Abstract
A total of 187 isolates from 470 clinical specimens were collected from three hospitals in El-Minia governorate and identified as 132 Staphylococcus aureus strains and 55 coagulase-negative staphylococci (CoNS) strains. Susceptibility of isolates to antimicrobial agents was tested by the agar dilution method. The isolated S. aureus strains showed low resistance to vancomycin (1.5%), amikacin (2.3%) and gatifloxacin (3.8%). Vancomycin was the most effective antibiotic against CoNS. The ampicillin-resistant isolates were tested for β-lactamase production where, 61.7% of S. aureus and 42.9% of CoNS were positive for β-lactamase enzyme. Beta-lactamase producing strains were screened for their plasmid profile using alkaline lysis method. Some of these strains carried at least one plasmid suggesting plasmid-mediated antibiotic resistance. When cells of these strains were exposed to curing agent ethidium bromide, the production of the β-lactamase was lost. Resistance by efflux was studied by a modified fluorometric assay. Addition of uncoupler carbonyl cyanide m-chlorophenylhydrazone (CCCP) increased norfloxacin accumulation in quinolone resistant S. aureus strains, suggesting endogenous energy-dependent efflux. Combinations of ciprofloxacin with four antimicrobial agents against methicillin resistant S.aureus (MRSA) strains were investigated using decimal assay for additivity (DAA) technique. Synergistic interaction was observed between ciprofloxacin and oxacillin. ciprofloxacin plus cefepime and gentamicin appeared to be additive, while ciprofloxacin plus erythromycin was antagonistic.
Keywords: β-lactamase, CCCP, fluorometric assay, efflux, DAA technique
INTRODUCTION
In the last decade, the staphylococci have again emerged as the predominant organisms causing infections in the hospital setting. Staphylococci are responsible for a variety of medical problems, including skin and soft-tissue infections, surgical site infections, endocarditis and hospital-acquired bacteraemia. An increasing number of infections are related to medical developments, including the use of joint prostheses, immunosuppressants and catheters (3,8).
Staphylococcus aureus is an important pathogen due to a combination of toxin-mediated virulence, invasiveness, and antibiotic resistance. This bacterium is a significant cause of nosocomial infections, as well as community-acquired diseases. The spectrum of staphylococcal infections ranges from folliculitis and furuncles to toxic shock syndrome and sepsis (20). Coagulase-negative staphylococci found in the normal skin flora and mucous membranes has recently got attention as a potential pathogen, specifically for nosocomial infections (34,37).
There is a significant increase in the methicillin-resistant staphylococci infections and these bacteria have recently started to gain resistance to many widely used antibiotics (15,19). In spite of the advancements in the antibacterial treatment field, there are still serious difficulties in the treatment of staphylococcal infections. In several countries, vancomycin-resistant staphylococci have been isolated (6,27).
Efflux systems are one of several mechanisms of resistance described for a variety of bacterial species, including S. aureus. They protect cells from antibiotics by actively transporting compounds out of the cytoplasm and thereby limit their steady-state accumulation at their site(s) of action. It would be valuable to use bacterial efflux pump inhibitors which is inhibitors of resistance mechanisms, able to potentiate the activity of existing antibiotics (1,22,24).
In this study, the incidence of staphylococcal infections in patients attending El-Minia governorate hospitals was detected. We studied the antibiogram of the isolated strains against different antimicrobial agents as an epidemiological indicator and possible mechanisms of resistance of the isolated strains to different antimicrobial agents. The main mechanisms involved in the bacterial resistance in our study were β-lactamase production, plasmid-mediated antibiotic resistance and efflux mechanism. We also studied some possible combinations for management of MRSA infections.
MATERIALS AND METHODS
Study design: A total of 470 clinical specimens were examined; 100 urine specimens (from patients with urinary tract infection), 195 specimens (from patients with respiratory tract infections), 140 specimens (from patients with skin infections) and 35 specimens (from patients with eye infections). The specimens were collected from different hospitals at El-Minia governorate. The present study was designed from July 2005 to January 2008. Identification of staphylococci was based on standard laboratory criteria (colony morphology, hemolytic zones, and production of catalase and coagulase) (11).
Antimicrobial Agents: The antimicrobial agents used in this study were obtained from the following manufacturers: ampicillin and chloramphenicol (Chemical Industries Development; CID, Egypt), amoxicillin and cefotaxime (Egyptian International Pharmaceutical Industries Company; EIPICO, Egypt), ampicillin/sulbactam, oxacillin, ciprofloxacin, ofloxacin, norfloxacin, levofloxacin, gatifloxacin, clindamycin and tetracycline from Sigma-Aldrich (St Louis, MO, USA), amoxycillin/clavulanic acid (Beecham Pharmaceuticals, England), cephalexin and cefuroxime (Glaxo Wellcome, Egypt), cefoperazone (Pfizer, Egypt), cefepime and amikacin (Bristol Myers Squibb; BMS, Egypt), gentamicin (Memphis for Pharmaceutical Chemical Industries Co., Egypt), erythromycin (WINLAB, Harborough), vancomycin (Lilly Pharma, Germany), and rifampicin (El Nasr , Egypt).
