisms implies that bacterial hydrocarbon tolerance cannot possibly be provided by one single mechanism. Only the combination of different mechanisms leads to hydrocarbon tolerance; a cascade of short-term and long-term mechanisms acts jointly to reach a complete adaptation. Hence, studying hydrocarbon-tolerant bacteria and, particularly, the molecular mechanisms enabling them to withstand such rather hostile environmental conditions, will not only contribute to further developing efficient two-phase biotransformation systems with whole cells but will also provide deep new insights into the general stress response of bacteria (28). The genome sequence divulges mechanisms contributing to metabolic fitness that allow these microorganisms to grow in a variety of ecosystems and begins to explain how these bacteria are able to survive in highly polluted environments (72).
This study is relevant for better understanding the adaptation of bacteria inhabiting polluted environments to environmental fluctuations of nutrients, for developing and implementing adequate bio-strategies for the remediation of oily sludge contaminated environments. Further studies will be carried out, as well as the genomic DNA will be screened by PCR for the presence of catabolic genes involved in known hydrocarbon biodegradative pathways.
ACKNOWLEDGMENTS
This study was supported by the grant of the Romanian Academy No. 75/2007, 72/2008.
REFERENCES
- 1.Abril, M.A.; Michan, C.; Timmis, K.N.; Ramos, J.L. (1989). Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 171, 6782-6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, O.R.; Gorrell, T.; Bergen, A.; Kruzansky, R.; Levandoswsky, M. (2001). Naked amoebas and bacteria in an oil-impacted salt marsh community. Microbial Ecology 42, 474-481. [DOI] [PubMed] [Google Scholar]
- 3.Aono, R.; Kobayashi, H. (1997). Cell surface properties of organic solvent-tolerant mutants of Escherichia coli K-12. Appl. Environ. Microbiol. 63, 3637-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbeau, C.; Deschenes, L.; Karamanev, D.; Comeau, Y.; Samson, R. (1997). Bioremediation of pentachlorophenol-contaminated soil by bioaugmentation using activated soil. Appl. Microbiol. Biotechnol. 48, 745-752. [DOI] [PubMed] [Google Scholar]
- 5.Benning, C.; Somerville, C.R. (1992). Isolation and genetic complementation of a sulfolipid-deficient mutant of Rhodobacter sphaeroides J. Bacteriol. 174, 352-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berekaa, M.M.; Steinbuchel, A. (2000). Microbial degradation of the multiply branched alkane 2,6,10,15,19,23-hexamethyltetracosane (squalane) by Mycobacterium fortuitum and Mycobacterium ratisbonense. Appl. Environ. Microbiol. 66, 4462-4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya, D.; Sarma, P.M.; Krishnan, S.; Mishra, S.; Lal, B. (2003). Evaluation of genetic diversity among Pseudomonas citronellolis strains isolated from oily sludge-contaminated sites. Appl. Envir. Microbiol. 69, 1435-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borges-Walmsley, M.I.; McKeegan, K.S.; Walmsley, A.R. (2003). Structure and function of efflux pumps that confer resistance to drugs. Biochem. J. 376, 313-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248-254. [DOI] [PubMed] [Google Scholar]
