Abstract
Ten out of fifty fresh and refrigerated samples of shrimp (Litopenaeus vannamei) collected from retailers in Natal (Rio Grande do Norte, Northeastern Brazil) tested positive for Vibrio parahaemolyticus. The Kanagawa test and multiplex PCR assays were used to detect TDH and TRH hemolysins and the tdh, trh and tlh genes, respectively. All strains were Kanagawa-negative and tlh-positive. Antibiotic susceptibility testing was done for seven antibiotics by the agar diffusion technique. Five strains (50%) presented multiple antibiotic resistance to ampicillin (90%) and amikacin (60%), while two strains (20%) displayed intermediate-level resistance to amikacin. All strains were sensitive to chloramphenicol. Intermediate-level susceptibility and/or resistance to other antibiotics ranged from 10 to 90%, with emphasis on the observed growing intermediate-level resistance to ciprofloxacin. Half our isolates yielded a multiple antibiotic resistance index above 0.2 (range: 0.14–0.29), indicating a considerable risk of propagation of antibiotic resistance throughout the food chain.
Keywords: Vibrio parahaemolyticus, Litopenaeus vannamei, multiple antibiotic resistance
INTRODUCTION
Brazilian marine shrimp farming started in the 1970s and became a consolidated industry by the end of the 1980s. The successful acclimation of the Pacific white shrimp (Litopenaeus vannamei) to Northeast Brazilian environmental conditions was among the main factors making the activity economically feasible (4).
Northeast Brazilian farmers generally cultivate shrimp in ponds with water drawn from adjacent estuaries. Such ponds are critical reservoirs for opportunistic vibrio species (3), some of which are potentially pathogenic to man. Thus, strains of Vibrio parahaemolyticus are commonly communicated to humans through inadequately cooked contaminated shrimp causing gastroenteritis (26). The Brazilian Agency of Health Surveillance (ANVISA) has accordingly included shrimp under item 22 (ready-to-eat food products) of the RDC 12 regulation (7).
The presence of pathogenic vibrios in shrimp ponds has led farmers to look for efficient control measures such as antibiotic therapy. However, overtime improper and constant use of antibiotics has resulted in the development of resistant strains. In addition, small amounts of antibiotics added to shrimp feed for purposes of growth promotion have favored the emergence of resistant strains (9).
The state of Rio Grande do Norte boasts the country’s largest output of pond-reared Pacific white shrimp. However, fresh shrimp is currently marketed with no special sanitary precautions through supermarkets and street peddlers in Natal, the state capital. To assess the microbiological condition of these products, we carried out a phenotypic and genotypic analysis of strains of Vibrio parahaemolyticus isolated from fresh and refrigerated pond-reared shrimp. The strains were tested for susceptibility to antibiotics commonly used in human therapy, including tetracycline, amikacin, ampicillin, chloramphenicol, sulfamethoxazole-trimethoprim, ciprofloxacin and nitrofurantoin.
MATERIALS AND METHODS
Samples
Fifty samples of Litopenaeus vannamei were obtained from various retailers (supermarkets: n=21; street peddlers: n=29) in Natal (Rio Grande do Norte, Brazil) between August 2005 and September 2007. Most samples (n=47) came from shrimp farms in the hinterland. The remaining samples (n=3) consisted of marine shrimp captured by professional fishermen.
Sample collection
Fresh and refrigerated whole shrimp (200 g) were collected in sterilized Beckers, labeled, accommodated in thermal boxes at 6−10°C and submitted to microbiological analysis at the Food Microbiology Laboratory at Potiguar University (UnP) within two hours of sampling.
Vibrio identification and phenotypic profiling
Each 200-g sample was macerated in a sterilized mortar. Then 25-g fractions were added to 225mL sterile alkaline peptone water (APW) supplemented with 1% NaCl (pH 8.6) and incubated for 24 hours at 37°C. Following primary enrichment, aliquots were streaked onto thiosulfate citrate bile sucrose (TCBS) agar and incubated for 24 hours at 37°C (21).