Determination of antimicrobial susceptibilities: Minimum inhibitory concentrations (MICs) were determined by the two-fold agar dilution method, according to the Clinical and Laboratory Standards Institute (CLSI) (2005) (10) guidelines with Mueller-Hinton agar (MHA). The overnight Mueller–Hinton broth (MHB) cultures of the bacterial strains were diluted with broth corresponding to a final concentration of about 107 CFU/ml. Inocula of about 105CFU per spot were applied with an inoculator to the surface of dry MHA plates containing twofold serial dilutions (from 0.25–512 mg/L) of the respective antibiotics. Plates were incubated at 37˚C for 18–24 h and spots with the lowest concentrations of antibiotic showing no growth were defined as the MIC.
Beta-lactamase test: After inoculation of tested isolates on nutrient agar plates and overnight incubation at 37˚C, the plates were overlaid with 1% molten agarose containing 0.2% soluble starch and 1% penicillin and incubated for 15 min at room temperature; iodine solution was poured onto the agar plates. After 10 s, the residual iodine solution was damped out and the plates were incubated at room temperature until a discolouration zone appeared around β-lactamase producing colonies (35).
Plasmid profiles: Beta-lactamase producing strains were screened for their plasmid profile using alkaline lysis method (2). Agarose gel electrophoresis was performed. The samples were run on 0.8% agarose gel with Tris-Borate (TBE) buffer and ethidium bromide (final concentration 0.5µg/ml). The gel was observed under UV light and a picture was captured using a digital camera.
Curing of β-lactamase: Beta-lactamase producing strains were inoculated in Luria-Bertani (LB) broth containing different concentrations of the curing agent, ethidium bromide ranging from 0.25 to 128 µg/ml and incubated at 37˚C for overnight. The cultures were diluted and subculture was done on nutrient agar plates. The growths obtained were tested for β-lactamase production (7). The plasmid profile of the cured derivatives was also analyzed.
Fluorometric assay: The accumulation of norfloxacin by S. aureus cells was determined using the modified fluorescence method (9). Carbonyl cyanide m-chlorophenylhydrazone was added to a parallel set of tubes to a final concentration of 100 µM. The concentration of norfloxacin accumulated was determined on a Perkin-Elmer fluorescence spectrophotometer. The norfloxacin excitation and emission wavelengths used were 277 and 443 nm respectively.
Combination studies: Combinations of ciprofloxacin with oxacillin, cefepime, gentamicin and erythromycin were investigated by the DAA with MRSA strains (32). For the combination decimal mixtures, the mean zone size was calculated from data obtained with combination mixtures, 95% confidence intervals (t distribution) were calculated for this mean. Results were considered indicative of synergism if and and the 95% confidence intervals for did not overlap those for or , antagonism if and and the 95% confidence intervals did not overlap and all other results were considered additive.
RESULTS
Incidence of staphylococcal species in clinical specimens
Out of the 470 clinical specimens, 187 (39.8%) staphylococcal strains were isolated and identified. Out of the 187 isolates, 132 were S. aureus and 55 were CoNS (70.6% and 29.4% respectively). The incidence of staphylococcal isolates was shown in Table 1.
Table 1.
Incidence of staphylococcal species in clinical specimens
| Infection/colonization | Number of samples (n=470) | Number of S. aureus isolates (n=132) | %* | Number of CoNS isolates (n=55) | %* |
|---|---|---|---|---|---|
| Skin | 140 | 60 | 42.9 | 20 | 14.3 |
| Respiratory tract | 195 | 53 | 27.2 | 10 | 5.1 |
| Urinary tract | 100 | 13 | 13 | 21 | 21 |
| Eye | 35 | 6 | 17.1 | 4 | 11.4 |
Percentage was correlated to the number of specimens collected from each type of infection/colonization.
Antibiotic susceptibility and determination of MICs
Tables 2 and 3 show the respective MIC distributions of different antibiotics for staphylococcal isolates. Figures 1 and 2 show whether the bacteria were sensitive, intermediately sensitive or resistant to each antibiotic. The isolated S. aureus strains showed lower resistance to vancomycin, amikacin and gatifloxacin. Vancomycin was the most effective antibiotic against CoNS.
Table 2.