- 10.Carney, B.F.; Leary, J.V. (1989). Novel alterations in plasmid DNA associated with aromatic hydrocarbon utilization by Pseudomonas putida R5-3. Appl. Environ. Microbiol. 55, 1523-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung, P.Y.; Kinkle, B.K. (2001). Mycobacterium diversity and pyrene mineralization in petroleum-contaminated soils. Appl. Environ. Microbiol. 67, 2222-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cruden, D.L.; Wolfram, J.H.; Rogers, R.D.; Gibson, D.T. (1992). Physiological properties of a Pseudomonas strain which grows with o-xylene in a two-phase (organic-aqueous) medium. Appl. Environ. Microbiol. 58, 2723-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Ley, J. (1976). The quick approximation of DNA base composition from absorbancy ratios. Antonie van Leeuwenhoek 33, 203-208. [DOI] [PubMed] [Google Scholar]
- 14.de Smet, M.J.; Kingma, J.; Witholt, B. (1978). The effect of toluene on the structure and permeability of the outer and cytoplasmic membranes of Escherichia coli Biochim. Biophys. Acta 506, 64-80. [DOI] [PubMed] [Google Scholar]
- 15.Dibble, T.; Bartha R. (1979). Effect of environmental parameters on the biodegradation of oil sludge. Appl. Envir. Microbiol. 37, 729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eriksson, M.; Dalhammar, G.; Borg-Karlson, A.-K. (1999). Aerobic degradation of hydrocarbon mixture in natural uncontaminated potting soil by indigenous microorganisms at 20°C and 6°C. Appl. Microbiol. Biotechnol. 51, 532-535. [DOI] [PubMed] [Google Scholar]
- 17.Espuny, M.; Egido, S.; Rodón, I.; Manresa, A.; Mercadé, M. (1996). Nutritional requirements of a biosurfactant producing strain Rhodococcus sp. 51T7. Biotechnol. Lett. 18, 521-526. [Google Scholar]
- 18.Garthright, W.E.; Blodgett, R.J. (2003). FDA's preferred MPN methods for standard, large or unusual tests, with a spreadsheet. Food Microbiology 20, 439-445. [Google Scholar]
- 19.Govindaswami, M.; Feldhake, D.J.; Kinkle, B.K.; Mindell, D.P.; Loper, J.C. (1995). Phylogenetic comparison of two polycyclic aromatic hydrocarbon-degrading mycobacteria. Appl. Environ. Microbiol. 61, 3221–3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grifoll, M.; Selifonov, S.A.; Gatlin, C.V.; Chapman, P.J. (1995). Actions of a versatile fluorene-degrading bacterial isolate on polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 61, 3711-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutierrez, J.A.; Nichols, P.; Couperwhite, I. (1999). Changes in whole cell-derived fatty acids induced by benzene and occurrence of the unusual 16:1 omega 6c in Rhodococcus sp 33. FEMS Microbiol Lett. 176, 213-218. [Google Scholar]
- 22.Haines, J.R.; Wrenn, B.A.; Holder, E.L.; Strohmeier, K.L.; Herrington, RT.; Venosa, A.D. (1996). Measurement of hydrocarbon-degrading microbial populations by a 96-well plate most-probable-number procedure. J. Ind. Microbiol. 16, 36-41. [DOI] [PubMed] [Google Scholar]
- 23.Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. (2005). Diversity and functional analysis of bacterial communities associated with natural hydrocarbon seeps in acidic soils at Rainbow Springs, Yellowstone National Park. Appl. Environ. Microbiol. 71, 5943-5950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamamura, N.; Olson, S.H.; Ward, D.M.; Inskeep, W.P. (2006). Microbial population dynamics associated with crude-oil biodegradation in diverse soils. Appl. Environ. Microbiol. 72, 6316-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harrop, A.J.; Hocknull, M.D.; Lilly, M.D. (1989). Biotransformations in organic solvents: a difference between Gram-positive and Gram-negative bacteria. Biotechnol. Lett. 11, 807-810. [Google Scholar]
