Abstract
The antimicrobial susceptibility of 212 Salmonella strains isolated from patients and foods was evaluated and 45% were found to be resistant to nalidixic acid. Nalidixic acid resistant strains showed a higher minimal inhibitory concentration for ciprofloxacin than sensitive strains. During the study an increase of strains with reduced susceptibility to ciprofloxacin was also observed.
Keywords: antimicrobial resistance, Minimal Inhibitory Concentration (MIC), quinolones, fluoroquinolones.
Salmonella spp. is recognized as a frequent cause of foodborne disease (22). In Parana State, Brazil, 55 out of 399 districts notified salmonellosis outbreaks between 1999 and 2006. The Enteritidis serovar was responsible for 82% of the outbreaks. Due to its self-limiting nature, gastroenteritis caused by Salmonella usually does not require antimicrobial therapy. However, the use of antimicrobials is necessary for immunocompromised patients, older persons and children or in cases of severe or systemic salmonellosis (3, 18).
The more commonly used drugs for salmonellosis treatment during the last decades were ampicillin, chloramphenicol and sulphametoxazole-trimethoprim. Growing resistance to these antimicrobials significantly reduced their use in clinical medicine and they were gradually substituted by fluoroquinolones (3, 14).
The non-therapeutic use of fluoroquinolones in veterinary medicine, as in prophylactic supplements or growth-promoting agents, can facilitate the selection of resistant bacteria or reduce susceptibility to these antimicrobials. Thus, the use of antimicrobial agents in animals raised for human consumption is, probably, the main cause for the increase and spread of resistant Salmonella strains (1, 9, 12).
The high prevalence of strains resistant to nalidixic acid, a 1st generation quinolone, constitutes a public health concern since most of these strains show reduced susceptibility to fluoroquinolones, and have been associated with treatment failures (8, 15, 18).
The present study was carried out to evaluate the antimicrobial susceptibility profile of 212 Salmonella spp. strains isolated from patients and foods related to outbreaks which occurred between 1999 and 2006 in Parana State, Brazil in order to alert about the more careful use of antibiotics in veterinary and human clinical treatment, especially fluoroquinolones.
All Salmonella strains were obtained from the Laboratório Central do Paraná (LACEN, Curitiba, Parana, Brazil) and serotyped and phagotyped at the Laboratório de Enterobactérias, Departamento de Bacteriologia da Fundação Oswaldo Cruz (FIOCRUZ, Rio de Janeiro, Brazil).
Sixteen different serovars were analysed, including 174 (82%) strains of S. Enteritidis, seven (3.3%) S. Infantis, six (2.9%) S. Typhimurium, six (2.9%) S. Derby, four (1.9%) S. Newport, three (1.4%) S. Johannesburg, two (0.9%) S. London, two (0.9%) S. Pomona and one strain of the following serovars: Anatum, Mbandaka, Oranienburg, Agona, Saintpaul, Heidelberg, 09:12 and Albany. The strains were maintained in Brain-Heart Infusion broth (BHI) (DIFCO®) containing 15% glycerol and stored at -15°C until testing.
Antimicrobial susceptibility was evaluated according to the Kirby-Bauer disk diffusion method as recommended by the Clinical and Laboratory Standards Institute (CLSI) (5). Antimicrobial agents tested were (OXOID®): ampicillin (10 µg); cefotaxime (30 µg), chloramphenicol (30 µg), sulphametoxazole-trimethoprim (25 µg), nalidixic acid (30 µg) and ciprofloxacin (5 µg). The minimal inhibitory concentration for ciprofloxacin was determined with the microdilution method using Müller-Hinton broth (DIFCO®) according to CLSI (5).
All Salmonella strains tested in the present study were susceptible to cefotaxime, chloramphenicol and ciprofloxacin. Ninety five (45%) strains were resistant to nalidixic acid; one (0.5%) to ampicillin and one (0.5%) to sulphametoxazole-trimethoprim. An increase in nalidixic acid resistance was observed from 15% in 1999 to 62.5% in 2006 (Figure 1.). Similar results were previously found by other researchers (6, 17).
Figure 1.
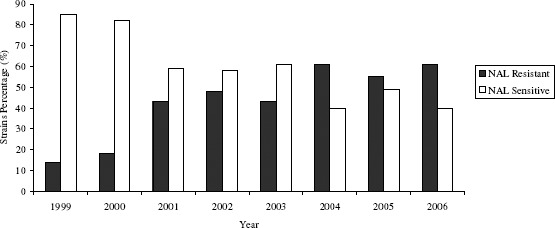
Nalidixic acid (NAL) susceptibility profile for strains associated with salmonellosis outbreaks occurred between 1999 and 2006 in Parana State, Brazil.
Quinolones have been increasingly used to treat human and animal infectious diseases since the introduction of nalidixic acid in the 1960s. The consequence of this widespread use has been an increase in bacterial resistance (11). An increase in nalidixic acid resistance is an indicator for the decrease in ciprofloxacin susceptibility, an antibiotic commonly used in the treatment of Salmonella infections (10, 11, 13).