Distribution of MICs of different antibiotics among S. aureus isolates (132 isolates)
| Antibiotic | Break Point* µg/ml | MIC in µg/ml | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ||
| β-lactams: | |||||||||||||
| a. Penicillins: | |||||||||||||
| Ampicillin | 0.25 | 17 | 15 | 13 | 9 | 14 | 12 | 14 | 9 | 13 | 8 | 6 | 2 |
| Amoxycillin | 0.25 | 24 | 16 | 6 | 7 | 15 | 18 | 10 | 13 | 9 | 7 | 6 | 1 |
| Oxacillin | 2 | 18 | 35 | 32 | 15 | 7 | 8 | 5 | 6 | 2 | 2 | 1 | 1 |
| b.Cephalosporins: | |||||||||||||
| Cephalexin | 16 | 2 | 3 | 5 | 7 | 5 | 8 | 16 | 28 | 19 | 23 | 12 | 4 |
| Cefuroxime | 8 | 3 | 5 | 6 | 8 | 13 | 11 | 17 | 24 | 20 | 14 | 8 | 3 |
| Cefoperazone | 16 | 5 | 5 | 6 | 3 | 21 | 1 | 12 | 14 | 23 | 14 | 7 | 0 |
| Cefotaxime | 8 | 2 | 2 | 6 | 12 | 24 | 22 | 14 | 6 | 28 | 17 | 12 | 1 |
| Cefepime | 8 | 17 | 28 | 13 | 12 | 3 | 8 | 12 | 23 | 11 | 0 | 0 | 0 |
| β-lactam β-lactamase | |||||||||||||
| inhibitor combinations: | |||||||||||||
| Amp/Sul** | 8 | 8 | 22 | 17 | 10 | 16 | 14 | 6 | 21 | 10 | 8 | 0 | 0 |
| Amox/Clav** | 4 | 10 | 15 | 17 | 21 | 26 | 19 | 12 | 8 | 4 | 0 | 0 | 0 |
| Aminoglycosides: | |||||||||||||
| Gentamicin | 4 | 11 | 18 | 28 | 32 | 20 | 8 | 7 | 5 | 3 | 0 | 0 | 0 |
| Amikacin | 16 | 24 | 30 | 43 | 24 | 5 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| Fluoroquinolones: | |||||||||||||
| Ciprofloxacin | 1 | 22 | 18 | 35 | 23 | 14 | 7 | 6 | 4 | 3 | 0 | 0 | 0 |
| Norfloxacin | 4 | 10 | 3 | 22 | 6 | 26 | 27 | 16 | 10 | 9 | 3 | 0 | 0 |
| Ofloxacin | 1 | 24 | 21 | 32 | 23 | 16 | 9 | 5 | 1 | 1 | 0 | 0 | 0 |
| Levofloxacin | 1 | 33 | 42 | 20 | 26 | 5 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| Gatifloxacin | 0.5 | 68 | 30 | 29 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Macrolides: Erythromycin | 0.5 | 20 | 9 | 12 | 14 | 16 | 25 | 14 | 8 | 6 | 5 | 2 | 1 |
| Lincosamides: Clindamycin | 0.5 | 36 | 30 | 21 | 9 | 6 | 10 | 8 | 5 | 4 | 2 | 1 | 0 |
| Tetracyclines: Tetracycline | 4 | 2 | 15 | 10 | 12 | 5 | 20 | 18 | 15 | 10 | 5 | 7 | 3 |
| Glycopeptides: Vancomycin | 4 | 21 | 20 | 29 | 25 | 17 | 8 | 10 | 2 | 0 | 0 | 0 | 0 |
| Phenicols: Chloramphenicol | 8 | 6 | 8 | 10 | 19 | 6 | 25 | 12 | 16 | 10 | 8 | 7 | 5 |
| Ansamycins: Rifampicin | 1 | 14 | 25 | 31 | 26 | 20 | 9 | 5 | 2 | 0 | 0 | 0 | 0 |
Break points of different antibiotics according to CLSI (2005).
Table 3.
Distribution of MICs of different antibiotics against CoNS isolates (55 isolates)
| Antibiotic | Break Point* µg/ml | MIC in Mg/ml | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| <0.25 | 0.5 | 1 | 2 | 4 | 8 | 16 | 32 | 64 | 128 | 256 | 512 | ||
| β-lactams: | |||||||||||||
| a. Penicillins: | |||||||||||||
| Ampicillin | 0.25 | 6 | 5 | 4 | 3 | 6 | 5 | 8 | 6 | 3 | 5 | 1 | 2 |
| Amoxycillin | 0.25 | 11 | 11 | 5 | 4 | 2 | 6 | 4 | 4 | 2 | 3 | 1 | 1 |
| Oxacillin | 2 | 10 | 15 | 12 | 5 | 2 | 3 | 2 | 2 | 1 | 1 | 2 | 0 |
| b. Cephalosporins: | |||||||||||||
| Cephalexin | 16 | 0 | 0 | 2 | 3 | 2 | 4 | 6 | 10 | 12 | 8 | 5 | 3 |