- 26.Hearn, E.M.; Dennis, J.J.; Gray, R.M.; Foght, J.M. (2003). Identification and characterization of the emhABC efflux system for polycyclic aromatic hydrocarbons in Pseudomonas fluorescens cLP6a. J. Bacteriol. 185, 6233-6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heipieper, H.J.; Diefenbach, R.; Keweloh, H. (1992). Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58, 1847-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heipieper, H.J.; Meulenbeld, G.; van Oirschot, Q.; de Bont, J.A.M. (1996). Effect of environmental factors on the trans/cis ratio of unsaturated fatty acids in Pseudomonas putida S12. Appl. Environ. Microbiol. 62, 2773-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heipieper, H.J.; Neumann, G.; Cornelissen, S.; Meinhardt, F. (2007). Solvent-tolerant bacteria for biotransformations in two-phase fermentation systems. Appl. Microbiol. Biotechnol. 74, 961-973. [DOI] [PubMed] [Google Scholar]
- 30.Heipieper, H.J.; Weber, F.J.; Sikkema, J.; Keweloh, H.; de Bont, J.A.M. (1994). Mechanisms behind resistance of whole cells to toxic organic solvents. Trends Biotechnol. 12, 409-415. [Google Scholar]
- 31.Inoue, A., Horikoshi, K. (1991). Estimation of solvent-tolerance of bacteria by the solvent parameter log P J. Ferment. Bioeng. 71, 194-196. [Google Scholar]
- 32.Inoue, A.; Horikoshi, K. (1989). A Pseudomonas thrives in high concentration of toluene. Nature 338, 264-266. [Google Scholar]
- 33.Inoue, A.; Yamamoto, M.; Horikoshi, K. (1991). Pseudomonas putida which can grow in the presence of toluene. Appl. Environ. Microbiol. 57, 1560-1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isken, S.; de Bont, J.A.M. (1998). Bacteria tolerant to organic solvents. Extremophiles 2, 229-238. [DOI] [PubMed] [Google Scholar]
- 35.Kalscheuer, R.; Stöveken, T.; Malkus, U.; Reichelt, R.; Golyshin, P.N.; Sabirova, J.S.; Ferrer, M.; Timmis, K.N.; Steinbüchel, A. (2007). Analysis of storage lipid accumulation in Alcanivorax borkumensis: evidence for alternative triacylglycerol biosynthesis routes in bacteria. J. Bacteriol. 189, 918-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kieboom, J.; Dennis, J.J.; Zylstra, G.J.; de Bont, J.A.M. (1998). Active efflux of organic solvents in Pseudomonas putida S12 is induced by solvents. J. Bacteriol. 180, 6769-6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, K.; Lee, L.; Lee, K.; Lim, D. (1998). Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J. Bacteriol. 180, 3692-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim, S.-J.; Jones, R.C.; Cha, C.-J.; Kweon, O.; Edmondson, R.D.; Cerniglia, C.E. (2004). Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics - Clinical Applications 4, 3899-3908. [DOI] [PubMed] [Google Scholar]
- 39.Lakshman, M.K. (2005). Synthesis of biologically important nucleoside analogs by palladium-catalyzed C-N bond-formation. Current Organic Synthesis 2, 83-112. [Google Scholar]
- 40.Leahy, J.G.; Colwell, R.R. (1990). Microbial degradation of hydrocarbons in the environment. Microbiol. Rev. 54, 305-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacNaughton, S.J.; Stephen, J.R.; Venosa, A.D.; Davis, G.A.; Chang, Y.J.; White, D.C. (1999). Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65, 3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Master, E.R.; Mohn, W.W. (2001). Induction of bphA, encoding biphenyl dioxygenase, in two polychlorinated biphenyl-degrading bacteria, psychrotolerant Pseudomonas strain Cam-1 and mesophilic Burkholderia strain LB400. Appl. Environ. Microbiol. 67, 2669-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Matsumoto, M.; de Bont, J.A.M.; Isken, S. (2002). Isolation and characterization of the solvent-tolerant Bacillus cereus strain R1. J. Biosci. Bioeng. 94, 45-51. [DOI] [PubMed] [Google Scholar]