Three to five colonies of greenish-blue saccharose-negative bacteria were transferred to screening media (Kligler iron agar and lysine iron agar) and trypticase soy agar slants containing 1.0% NaCl and incubated for 24 hours at 37°C for purification. Cytochrome oxidase-positive isolates were subsequently selected for phenotypic profiling (21).
Kanagawa test
Purified colonies of V. parahaemolyticus were seeded in 1-cm circles on Wagatsuma agar containing sheep erythrocytes (35). Following incubation for 24 hours at 35–37°C, the cultures were analyzed for the presence of inhibition halos indicative of thermostable direct hemolysin. A V. parahaemolyticus K+ isolate from an outbreak in Cascavel (a town in Northeastern Brazil) was used as positive control (14).
Susceptibility to antibiotics
Colonies were seeded on Mueller-Hinton agar on antibiotic-impregnated paper discs (5) (Cecon, São Paulo, Brazil) and incubated for 24 hours at 37°C. The discs were impregnated with 30ug tetracycline, 30ug amikacin, 10ug ampicillin, 30ug chloramphenicol, 25ug sulfamethoxazole-trimethoprim, 5ug ciprofloxacin or 300ug nitrofurantoin. Inhibition halos were measured with a caliper. A strain of Escherichia coli ATCC 25922 was used as control.
Multiple antibiotic resistance index
The multiple antibiotic resistance (MAR) index is calculated by dividing the number of antibiotics to which the strain is resistant by the number of antibiotics to which the strain has been exposed. A MAR index above 0.2 is defined as multiple antibiotic resistance (24).
Hemolysin detection
The strains were submitted to multiplex PCR assay for detection of the pathogenic genes tdh (thermostable direct hemolysin gene) and trh (thermostable direct hemolysin-related hemolysin gene), followed by detection of the V. parahaemolyticus-specific gene tlh (thermolabile hemolysin gene) (32). The procedure included DNA extraction, selection of primers and multiplex PCR amplification.
DNA extraction: Genomic DNA extraction was performed with a commercially available extraction kit (DNeasy Blood & Tissue Kit, Qiagen) in accordance with the manufacturer’s instructions.
Selection of primers: The nucleotide sequences and amplicon sizes of the tlh, tdh and trh-specific primers are shown in Table 1.
Table 1.
Nucleotide sequences and amplicon sizes of tlh, tdh and trh-specific primers of strains of Vibrio parahaemolyticus isolated from fresh and refrigerated samples of Litopenaeus vannamei collected from retailers in Natal, Rio Grande do Norte, Brazil.
| Gene | Sequence | Amplicon size (pb) | Source |
| Tl | 5’–AAA GCG GAT TAT GCA GAA GCA CTG – 3’ 5’- GCT ACT TTC TAG CAT TTT CTC TGC – 3’ | 450 | Tanigushi et al.(33) |
| Tdh | 5’-GTA AAG GTC TCT GAC TTT TGG AC–3’ 5’-TGG AAT AGA ACC TTC ATC TTC ACC-3’ | 269 | Nishibuchi, Kaper, (29) |
| Trh | 5’- TTG GCT TCG ATA TTT TCA GTA TCT-3’ 5’- CAT AAC AAA CAT ATG CCC ATT TCC-3’ | 500 | H Honda et al.(18) |
Multiplex PCR amplification: The amplification reaction solution for the simultaneous use of the three primers was prepared in 200-µL microtubes with a final reaction volume of 25µL, containing: 2.5µL reaction buffer (50mM KCl, 1.5mM MgCl2, 10mM Tris-HCl, pH 9.0; Amersham Pharmacia Biotech, USA), 2.0µL dNTP solution (200µM of each deoxynucleotide triphosphate [dATP, dCTP, dGTP, dTTP], Amersham Pharmacia Biotech, USA), 0.5µL (10 pmol) of each primer, 0.25µL Taq DNA polymerase (1U, Amersham Pharmacia Biotech, USA), 16.75µL sterilized deionized water and 0.5µL genomic DNA.