| Cefuroxime | 8 | 0 | 0 | 3 | 4 | 1 | 8 | 9 | 14 | 9 | 5 | 1 | 1 |
| Cefoperazone | 16 | 0 | 0 | 5 | 7 | 2 | 8 | 5 | 6 | 11 | 9 | 2 | 0 |
| Cefotaxime | 8 | 0 | 0 | 2 | 2 | 3 | 5 | 7 | 8 | 13 | 9 | 3 | 3 |
| Cefepime | 8 | 6 | 7 | 5 | 3 | 4 | 7 | 5 | 8 | 6 | 4 | 0 | 0 |
| β-lactam β-lactamase | |||||||||||||
| inhibitor combinations: | |||||||||||||
| Amp/Sul** | 8 | 2 | 3 | 8 | 9 | 5 | 8 | 4 | 7 | 7 | 2 | 0 | 0 |
| Amox/Clav** | 4 | 7 | 6 | 5 | 3 | 7 | 9 | 7 | 3 | 4 | 3 | 1 | 0 |
| Aminoglycosides: | |||||||||||||
| Gentamicin | 4 | 9 | 12 | 10 | 9 | 7 | 2 | 3 | 2 | 1 | 0 | 0 | 0 |
| Amikacin | 16 | 16 | 13 | 6 | 11 | 5 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| Fluoroquinolones: | |||||||||||||
| Ciprofloxacin | 1 | 0 | 8 | 12 | 13 | 9 | 7 | 3 | 1 | 2 | 0 | 0 | 0 |
| Norfloxacin | 4 | 0 | 4 | 3 | 7 | 3 | 9 | 12 | 8 | 5 | 4 | 0 | 0 |
| Ofloxacin | 1 | 3 | 12 | 10 | 8 | 11 | 6 | 3 | 1 | 1 | 0 | 0 | 0 |
| Levofloxacin | 1 | 13 | 21 | 10 | 8 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gatifloxacin | 0.5 | 25 | 20 | 8 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 |
| Macrolides: Erythromycin | 0.5 | 5 | 6 | 8 | 9 | 6 | 7 | 2 | 3 | 3 | 4 | 1 | 1 |
| Lincosamides: Clindamycin | 0.5 | 10 | 12 | 11 | 8 | 5 | 2 | 3 | 2 | 1 | 1 | 0 | 0 |
| Tetracyclines: Tetracycline | 4 | 0 | 6 | 8 | 4 | 5 | 7 | 9 | 6 | 5 | 2 | 2 | 1 |
| Glycopeptides: Vancomycin | 4 | 12 | 6 | 9 | 11 | 5 | 7 | 4 | 4 | 1 | 0 | 0 | 0 |
| Phenicols: Chloramphenicol | 8 | 0 | 5 | 2 | 8 | 6 | 7 | 3 | 9 | 6 | 2 | 5 | 2 |
| Ansamycins: Rifampicin | 1 | 6 | 8 | 11 | 14 | 10 | 4 | 2 | 0 | 0 | 0 | 0 | 0 |
Break points of different antibiotics according to CLSI (2005).
Figure 1.
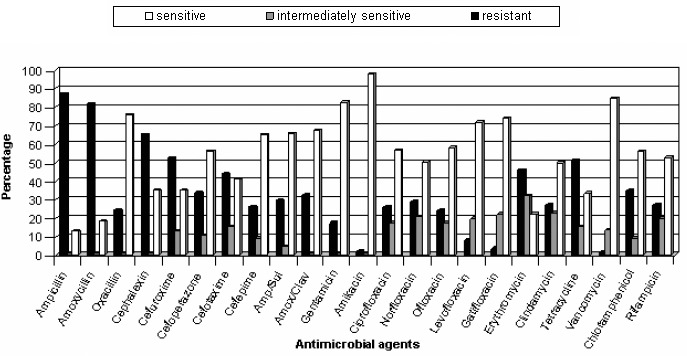
Antibiotic susceptibility ofS. aureus isolates
Figure 2.
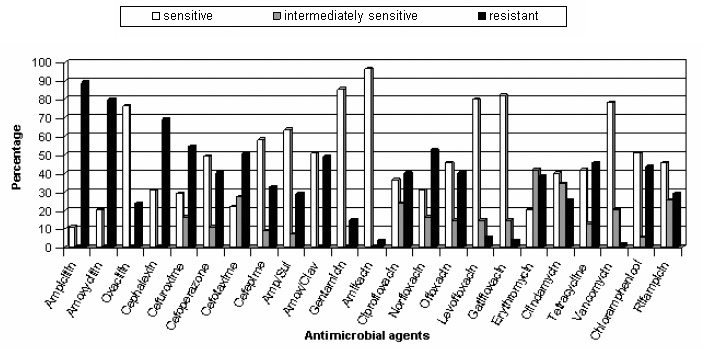
Antibiotic susceptibility of CoNS isolates
Resistance through b-lactamase production
It was found that 61.7% of ampicillin-resistant S. aureus strains and 42.9% of ampicillin-resistant CoNS strains were β-lactamase producers (Figure 3).
Figure 3.
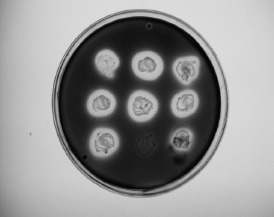
Beta-lactamase production by the isolated staphylococci
Plasmid-mediated antibiotic resistance
Some β-lactamase producing strains carried at least one plasmid and others don't (Figure 4.). Results of curing revealed that when these cells were exposed to curing agent ethidium bromide, the production of the β-lactamase was lost (Figure 5). When the plasmid profile of the cured derivatives was analyzed, none of the isolates harbored plasmids. This indicates that β-lactamase production by these strains were plasmid-mediated. On the other hand, strains which were resistant to ampicillin, β-lactamase producers and did not show plasmid bands were not cured and did not loose their enzyme activity. This evidence that β-lactamase production by these strains might be chromasomally mediated.
Figure 4.
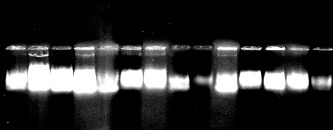
Agarose gel electrophoresis of plasmid DNA of the selected ampicillin-resistant isolates.
Figure 5.