- 44.McLellan, T.; Ames, G.F.-L.; Nikaido, K. (1983). Genetic variation in proteins: comparison of onedimensional and two-dimensional gel electrophoresis. Genetics 104, 381-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meguro, N.; Kodama, Y.; Gallegos, M.T.; Watanabe, K. (2005). Molecular characterization of resistance-nodulation-division transporters from solvent- and drug-resistant bacteria in petroleum-contaminated soil. Appl. Environ. Microbiol. 71, 580-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller, J.H. (1972). Assay of β-galactosidase. In: Experiments in Molecular Genetics Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, p.352-355. [Google Scholar]
- 47.Mishra, S.; Jyot, J.; Kuhad, R.C.; Lal, B. (2001). Evaluation of inoculum addition to stimulate in situ bioremediation of oily sludge-contaminated soil. Appl. Environ. Microbiol. 67, 1675-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moriya, K.; Horikoshi, K. (1993). A benzene-tolerant bacterium utilizingsulfur compounds isolated from deep sea. J. Ferment. Bioeng., 76: 397-399. [Google Scholar]
- 49.Nakajima, H.; Kobayashi, H.; Aona, R.; Horikoshi, K. (1992). Effective isolation and identification of toluene-tolerant Pseudomonas strains. Biosci. Biotechnol. Biochem. 56, 1872-1873. [Google Scholar]
- 50.Nielsen, L.E.; Kadavy, D.R.; Rajagopal, S.; Drijber, R.; Nickerson, K.W. (2005). Survey of extreme solvent tolerance in Gram-positive cocci: membrane fatty acid changes in Staphylococcus haemolyticus grown in toluene. Appl. Environ. Microbiol. 71, 5171-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishino, K.; Yamaguchi, A. (2004). Role of histone-like protein H-NS in multidrug resistance of Escherichia coli J. Bacteriol. 186, 1423-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Norman, R.S.; Moeller, P.; McDonald, T.J.; Morris, P.J. (2004). Effect of pyocyanin on a crude-oil-degrading microbial community. Appl. Envir. Microbiol. 70, 4004-4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ogino, H.; Miyamoto, K.; Ishikawa, H. (1994). Organic solvent-tolerant bacterium which secretes an organic solvent-stable lipolytic enzyme. Appl. Environ. Microbiol. 60, 3884-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ohta, Y.; Maeda, M.; Kudo, T.; Horikoshi, K. (1996). Isolation and characterization of solvent-tolerant bacteria which can degrade biphenyl/polychlorinated biphenyls. J. Gen. Appl. Microbiol. 42, 349-354. [Google Scholar]
- 55.Okoh, A.I. (2006). Biodegradation alternative in the cleanup of petroleum hydrocarbon pollutants. Biotechnol. Molec. Biol. Rev. 1, 38-50. [Google Scholar]
- 56.Olivera, N.L.; Esteves, J.L.; Commendatore, M.G. (1997). Alkane biodegradation by a microbial community from contaminated sediments in Patagonia, Argentina. Int. Biodeterior. Biodegrad. 40, 75-79. [Google Scholar]
- 57.Osborne, S.J.; Leaver, J.; Turner, M.K.; Dunnill, P. (1990). Correlation of biocatalytic activity in an organic-aqueous two-liquid phase system with solvent concentration in the cell membrane. Enzyme. Microb. Technol. 12, 281-291. [DOI] [PubMed] [Google Scholar]
- 58.Paje, M.L.; Neilan, B.A.; Couperwhite, I. (1997). A Rhodococcus species that thrives on medium saturated with liquid benzene. Microbiology 143, 2975-2981. [DOI] [PubMed] [Google Scholar]