The DNA fragments were amplified in a thermal cycler (Px2, Thermo Electron Corporation) set to 30 cycles and starting with DNA template degradation for 1 minute at 94°C followed by primer annealing in the target region of the template for 1 minute at 58°C and strand extension for 1 minute at 72°C. To make sure template degradation was complete and the new strand was fully extended the solution was heated initially to 94°C for 3 minutes and finally to 72°C for 5 minutes, respectively.
The amplified product was submitted to 1% agarose gel electrophoresis in 0.5X TBE buffer for approximately one hour. A molecular weight standard (Amersham Pharmacia Biotech, USA) was added to each gel.
Following migration, the products were stained by immersion of the gel in 10mg/mL ethidium bromide solution and analyzed under ultraviolet light using an image analysis system (ImageQuant 300, GE) (Figure 1).
Figure 1.
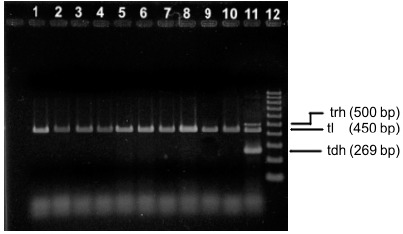
Multiplex PCR patterns of Vibrio parahaemolyticus obtained from shrimp samples. Lanes 1–10 represented different isolates from different samples. Lane 11 : positive control. Lane 12 molecular size standard.
RESULTS AND DISCUSSION
Ten out of 50 shrimp samples (10/50; 20%) tested positive for Vibrio parahaemolyticus, all of which were collected at supermarkets in Natal. This detection rate is much higher than that reported by Chen et al. (10) in a study using 112 samples of tunafish fillet from various retailers in São Paulo (n=3; 2.68%). The 10 isolates were identified phenotypically and genotypically as V. parahaemolyticus, including detection of the species-specific tlh gene (6) (Figure 1).
Few strains were isolated in our study, possibly due to the ice storage practices adopted by the sampled supermarkets. Cryoconservation is commonly used in the fishing industry to control growth and survival of spoiling and pathogenic bacteria (25). Although vibrios are not thought to survive at temperatures below growth optima for long (13), a number of authors have isolated V. parahaemolyticus from fresh, refrigerated and frozen seafoods (35,36,37,38). Muntada-Garrida et al. (28) observed the inactivation of V. parahaemolyticus in oyster meat at various commercial cold storage temperatures and found die-off to be faster at temperatures below zero.
The strains isolated in our study were Kanagawa phenomenon-negative. Strains producing thermostable direct hemolysin are referred to as Kanagawa phenomenon-positive and can be identified by β-type hemolysis on Wagatsuma blood agar (20,35). Some authors believe that on the average 99% of V. parahaemolyticus isolates from marine environments and seafoods are Kanagawa-negative, even when collected from sources associated with intoxication and infection (15, 19, 31). Other studies have shown that Kanagawa-negative strains of V. parahaemolyticus isolated from seafoods and marine environments can be cytotoxic or cytotonic and lethal to mice (36), suggesting that the Kanagawa phenomenon is not an absolute indicator of pathogenicity (16,17). According to Bej et al. (6), in addition to multiplex PCR-based detection targeting the tlh gene, comprehensive detection of this pathogen should include both tdh and trh for hemolysin-producing pathogenic strains of V. parahaemolyticus. The absence of the tdh and trh genes, as observed in our isolates, reduces the risk of developing gastroenteritis.
The 10 isolated strains of V. parahaemolyticus were strongly resistant to ampicillin (90%) and amikacin (60%), and without exception sensitive to chloramphenicol (100%). Intermediate-level susceptibility and/or resistance to other antibiotics ranged from 10 to 90%, with emphasis on the observed growing intermediate-level resistance to ciprofloxacin. Half the samples were resistant to both ampicillin and amikacin (Table 2).