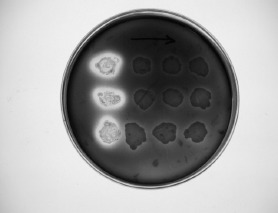
Beta-lactamase detection after plasmid curing.
Resistance through the efflux system
A biphasic pattern of accumulation of norfloxacin was observed with an initial rapid phase of accumulation seen within the first 5 minutes. The rapid phase was followed by a plateau phase. Norfloxacin-sensitive strain 1 took up higher amount of norfloxacin than did any of the resistant strains tested (2, 3, and 4) and the addition of CCCP did not affect the accumulation (figure 6). In norfloxacin-resistant strains, norfloxacin uptake was reduced when compared with that in norfloxacin-sensitive strains, and the uptake of norfloxacin by these strains increased rapidly after the addition of CCCP (137%, 191% and 218% for 3 resistant strains) (Table 4). Thus addition of uncoupler CCCP increased norfloxacin accumulation in quinolone resistant S. aureus, suggesting endogenous, energy-dependent efflux (Figure 6, 7, 8, and 9).
Figure 6.
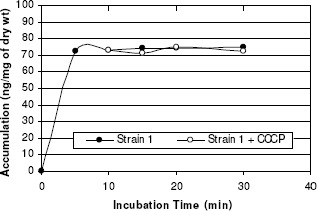
Norfloxacin accumulation in case of strain 1.
Table 4.
Concentration of norfloxacin ± 100µM CCCP accumulated by staphylococcal strains
| Staphylococcus aureus strains | MIC (µg/mg) | Susceptibility to Norfloxacin | Accumulation (ng/mg dry cells) | Percentage of increase in accumulation | |
|---|---|---|---|---|---|
| - CCCP | + CCCP | ||||
| Strain 1 | 2 | Sensitive | 72 | 71 | ┄ |
| Strain 2 | 16 | Resistant | 57 | 78 | 137% |
| Strain 3 | 128 | Resistant | 47 | 90 | 191% |
| Strain 4 | 128 | Resistant | 39 | 85 | 218% |
Figure 7.
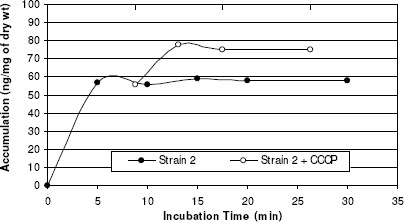
Norfloxacin accumulation in case of strain 2.
Figure 8.
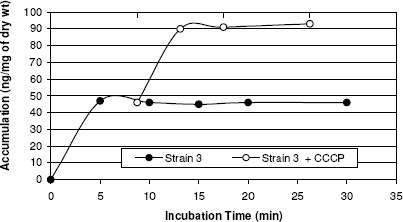
Norfloxacin accumulation in case of strain 3.
Figure 9.
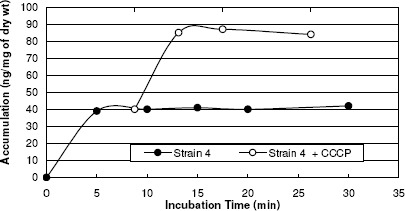
Norfloxacin accumulation in case of strain 4.
Decimal assay for additivity method
For each pair of antibiotics tested, the selected target size for zones of inhibition, the BEF, and the mean diameters of the inhibition zones (with 95% confidence intervals) obtained are provided in table 5. For each antibiotic the BEF was calculated by using the linear regression equation for the standard dose-response curve, similar to that illustrated in figure 10 for ciprofloxacin.
Table 5.
Interactions between pairs of antibiotics with MRSA strain
| Antibiotics | Target zone diameter (mm) | BEF (µg) | Mean diameter of zone of inhibition attained (mm) | 95% CIa (mm) | Type of interaction |
|---|---|---|---|---|---|
| Ciprofloxacin | 14 | 25.06 | 14.3 | 13.8-14.8 | Synergism |
| Oxacillin | 14 | 309.03 | 13.3 | 12.6-14.0 | |
| Ciprofloxacin+Oxacillin | 18.2 | 17.4-19.0 | |||
| Ciprofloxacin | 13 | 21.53 | 14.3 | 13.8-14.8 | Additive |
| Cefepime | 13 | 918.33 | 12.9 | 12.4-13.4 | |
| Ciprofloxacin+Cefepime | 13.7 | 13.1-14.3 | |||
| Ciprofloxacin | 18 | 46.03 | 17.5 | 17.0-18.0 | Additive |
| Gentamicin | 18 | 67.14 | 16.5 | 16.2-16.8 | |
| Ciprofloxacin+Gentamicin | 17.1 | 16.6-17.6 | |||
| Ciprofloxacin | 18 | 46.03 | 17.4 | 17.1-17.7 | Antagonism |
| Erythromycin | 18 | 74.64 | 17.1 | 16.6-17.6 | |
| Ciprofloxacin+Erythromycin | 14.1 | 13.2-15.0 |
Confidence interval.
Figure 10.