- 59.Pinkart, H.C.; White, D.C. (1997). Phospholipid biosynthesis and solvent tolerance in Pseudomonas putida strains. J. Bacteriol. 179, 4219-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poole, K. (2005). Efflux-mediated antimicrobial resistance. J. Antimicrob. Chemother. 56, 20-51. [DOI] [PubMed] [Google Scholar]
- 61.Propst, T.L.; Lochmiller, R.L.; Qualis, C.W.Jr.; McBee, K. (1999). In situ (mesocosm) assessment of immunotoxicity risks to small mammals inhabiting petrochemical waste sites. Chemosphere 38, 1049-1067. [DOI] [PubMed] [Google Scholar]
- 62.Ramos, J.L.; Duque, E.; Gallegos, M.T.; Godoy, P.; Ramos-Gonzalez, M.I.; Rojas, A. (2002). Mechanisms of solvent tolerance in Gram-negative bacteria. Annu. Rev. Microbiol. 56, 743-768. [DOI] [PubMed] [Google Scholar]
- 63.Ramos, J.L.; Duque, E.; Huertas, M.J.; Haídour, A. (1995). Isolation and expansion of the catabolic potential of a Pseudomonas putida strain able to grow in the presence of high concentrations of aromatic hydrocarbons. J. Bacteriol. 177, 3911-3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramos, J.L.; Duque, E.; Rodriguez-Hervas, J.J.; Godoy, P.; Haidour, A.; Reyes, F.; Fernández-Barrero, A. (1997). Metabolism for solvent tolerance in bacteria. J. Biol. Chem. 272 3887-3890. [DOI] [PubMed] [Google Scholar]
- 65.Reva, O.N.; Weinel, C.; Weinel, M.; Bohm, K.; Stjepandic, D.; Hoheisel, J.D.; Tummler, B. (2006). Functional genomics of stress response in Pseudomonas putida KT2440. J. Bacteriol. 188, 4079-4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sambrook, J.; Fritsch, E.F.; Maniatis, T. (1989). Molecular Cloning, A Laboratory Manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York. [Google Scholar]
- 67.Santos, P.M.; Benndorf , D.; Sa-Correia, I. (2004). Insights into Pseudomonas putida KT2440 response to phenol-induced stress by quantitative proteomics. Proteomics 4, 2640-2652. [DOI] [PubMed] [Google Scholar]
- 68.Sardessai, Y.; Bhosle, S. (2002). Organic solvent tolerant bacteria in mangrove ecosystem. Curr. Sci. 82, 622-623 [Google Scholar]
- 69.Segura, A.; Duque, E.; Mosqueda, G.; Ramos, J.L.; Junker, F. (1999). Multiple responses of Gram-negative bacteria to organic solvents. Environ. Microbiol. 1, 191-198. [DOI] [PubMed] [Google Scholar]
- 70.Segura, A.; Godoy, P.; van Dillewijn, P.; Hurtado, A.; Arroyo, N.; Santacruz, S.; Ramos, J.L. (2005). Proteomic analysis reveals the participation of energy- and stress-related proteins in the response of Pseudomonas putida DOT-T1E to toluene. J. Bacteriol. 187, 5937-5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Segura, A.; Hurtado, A.; Rivera, B.; Lăzăroaie, M.M. (2008). Isolation of new toluene-tolerant marine strains of bacteria and characterization of their solvent-tolerance properties. J. Appl. Microbiol. 104, 1408-1416. [DOI] [PubMed] [Google Scholar]
- 72.Seshadri, R.; Joseph, S.W.; Chopra, A.K.; Sha, J.; Shaw, J.; Graf, J.; Haft, D.; Wu, M.; Ren, Q.; Rosovitz, M.J.; Madupu, R.; Tallon, L.; Kim, M.; Jin, S.; Vuong, H.; Stine, O.C.; Ali, A.; Horneman, A.J.; Heidelberg, J.F. (2006). Genome sequence of Aeromonas hydrophila ATCC 7966T: jack of all trades. J. Bacteriol. 188, 8272-8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sikkema, J.; de Bont, J.A.M.; Poolman, B. (1994). Interactions of cyclic hydrocarbons with biological membranes. J. Biol. Chem. 269, 8022-8028. [PubMed] [Google Scholar]