Table 2.
Antibiotic susceptibility profile of strains of Vibrio parahaemolyticus isolated from fresh and refrigerated samples of Litopenaeus vannamei collected from retailers in Natal, Rio Grande do Norte, Brazil.
| Sample | Antibiotic susceptibility profile | ||||||
|---|---|---|---|---|---|---|---|
| AMP 10µg | NIT 300µg | TCY 30µg | CHL 30µg | AMK 30µg | CIP 5µg | SFT 25µg | |
| 1. | R | S | I | S | I | I | S |
| 2. | R | S | I | S | R | I | S |
| 3. | R | S | I | S | R | I | I |
| 4. | R | S | S | S | R | S | S |
| 5. | R | S | S | S | S | I | S |
| 6. | S | S | S | S | R | I | S |
| 7. | R | I | S | S | R | I | S |
| 8. | R | I | I | S | R | I | S |
| 9. | R | I | S | S | S | I | S |
| 10 | R | S | S | S | I | I | S |
S=Sensitive; R=Resistant; I=Intermediate
AMP=Ampicilin; NIT=Nitrofurantoin; TCY=Tetracycline; CLH=Chloramphenicol; AMK=Amikacin; CIP= Ciprofloxacin; SFT=Sulfamethoxazole-trimethoprim
In contrast, in a study by Molitoris et al. (27) isolating V. parahaemolyticus from seawater and various seafoods (fish, crabs and shrimps), a total of 92 different antibiotic resistance patterns were observed, 3.5% of which included ampicillin. Another study (30), however, reported over 50% of V. parahaemolyticus strains isolated from fresh and frozen seafood to be resistant to ampicillin.
Antibiotic resistance is the acquired ability of an organism to tolerate the effect of antibiotics to which it is normally susceptible. Antibiotic-producing bacteria are capable of transmitting naturally occurring resistance genes to other bacteria through genetic exchange, enabling them to neutralize or destroy the antibiotics with which they are challenged (9). The proportion of isolates presenting multiple antibiotic resistance (Table 2) is a disquieting finding in view of the possibility of propagation of resistance factors to other microorganisms and to livestock and humans.
Worldwide pond shrimp production is increasing at about 9.25% per year (12). Previous estimates suggested half the world’s seafood demand would be met by aquaculture in 2020 (11). However, widespread bacterial infections, especially involving the genera Aeromonas, Vibrio, Pseudomonas and Flavobacterium, have become a major challenge to producers (1), many of whom have resorted to treatment with an array of antibiotics without regulation by local government agencies. For example, in Brazil no antibiotics are registered for use in shrimp farming and their use is illegal. According to Chythanya et al. (9), although antibiotics have played an important role in the treatment of disease in humans and aquaculture livestock, their indiscriminate use is associated with serious consequences to public health.
The MAR index of the V. parahaemolyticus strains isolated from our shrimp samples ranged from 0.14 to 0.29. Half our isolates yielded a MAR index above 0.2, indicating a considerable risk of propagation of antibiotic resistance throughout the food chain.
The growing intermediate-level resistance to ciprofloxacin observed in our strains matches findings from a study on the same bacterial species using samples of tunafish fillet from retailers in São Paulo (10). Ciprofloxacin―the most potent quinolone available for the treatment of Gram-negative microorganisms―is four to eight times more effective than norfloxacin against enterobacteria and Pseudomonas (34). Strains with intermediate-level resistance (10–90% in our study) are of particular concern as they can have a negative impact on treatment efficacy in spite of behaving like sensitive organisms. In other words, some of the bacteria in the microbiota may be particularly sensitive a given antibiotic while others are genetically immune (22). In our study the profile of intermediate-level resistance included tetracycline―the antibiotic most commonly used as growth promoter and therapeutic agent in livestock. Approximately 65% of the antibiotics prescribed in EU countries for veterinary therapeutic use are tetracyclines (23). In general, bacteria become resistant to tetracycline through the acquisition of plasmids containing resistant genes. The mechanism involves Tet proteins (A, B, C and D) which, once formed, localize to the cytoplasmic membrane, inducing the cell to excrete the antibiotic almost immediately (2). Carvalho et al. (8) also isolated tetracycline-resistant Salmonella strains from shrimp cultures (water, sediments and shrimp).