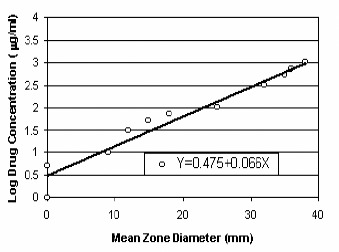
Standard dose-response curve generated in disk diffusion assays with S. aureus strain and ciprofloxacin.
Interactions between ciprofloxacin and oxacillin, cefepime, gentamicin and erythromycin were shown in figures 11, 12, 13 and 14.
Figure 11.
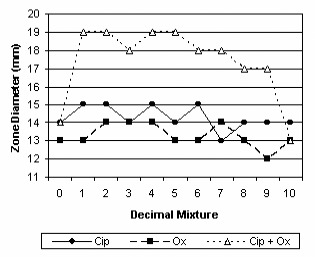
Synergy between ciprofloxacin and oxacillin.
Figure 12.
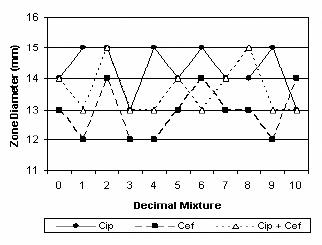
Additivity between ciprofloxacin and cefepime.
Figure 13.
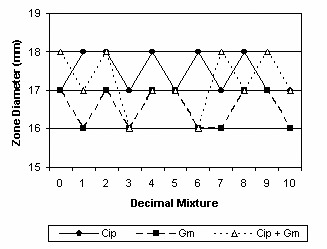
Additivity between ciprofloxacin and gentamicin.
Figure 14.
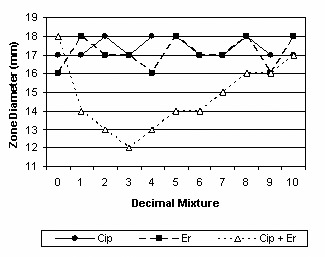
Antagonism of ciprofloxacin by erythromycin.
DISCUSSION
Staphylococcal infections are a common and significant clinical problem in medical practice (30). In the present work, the incidence of staphylococci was studied among the clinical specimens isolated in Minia. Out of the 470 clinical specimens, 187 (39.8%) staphylococcal strains were isolated and identified. Out of the isolated staphylococci, 70.6% were S. aureus and 29.4% were CoNS. Staphylococcus aureus was more prevalent than CoNS in skin, respiratory and eye infections, while CoNS were the most common isolates in urinary tract infections.
Our study revealed high activity of vancomycin and amikacin towards staphylococci. The effectiveness of vancomycin against staphylococci is corroborated by data from other groups (5,12,18).
In order to define the main mechanisms used by staphylococci to resist antibiotics, we tested for β-lactamase production and for possess of plasmid and efflux-mediated resistance. Liang et al. (21) reported that one of the major mechanisms of resistance to β-lactams was the expression of ß-lactamases, such as penicillinase and cephalosporinase. We observed high levels of β-lactamase production in staphylococcal isolates (61.7% in S. aureus and 42.9% in CoNS). High levels of β-lactamase production in staphylococci had been reported elsewhere (4).
Other mechanisms of staphylococcal resistance to β-lactams were reported; such as intrinsic mechanism, which is not due to drug inactivation, and accounts for methicillin-resistance; and tolerance, in which there is a dissociation of the inhibitory and killing actions of beta-lactam antibiotics (31).
Beta-lactamase producing staphylococcal strains were screened for their plasmid profile, results of the screening revealed that some strains were carrying at least one plasmid suggesting plasmid-mediated antibiotic resistance and others did not. Plasmid-mediated antibiotic resistance was reported in several studies (14,25). Results of β-lactamase curing were compatible with other studies (7,23). Also, Shakibaie et al. (33) reported that the β-lactamase-cured cells did not exhibit any enzyme activity.
In this study the efflux mechanism in S. aureus strains was measured by a modified fluorometric assay. Addition of uncoupler CCCP increased norfloxacin accumulation in quinolone resistant S. aureus strains. Numerous reports involved fluorimetric method to study the efflux mechanisms and the effect of CCCP on the accumulation (28,29). Kaatz et al. (17) and Aeschlimann et al. (1) had shown increased activity or accumulation of the hydrophilic quinolones, norfloxacin and ciprofloxacin, in the presence of efflux inhibitors. Takenouchi et al. (36) demonestrated an increase in norfloxacin accumulated by S. aureus strains in the presence of CCCP.
Our study revealed that, using DAA technique with MRSA strain, synergistic interaction was observed between ciprofloxacin and oxacillin, ciprofloxacin plus cefepime and gentamicin appeared to be additive, while ciprofloxacin plus erythromycin was antagonistic. Combinations of fluoroquinolones with other antimicrobial agents against S. aureus strains have been previously investigated (13,16). Also the validity of DAA had been demonstrated (26,32).
CONCLUSION
This study examined the incidence of staphylococci, and its susceptibility patterns to different antibiotics. Drug resistance mediated by β-lactamases, plasmid and efflux pumps were important modes of cellular resistance to antimicrobial agents in our study. It would be valuable to use efflux pump inhibitors. Also, combination of antimicrobials which potentiate the activity of the existing antibiotics may be useful to decrease the resistance rate.