- 74.Sikkema, J.; de Bont, J.A.M.; Poolman, B. (1995). Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59, 201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sokolovská, I.; Rozenberg, R.; Riez, C.; Rouxhet, P.G.; Agathos, S.N.; Wattiau, P. (2003). Carbon-induced modifications in the mycolic acid content and cell wall permeability of Rhodococcus erythropolis E1. Appl. Environ. Microbiol. 69, 7019-7027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Solano-Serena, F.; Marchal, R.; Casaregola, S.; Vasnier, C.; Lebeault, J.M.; Vandecasteele, J.P. (2000). A Mycobacterium strain with extended capacities for degradation of gasoline hydrocarbons. Appl. Environ. Microbiol. 66, 2392-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Song, H.-G.; Wang, X.; Bartha, R. (1990). Bioremediation potential of terrestrial fuel spills. Appl. Envir. Microbiol. 56, 652-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van Hamme, J.D.; Singh, A.; Ward, O.P. (2003). Recent advances in petroleum microbiology. Microbiol. Mol. Biol. Rev. 67, 503-549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vermuë, M.; Sikkema, J.; Verheul, A.; Bakker, R.; Tramper, J. (1993). Toxicity of homologous series of organic solvents for the Gram-positive bacteria Arthrobacter and Nocardia sp. and Gram-negative bacteria Acinetobacter and Pseudomonas sp. Biotechnol. Bioeng. 42, 747-758. [DOI] [PubMed] [Google Scholar]
- 80.Vinas, M.; Sabate, J.; Espuny, M.J.; Solanas, A.M. (2005). Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 71, 7008-7018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viveiros, M.; Dupont, M.; Rodrigues, L.; Couto, I.; Davin-Regli, A.; Martins, M.; Pagès, J.-M.; Amaral, L. (2007). Antibiotic stress, genetic response and altered permeability of E. coli. PLoS ONE2 (4), e365. 10.1371/journal.pone.0000365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Volkers, R.J.M.; de Jong, A.L.; Hulst, A.G.; van Baar, B.L.M.; de Bont, J.A.M.; Wery, J. (2006). Chemostat-based proteomic analysis of toluene affected Pseudomonas putida S12. Environ. Microbiol. 8, 1674-1679. [DOI] [PubMed] [Google Scholar]
- 83.von Wallbrunn, A.; Heipieper, H.J.; Meinhardt, F. (2002). Cis/trans isomerisation of unsaturated fatty acids in a cardiolipin synthase knock-out mutant of Pseudomonas putida P8. Appl. Microbiol. Biotechnol. 60, 179-185. [DOI] [PubMed] [Google Scholar]
- 84.Weber, F.J.; de Bont, J.A.M. (1996). Adaptation mechanisms of microorganisms to the toxic effects of organic solvents on membranes. Biochim. Biophys. Acta 1286, 225-245. [DOI] [PubMed] [Google Scholar]
- 85.Weber, F.J.; Ooijkaas, L.P.; Schemen, R.M.W.; Hartmans, S.; de Bont, J.A.M. (1993). Adaptation of Pseudomonas putida S12 to high concentrations of styrene and other organic solvents. Appl. Environ. Microbiol. 59, 3502-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei, S.-J.C.; Desai, S.M.; Harvey, R.G.; Weiss, S.B. (1984). Use of short DNA oligonucleotides for determination of DNA sequence modifications induced by benzo[a]pyrene diol epoxide. Proc. Nati. Acad. Sci. USA 81, 5936-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Whyte, L.G.; Greer, C.W.; Inniss, W.E. (1995). Assessment of the biodegradation potential of psychrotrophic microorganisms. Can. J. Microbiol. 42, 99-106. [DOI] [PubMed] [Google Scholar]
- 88.Wrenn, B.; Venosa, A.D. (1996). Selective enumeration of aromatic and aliphatic hydrocarbon degrading bacteria by a most probable number procedure. Can. J. Microbiol. 42, 252-259. [DOI] [PubMed] [Google Scholar]
- 89.Zahir, Z.; Seed, K.D.; Dennis, J.J. (2006). Isolation and characterization of novel organic solvent-tolerant bacteria. Extremophiles 10, 129-138. [DOI] [PubMed] [Google Scholar]