Although seafoods consumed by humans traditionally come from the ocean and estuaries rather than from artificial aquaculture, investments over the past few decades have shifted to aquaculture, especially shrimp farming. However, artificial environments are far more easily exposed to contamination by resistant Vibrio strains. Farmers and authorities are therefore advised to adopt appropriate risk management policies and measures, including a) greater sanitary/microbiological control of farm products from harvesting to marketing, b) strict routine on-farm sanitary practices with microbiological monitoring of livestock, feed and water, c) awareness campaigns informing consumers of the risk associated with the consumption of raw or inadequately cooked aquaculture products regardless of physical appearance, d) sanitary control and education of fishermen and seafood handlers (with emphasis on the microbiological quality of ice) exposed to the risk of skin infections during the cleaning and processing of aquaculture products and e) proper use of antibiotic therapy and quarantine on shrimp farms.
32 REFERENCES
- 1.Akinbowale O.L., Peng H., Barton M.D. Antimicrobial resistance in bacteria isolated from aquaculture sources in Australia. J. Appl. Microbiol. 2006;100:1103–1113. doi: 10.1111/j.1365-2672.2006.02812.x. [DOI] [PubMed] [Google Scholar]
- 2.Alterthum F. Mecanismo de ação dos antimicrobianos e mecanismos de resistência. In: Trabulsi L.R., Alterthum F, editors. Microbiologia. Atheneu: São Paulo; 2008. pp. 79–85. [Google Scholar]
- 3.Barbieri E., Falzano L., Fiorentini C., Pianetti A., Baffone W., Fabbri A., Matarrese P., Casiere A., Katouli M., Kühn I., Möllby R., Bruscolini F., Donelli G. Occurrence, diversity, and pathogenicity of halophilic Vibrio spp. and non-O1 Vibrio cholerae from estuarine waters along the Italian Adriatic coast. J. Appl. Environ. Microbiol. 1999;65(6):2748–2753. doi: 10.1128/aem.65.6.2748-2753.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbieri Junior R.C., Ostresnky Neto A. Aprenda Fácil. Viçosa; 2002. Camarões Marinhos - Engorda; p. 2. [Google Scholar]
- 5.Bauer A.W., Kirby W.M.M., Sherris J.C., Turck M. Antibiotic susceptibility testing by standardized single disk method. Am. J. Clin. Pathol. 1966;36:493–496. [PubMed] [Google Scholar]
- 6.Bej A.K., Patterson D.P., Brasher C.W., Vickery M.C.L., Jones D.D., Kaysner C.A. Detection of total and hemolysin –producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl , tdh and trh. J. Microbiol. Methods. 1999;36:215–225. doi: 10.1016/s0167-7012(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 7.Brasil . Diário Oficial da República Federativa do Brasil. Brasília; 2001. Agência Nacional de Vigilância Sanitária. Resolução—RDC N° 12, de 2 de janeiro de 2001. Regulamento técnico sobre padrões microbiológicos para alimentos. [Google Scholar]
- 8.Carvalho F.C.T., Barreto N.S.R., Reis C.M.F., Hofer E., Vieira R.H.S.F. Antimicrobial susceptibility of Salmonella isolated from shrimp farms in Ceará State, Brazil. Rev. Ciên. Agron. 2009;40:549–556. [Google Scholar]
- 9.Chythanya R., Nayak D.K., Venugopal M.N. Antibiotic resistance in aquaculture. Aquaculture. 1999;6:30–32. [Google Scholar]