ACKNOWLEDGEMENTS
The author gratefully acknowledges the referee, who made useful suggestions and remarks which helped me improve the paper.
REFERENCES
- 1.Aeschlimann J.R., Kaatz G.W., Rybak M.J. The effects of NorA inhibition on the activities of levofloxacin, ciprofloxacin and norfloxacin against two genetically related strains of Staphylococcus aureus in an in-vitro infection model. J Antimicrob Chemother. 1999;44(3):343–349. doi: 10.1093/jac/44.3.343. [DOI] [PubMed] [Google Scholar]
- 2.Alexander S.T., Strele D., Niles M.J. USA: Mc. Grow Hill; 2004. Laboratory exercises in organismal and molecular microbiology. [Google Scholar]
- 3.Archer G.L. Staphylococcus aureus: A well-armed pathogen. Clin Infect Dis. 1998;26:1179–1181. doi: 10.1086/520289. [DOI] [PubMed] [Google Scholar]
- 4.Arslan S., Ozkarde F. Slime production and antibiotic susceptibility in staphylococci isolated from clinical samples. Mem Inst Oswaldo Cruz. 2007;102(1):29–33. doi: 10.1590/s0074-02762007000100004. [DOI] [PubMed] [Google Scholar]
- 5.Bashir A., Mujahid T.Y., Jehan N. Antibiotic resistance profile: isolation and characterization of clinical isolates of staphylococci from patients with community-acquired skin infections. Pak J Pharm Sci. 2007;20(4):299–304. [PubMed] [Google Scholar]
- 6.Boneca I.G., Chiosis G. Vancomycin resistance: occurrence, mechanisms and strategies to combat it. Expert Opin Ther Targets. 2003;7:311–328. doi: 10.1517/14728222.7.3.311. [DOI] [PubMed] [Google Scholar]
- 7.Bonfiglio G., Livermore D.M. P-lactamase types amongst Staphylococcus aureus isolates in relation to susceptibility to P-lactam inhibitor combinations. Antimicrob. Chemother. 1994;33:465–481. doi: 10.1093/jac/33.3.465. [DOI] [PubMed] [Google Scholar]
- 8.Casey A.L., Lambert P.A., Elliott T.S.J. Staphylococci. Int J Antimicrob Agents. 2007;29(Suppl.3):23–32. doi: 10.1016/S0924-8579(07)72175-1. [DOI] [PubMed] [Google Scholar]
- 9.Chapman J.S., Georgopapadakou N.H. Fluorometric assay for fleroxacin uptake by bacterial cells. Antimicrob. Agents Chemother. 1989;33(1):27–29. doi: 10.1128/aac.33.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clinical and Laboratory Standards Institute (CLSI) Performance standards for antimicrobial disk susceptibility tests. Clin. Lab. Stand. Inst. (9th) 2006;26(1):1–172. Approved. M2-A9. [Google Scholar]
- 11.Collee J.G., Fraser A.G., Marmion B.P., Simmons A. Tests for identification of bacteria. In: Mackie, McCartney, editors. New York, USA: Practical medical microbiology; 1996. [Google Scholar]
- 12.Elouennass M., Sahnoun I., Zrara A., Bajjou T., Elhamzaoui S. Epidemiology and susceptibility profile of blood culture isolates in an intensive care unit (2002–2005) Med Mal Infect. 2008;38(1):18–24. doi: 10.1016/j.medmal.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Ferrara A., Dos-Santos C., Cimbro M., Gialdroni Grassi G. Effect of different combinations of sparfloxacin, oxacillin, and fosfomycin against methicillin-resistant staphylococci. Eur J Clin Microbiol Infect Dis. 1997;16(7):535–537. doi: 10.1007/BF01708239. [DOI] [PubMed] [Google Scholar]
- 14.Firth N., Apisiridej S., Berg T., O’Rourke B.A., Curnock S., Dyke G.H.K., Skurray R.A. Replication of Staphylococcal Multiresistance Plasmids. J. Bacteriol. 2000;182:2170–2178. doi: 10.1128/jb.182.8.2170-2178.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain A., Agarwa J., Bansal S. Prevalence of methicillin-resistant, coagulase-negative staphylococci in neonatal intensive care units: findings from a tertiary care hospital in India. J Med Microbiol. 2004;53:941–944. doi: 10.1099/jmm.0.45565-0. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins S.G., Lewis J.W. Synergistic interaction between ofloxacin and cefotaxime against common clinical pathogens. Infection. 1995;23(4):245. doi: 10.1007/BF01793856. [DOI] [PubMed] [Google Scholar]
- 17.Kaatz G.W., Seo S.M., Ruble C.A. Mechanisms of fluoroquinolone resistance in Staphylococcus aureus. Journal of infectious diseases. 1991;163:1080–1086. doi: 10.1093/infdis/163.5.1080. [DOI] [PubMed] [Google Scholar]
- 18.Khosravi A.D., Mehdinejad M., Heidari M. Bacteriological findings in patients with ocular infection and antibiotic susceptibility patterns of isolated pathogens. Singapore Med J. 2007;48(8):741–743. [PubMed] [Google Scholar]