Although no fluoroquinolone resistant strains were observed in the present study, a significant increase in minimal inhibitory concentration for ciprofloxacin (CipMIC) was verified for the strains resistant to nalidixic acid (Figure 2.). All nalidixic acid resistant strains had higher CipMIC than the sensitive ones. Most of them had CipMIC’s eight times higher, demonstrating the correlation between nalidixic acid resistance increase and fluoroquinolone susceptibility decrease.
Figure 2.
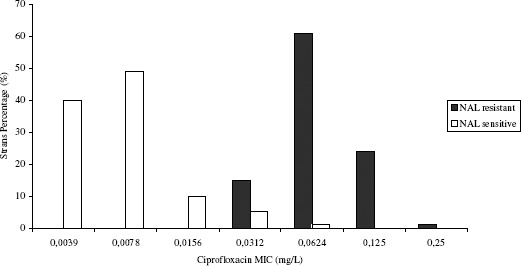
Ciprofloxacin MIC distribution according to nalidixic acid (NAL) susceptibility for Salmonella strains associated with outbreaks occurred between 1999 and 2006 in Parana State, Brazil
Earlier studies showed that strains with MIC 0.125 mg/L had reduced ciprofloxacin susceptibility and were associated with treatment failures (15, 16). In this study, 25% of the strains resistant to nalidixic acid had CipMIC ranging from 0.125 mg/L to 0.250 mg/L.
MIC50 values are important indicators for in vitro antimicrobial activity. Although these values do not necessarily predict in vivo activity, an increase in CipMIC50 from 0.0039 mg/L to 0.0625 mg/L observed during this study indicates a possible reduction in ciprofloxacin potency.
Strains with high MIC for ciprofloxacin have been associated with single point mutations in the quinolone resistant determining region (QRDR) of the gyr A gene that codifies DNA-gyrase enzyme, which is the primary target of fluoroquinolone action (9, 15). Single point mutations may be sufficient to generate high levels of resistance to such non-fluorated quinolones as nalidixic acid. However, apparently additional mutations are necessary to achieve resistance to fluoroquinolones such as ciprofloxacin (7, 12). It is also known that the antimicrobial concentration reduction within the cell due to hyperexpression of the efflux pump system contributes to a heightened quinolone MIC in Salmonella spp. but is insufficient to generate high fluoroquinolone resistance levels (2, 19).
The emergence of Salmonella strains with fluoroquinolones reduced susceptibility is a potential public health risk and may compromise effective antibiotic therapy restricting therapeutic options against infectious diseases (1, 9, 12).
The serovar and phagotype can also influence the antimicrobial resistance levels in Salmonella spp. It has been reported that serovars Hadar, Virchow, Blockley and Typhimurium showed higher resistance fluoroquinolone levels than Enteritidis, the most prevalent serovar in the present study (4, 21).
Threfall (30) found that Enteritidis PT1 isolated in England and Wales exhibited higher reduction in ciprofloxacin susceptibility than other phagotypes. The Enteritidis phagotype PT1 analysed in this study showed higher reduction in ciprofloxacin susceptibility than other phagotypes, such as PT4 (13, 20). An increased incidence of S. Enteritidis PT1 from 52.5% in 1999 to 72.9% in 2006 probably explains the increase of NAL-resistant strains prevalence during this period (Figure 1.).
Regarding the importance of fluoroquinolones for salmonellosis treatment, the results obtained in the present study alert to a more careful use of these antimicrobials in order to maintain their clinical efficacy and broad spectrum.
ACKNOWLEDGEMENTS
The authors thank LACEN for the donation of Salmonella strains, FIOCRUZ for the Salmonella serotyping and phagotyping and Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil (CNPq) for R.B.S.; R.G.F. and M.M graduate scholarships. This work was partially supported by CNPq (Process 476834/2004-0).