- 10.Chen J., Balian S.C., Telles E.O. Vibrio parahaemolyticus in tuna (Thunnus spp traded in the city of São Paulo. Veterinária e Zootecnia. 2005;12:89–95. [Google Scholar]
- 11.FAO (Food and Agricultural Organisation). Rome: 1995. The state of the world Fisheries and aquaculture; p. 27. [Google Scholar]
- 12.FAO (Food and Agricultural Organisation ). Review of the State of the World Aquaculture. FAO Fish Circular. 1997;886:1. [Google Scholar]
- 13.FAO/WHO Matters arising from FAO and WHO: Microbiological risk assessment of Vibrio spp. CX/FFP 03/2-Add.1. Joint FAO/WHO Food Standards Programme, Codex Alimentarius Commission. Food and Agriculture Organization of the United Nations/World Health Organization; 13–17 October; Alesund, Norway. 2003. [Google Scholar]
- 14.Hofer E. Primeiro isolamento e identificação de Vibrio parahaemolyticus no Brasil de infecção gastrointestinal humana. Rev. Microbiol. 1983;14:174–175. [Google Scholar]
- 15.Hofer E., Silva C.H.D. Caracterização sorológica de amostras de Vibrio parahaemolyticus isoladas de peixes capturados no litoral brasileiro. Rev. Microbiol. 1986;17:327–331. [Google Scholar]
- 16.Hoashi K., Ogata K., Taniguchi H., Yamashita H., Tsuji K., Mizuguchi Y., Othomo N. Pathogenesis of Vibrio parahaemolyticus: intraperitoneal and orogastric challenge experiments in mice. Microbiol. and Immunol. J. 1990;34:355–366. doi: 10.1111/j.1348-0421.1990.tb01016.x. [DOI] [PubMed] [Google Scholar]
- 17.Hondo S., Goto I., Minematsu I., Ikeda N., Asano S., Ishibashi M., Kinoshita Y., Nishibuchi M., Honda T., Miwatani T. Gastroenteritis due to Kanagawa negative Vibrio parahaemolyticus. Lancet. 1987;1:331–332. doi: 10.1016/s0140-6736(87)92062-9. [DOI] [PubMed] [Google Scholar]
- 18.Honda T., Abad-Lapuebla M.A., Ni Y., Yamamoto K., Miwatani T. Characterization of a new thermostable direct haemolysin produced by a Kanagawa-phenomenon-negative clinical isolate of Vibrio parahaemolyticus. J. Gen.Microbiol. 1991;137:253–259. doi: 10.1099/00221287-137-2-253. [DOI] [PubMed] [Google Scholar]
- 19.Kaysner C.A., Abeyta C., Stott R.F., Krane M.H., Wekell M.M. Enumeration of Vibrio species, including Vibrio cholera, from samples of an oyster growing area, Grays Harbor, Washington. J. Food Protection. 1990;53:300–301. doi: 10.4315/0362-028X-53.4.300. [DOI] [PubMed] [Google Scholar]
- 20.Kaysner C.A., Tamplin M.L., Twedt R.M. Vanderzant C, Splittstoesser D.F. Vibrio. Washington, DC: 1992. Compendium of Methods for the Microbiological Examination of foods, American Public Health Association; pp. 451–473. [Google Scholar]
- 21.Kaysner C.A., DePaola J.A. 2004. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and Other Vibrio spp., Bacteriological Analytical Manual, 8th Edition. http://www.cfsan.fda.gov/ebam/bam-9.html. [Google Scholar]
- 22.Krempels D. 2006. [(acessed January 2010)]. Culture and sensitivity Test. In: Case of Infection.8. Disponível em: http://www.bio.miami.edu/hare/warrenpeace06.pdf. [Google Scholar]
- 23.Kuhne M., Wegmann S., Kobe A., Fries R. Tetracycline residues in bones of slaughtered animals. Food Control. 2000;11:175–180. [Google Scholar]