- 19.Knauer A., Fladerer P., Strempfl C. Effect of hospitalization and antimicrobial therapy on bantimicrobial resistance of colonizing Staphylococcus epidermidis. Wien Klin Wochenschr. 2004;116:489–494. doi: 10.1007/BF03040945. [DOI] [PubMed] [Google Scholar]
- 20.Le Loir Y., Baron F., Gautier M. Staphylococcus aureus and food poisoning. Genet Mol Res. 2003;2(1):63–76. [PubMed] [Google Scholar]
- 21.Liang W., Huang H., Lin R., Hou W. Screening for natural inhibitors of penicillinase by copolymerization of hydrolyzed starch or glycogen in sodium dodecylsulfate polyacrylamide gels for detecting penicillinase activity. Bot. Bull. Acad. Sin. 2003;44:187–191. [Google Scholar]
- 22.Lynch A.S. Efflux systems in bacterial pathogens: an opportunity for therapeutic intervention? An industry view. Biochem Pharmacol. 2006;71(7):949–956. doi: 10.1016/j.bcp.2005.10.021. [DOI] [PubMed] [Google Scholar]
- 23.Mansouri S., Khalegi M. Antibacterial resistance pattern and frequency of methicillin resistant Staphylococcus aureus isolated from different sources in Southern Iran. Am. J. Med. Sci. 1997;22:89–93. [Google Scholar]
- 24.Marquez B. Bacterial efflux systems and efflux pumps inhibitors. Biochimie. 2005;87(12):1137–1147. doi: 10.1016/j.biochi.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 25.Nayak N., Satpathy G., Vajpayee R.B., Mrudula S. Phenotypic and plasmid pattern analysis of Staphylococcus epidermidis in bacterial keratitis. Indian J Ophthalmol. 2007;55(1):9–13. doi: 10.4103/0301-4738.29488. [DOI] [PubMed] [Google Scholar]
- 26.Nworu C.S., Esimone C.O. Comparative Evaluation of Three In Vitro Techniques in the Interaction of Ampicillin and Ciprofloxacin against Staphylococcus aureus and Escherichia coli. Tropical Journal of Pharmaceutical Research. 2006;5(2):605–611. [Google Scholar]
- 27.Palazzo I.C.V., Araujo M.L.C., Darini A.L.C. First report of vancomycin-resistant Staphylococci isolated from healthy carriers in Brazil. J Clin Microbiol. 2005;43:179–185. doi: 10.1128/JCM.43.1.179-185.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Piddock L.J., Jin Y.F., Griggs D.J. Effect of hydrophobicity and molecular mass on the accumulation of fluoroquinolones by Staphylococcus aureus. J Antimicrob Chemother. 2001;47(3):261–270. doi: 10.1093/jac/47.3.261. [DOI] [PubMed] [Google Scholar]
- 29.Piddock L.J., Jin Y.F., Webber M.A., Everett M.J. Novel ciprofloxacin-resistant, nalidixic acid-susceptible mutant of Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46(7):2276–2278. doi: 10.1128/AAC.46.7.2276-2278.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rayner C., Munckhof W.J. Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern Med J. 2006;36(2):142–143. doi: 10.1111/j.1444-0903.2005.00976.x. [DOI] [PubMed] [Google Scholar]
- 31.Sabath L.D. Mechanisms of resistance to beta-lactam antibiotics in strains of Staphylococcus aureus. Ann Intern Med. 1982;97(3):339–344. doi: 10.7326/0003-4819-97-3-339. [DOI] [PubMed] [Google Scholar]
- 32.Sanders C.C., Sanders W.E., Moland E.S. Decimal assay for additivity of drugs permits delineation of synergy and antagonism. Antimicrob Agents Chemother. 1993;37(2):260–264. doi: 10.1128/aac.37.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shakibaie M.R., Mansouri S., Hakak S. Plasmid pattern of antibiotic resistance in beta-lactamase producing Staphylococcus aureus strains isolated from hospitals in Kerman, Iran. Am. J. Med. Sci. 2002;27:80–83. [Google Scholar]
- 34.Skov R., Frimodt-Moller N., Espersen F. Tentative interpretative zone diameters for fusidic Neosensitabs† on Mueller Hinton agar and three blood media. Int J Antimicrob Agents. 2003;22:502–507. doi: 10.1016/s0924-8579(03)00123-7. [DOI] [PubMed] [Google Scholar]
- 35.Sykes R.B. Methods for detecting P-lactamases. Laboratory methods in Antimicrobial Chemotherapy. In: Reeves D.S., Phillips I, Williams J.D., Wise R., editors. Mulligan: Churchill Livingstone; 1978. pp. 64–69. [Google Scholar]
- 36.Takenouchi T., Tabata F., Iwata Y., Hanzawa H., Sugawara M., Ohya S. Hydrophilicity of quinolones is not an exclusive factor for decreased activity in efflux-mediated resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 1996;40:1835–1842. doi: 10.1128/aac.40.8.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Winn W.C., Allen S.D., Janda W.M., Koneman E.W., Procop G.W., Schreckenberger P.C., Woods G.L. Color Atlas Textb. Diagn. Microbiol. New York, USA: 2006. [Google Scholar]