REFERENCES
- 1.Angulo F., Johnson K., Tauxe R., Cohen M. Significance and sources of antimicrobial-resistant nontyphoidal Salmonella infections in the United States. Microb. Drug Resist. 2000;1:77–83. doi: 10.1089/mdr.2000.6.77. [DOI] [PubMed] [Google Scholar]
- 2.Biedenbach D.J., Toleman M., Walsh T.R., Jones R.N. Analysis of Salmonella spp. with resistance to extended-spectrum cephalosporin and fluoroquinolones isolated in North America and Latin America: report from the SENTRY Antimicrobial Surveillance Program (1997–2004) Diagn. Microbiol. Infect. Dis. 2006;54:13–21. doi: 10.1016/j.diagmicrobio.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 3.Butaye P., Michael G.B., Schwarz S., Barrett T.J., Brisabois A., White D.G. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 2006;8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 4.Cebrian L., Sirvent E.R., Díaz J.C. Characterization of Salmonella spp. mutants produced by exposure to various fluoroquinolones. Int. J. Antimicrob. Agents. 2003;22:134–139. doi: 10.1016/s0924-8579(03)00099-2. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. PA, USA: CLSI, Wayne; 2006. Performance Standards for Antimicrobial Susceptibility Testing: Fifteenth Informational Supplement M100-S16. [Google Scholar]
- 6.Duarte D.A.M., Ribeiro A.R., Vasconcelos A.M.M., Santos S.B., Silva J.V.D., Patrícia Andrade P.L.A., Falcão L.S.P.C.A. Occurrence of Salmonella spp. in broiler chicken carcasses and their susceptibility to antimicrobial agents. Braz. J. Microbiol. 2009;40:569–573. doi: 10.1590/S1517-838220090003000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eaves D.J., Randall L., Gray D.T., Buckley A., Woodward M.J., White A.P., Piddock L.J.V. Prevalence of Mutations within the Quinolone Resistance-Determining Region of gyrA, gyrB, parC e parE and Association with Antibiotic Resistance in Quinolone-Resistant Salmonella enterica. Antimicrob. Agents Chemother. 2004;48(10):4012–4015. doi: 10.1128/AAC.48.10.4012-4015.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Escribano I., Rodríguez J. C., Cebrian L., Royo G. The importance of active efflux systems in the quinolone resistance of clinical isolates of Salmonella spp. Int. J. Antimicrob. Agents. 2004;24:428–432. doi: 10.1016/j.ijantimicag.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Giraud E., Baucheron S., Cloeckaert A. Resistance to fluoroquinolones in Salmonella: emerging mechanisms and resistance prevention strategies. Microb. Infect. 2006;8:1937–1944. doi: 10.1016/j.micinf.2005.12.025. [DOI] [PubMed] [Google Scholar]
- 10.Hakanen A., Kotilainen P., Huovinen P. Reduced fluoroquinolone susceptibility in Salmonella enterica serotypes in travelers returning from Southeast Asia. Emerging Infect. Dis. 2001;7:996–1003. doi: 10.3201/eid0706.010613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hooper D.C. Emerging Mechanism of Fluoroquinolone Resistance. Emerging Infect. Dis. 2001;7:337–341. doi: 10.3201/eid0702.010239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hopkins K.L., Davies R.H., Threlfall E.J. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: Recent developments. Int. J. Antimicrob. Agents. 2005;25:358–73. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 13.Murray A., Coia J.E., Mather H., Brown D.J. Ciprofloxacin resistance in non-typhoidal Salmonella serotypes in Scotland, 1993–2003. J. Antimicrob. Chemother. 2005;56:110–114. doi: 10.1093/jac/dki164. [DOI] [PubMed] [Google Scholar]
- 14.Piddock L.J.V. Fluoroquinolone resistance: Overuse of fluoroquinolones in human and veterinary medicine can breed resistance. BMJ. 1998;317:1029–1030. doi: 10.1136/bmj.317.7165.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piddock L.V.J. Fluoroquinolone resistance in Salmonella serovars isolated from humans and food animals. FEMS Microbiol.Rew. 2002;26:3–16. doi: 10.1111/j.1574-6976.2002.tb00596.x. [DOI] [PubMed] [Google Scholar]
- 16.Reller M., McClellan J., Joyce K., Polyak C., Mintz E., Angulo F., NARMS WORKING GROUP Emerging resistance to quinolones among Salmonella Typhi isolates in the United States, 1999–2001. Infect. Dis. Soc. Am. 2002 [Google Scholar]
- 17.Ribeiro A.R., Kellermann A., Santos L.R., Fittél A.P., Nascimento V.P. Salmonella spp. in Raw Broiler Parts: Occurrence, Antimicrobial Resistance Profile and Phage Typing of The Salmonella Enteritidis Isolates. Braz. J. Microbiol. 2007;38:296–299. [Google Scholar]
- 18.San-Martín B., Lapierre L., Toro C., Bravo V., Cornejo J., Hormazabal J.C., Borie C. Isolation and molecular characterization of quinolone resistant Salmonella spp. from poultry farms. Vet. Microbiol. 2005;110:239–244. doi: 10.1016/j.vetmic.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Soto S.M., Ruíz J., Mendoza M.C., Vila J. In vitro fluoroquinolone-resistant mutants of Salmonella enterica serotype Enteritidis: analysis of mechanisms involved in resistance. Int. J. Antimicrob. Agents. 2003;22:537–540. doi: 10.1016/s0924-8579(03)00241-3. [DOI] [PubMed] [Google Scholar]
- 20.Threlfall E.J. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiol. Rev. 2002;26:141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 21.Threlfall E.J., Fisher I.S.T., Berghold C. Antimicrobial drug resistance in isolates of Salmonella enterica from cases of salmonellosis in humans in Europe in 2000: results of international multicentre surveillance. Eurosurveillance - European Communicable Disease Bolletin: Salmonella. 2003;82(2):41–45. doi: 10.2807/esm.08.02.00400-en. [DOI] [PubMed] [Google Scholar]
- 22.WHO Global Salm-Surv. Progress report (2000–2005): building capacity for laboratory-based foodborne disease surveillance and outbreak detection and response. 2006 [Google Scholar]