- 24.Krumperman P.H. Multiple antibiotic indexing of Escherichia coli to identify high risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin C., Yu R.C., Chou C.C. Susceptibility of Vibrio parahaemolyticus to various environmental stresses after cold shock treatment. Food Microbiol. 2004;92:207–215. doi: 10.1016/j.ijfoodmicro.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto C., Okuda J., Ishibashi M., Iwanaga M., Garg P., Rammamurthy T., Wong H.C., Depaola A., Kim Y.B., Albert M.J., Nishibuchi M. Pandemic spread of an O3: K6 clone of Vibrio parahamolyticus and emergence of related strains evidenced by arbitratily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 2000;38:578–585. doi: 10.1128/jcm.38.2.578-585.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Molitoris E., Joseph S.W., Krichevsky M.I., Sindhuhardja W., Colwell R.R. Characterization and distribution of Vibrio alginolyticus and V. parahaemolyticus isolated in Indonesia. Appl. Environ. Microbiol. 1985;50(6):1388–1394. doi: 10.1128/aem.50.6.1388-1394.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muntada-Garriga J.M., Rodriguez-Jerez J.J., Lopez-Sabater E.I, Mora-Ventura M.T. Effect of chill and freezing temperatures on survival of Vibrio parahaemolyticus inoculated in homogenates of oyster meat. Lett. Appl. Microbiol. 1995;20:225–227. doi: 10.1111/j.1472-765x.1995.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 29.Nishibuchi M., Kaper J.B. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 1985;162:558–564. doi: 10.1128/jb.162.2.558-564.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ottaviani D., Bacchiocchi I., Masini L., Leoni F., Carraturo A., Giammarioli M., Sbaraglia G. Antimicrobial susceptibility of potentially pathogenic halophilic vibrios isolated from seafood. Int. J. Antimicrobial Agents. 2001;18:135–140. doi: 10.1016/s0924-8579(01)00358-2. [DOI] [PubMed] [Google Scholar]
- 31.Rodrigues D.P., Hofer E. Vibrio species from the water-oyster ecosystem of Sepetiba Bay in Rio de Janeiro State, Brazil. Rev. de Microbiol. 1986;17:332–338. [Google Scholar]
- 32.Su Y.C., Chengchu L. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol. 2007;24(6):549–558. doi: 10.1016/j.fm.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 33.Taniguchi H., Ohta H., Ogawa M., Mizuguchi Y. Cloning and expression of Escherichia coli of Vibrio parahaemolyticus thermostable direct hemolysin and thermolabile hemolysin genes. J. Bacteriol. 1985;162:510–515. doi: 10.1128/jb.162.2.510-515.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tavares W. Manual de antibióticos e quimioterápicos antiinfecciosos. Atheneu, São Paulo; 2002. [Google Scholar]
- 35.Wagatsuma S. A medium for the test of the haemolytic activity of Vibrio parahaemolyticus. Media Circle. 1968;13:159–161. [Google Scholar]
- 36.Wong H.C., Ting S.H., Shich W.R. Incidence of toxigenic vibrios in foods available in Taiwan. J. Appl. Bacteriol. 1992;73:197–202. doi: 10.1111/j.1365-2672.1992.tb02978.x. [DOI] [PubMed] [Google Scholar]
- 37.Wong H.C., Shich W.R., Lee Y.S. Toxigenic characterization of vibrios isolated in foods available in Taiwan. J. Food Protection. 1993;56:980–982. doi: 10.4315/0362-028X-56.11.980. [DOI] [PubMed] [Google Scholar]
- 38.Wong H.C., Chen L.L., Yu C.M. Occurrence of vibrios in frozen seafoods and survival of psychrotrophic Vibrio cholerae in broth and shrimp homogenate at low temperatures. J. Food Protection. 1995;58:263–267. doi: 10.4315/0362-028X-58.3.263. [DOI] [PubMed] [Google Scholar]


