Abstract
Out of the vast pool of enzymes, proteolytic enzymes from microorganisms are the most widely used in different industries such as detergent, food, peptide production etc. Several marine microorganisms are known to produce proteases with commercially desirable characteristics. We have isolated nine different cultures from marine samples of the Indian Ocean. All of them were i) motile ii) rod shaped iii) non spore forming iv) catalase and amylase positive v) able to grow in presence of 10 % NaCl. They produced acid from glucose, fructose and maltose and grew optimally at 30 °C temperature and pH 7.0–8.0. None of them could grow above 45 °C and below 15 °C. Only one of them (MBRI 7) exhibited extracellular protease activity on skim milk agar plates. Based on 16S rDNA sequencing, it belonged to the genus Marinobacter (98% sequence similarity, 1201 bp). The cell free extract was used to study effects of temperature and pH on protease activity. The optimum temperature and pH for activity were found to be 40 °C and 7.0 respectively. The crude enzyme was stable at temperature range of 30–80 °C and pH 5.0–9.0. It retained 60 % activity at 80 °C after 4 h and more than 70 % activity at 70 °C after 1 h. D value was found to be 342 minutes and 78 minutes for 40 °C and 80 °C respectively. Interestingly the enzyme remained 50 % active at pH 9.0 after 1 h. Comparison with other proteases from different microbial sources indicated that the neutral protease from the halotolerant marine isolate MBRI 7 is a novel enzyme with high thermostability.
Keywords: Halotolerant, mesophilic, neutral protease, thermostable
INTRODUCTION
With the recent advent of biotechnology, there has been a growing interest and demand for enzymes with novel properties. Considerable efforts have been directed towards the selection of microorganisms producing enzymes with new physiological properties and tolerance to extreme conditions used in industrial processes such as temperature, salts, pH etc. Proteases represent the class of enzymes that occupy a pivotal position with respect to their physiological roles as well as to their commercial applications. More than 75 % of industrial enzymes are observed as hydrolases (17). Proteases constitute one of the most important groups of hydrolytic enzymes which act upon native proteins to disintegrate them into small peptides and amino acids. They have applications in different industries like detergent, food, pharmaceuticals, silk and leather, and also for recovery of silver from used X-ray films (2, 22, 26, 33, 37). They are also used for meat tenderization and in some medical applications (11, 34). Proteases account for 60 % of the total worldwide enzyme sales (26). Although proteases are widespread in nature, microbes serve as a preferred source of these enzymes presently. Microbes are characterized on their rapid growth, the limited space required for their cultivation and the ease of their generating capability. They can be genetically manipulated to generate new enzymes with altered properties that are desirable for their various applications. Presently simultaneous efforts are being made to increase the protease production by i) optimizing media composition ii) genetic manipulation iii) looking for new strains. Marine microorganisms have recently emerged as a rich source for the isolation of industrial enzymes compatible solutes, polyhydroxyalkanote and also for enhanced oil recovery (20) and generally they are non pathogenic. Marine bacterial enzymes have several advantages for industrial utilization. Their optimum activity usually occurs at high salinity, making these enzymes utilizable in many harsh industrial processes, where the concentrated salt solutions used would otherwise inhibit many enzymatic transformations. In addition, enzymes from halophilic organisms are reported to be stable at room temperature over long periods (10). We have earlier reported halotolerant organisms from wall scrapings of historical building in India (13) and also from some food samples (14) with an ability to produce industrially useful enzymes. Recently we have characterized some marine isolates from the Antarctic Ocean as well as from the Indian Ocean with respect to their fatty acid profiles (15, 32). Comparatively few reports are available on the saline- tolerant proteases from microbial sources. So, it is meaningful to isolate microorganisms producing protease with greater NaCl tolerance. Therefore the aim of the present study was (i) to screen different marine samples from the Indian Ocean for the microbes producing proteases (ii) to characterize the enzyme.
MATERIALS AND METHODS
The chemicals used were procured from Hi-Media (Mumbai, India), Merck (Mumbai, India), s d FINE-chem Ltd (Mumbai, India) and all were of analytical grade.
Sampling
Marine seawater samples from the Indian Ocean were collected from different locations. (Table 1)
Table 1.
Sampling location
| Sample | Latitude | Longitude | Depth(m) | pH | Temp (°C) | Salinity(ppt) | Conductivity(ms) | D.O.(mg/l) | Isolates |
|---|---|---|---|---|---|---|---|---|---|
| MBRI 3 | |||||||||
| 1 | 18°50’9.0”N | 72 °50’0.0”E | 13.5 | 8.48 | 30.1 | 71.73 | 978.4 | 4.14 | MBRI 4 |
| MBRI 5 | |||||||||
| 2 | 15°33.01”6”N | 73°56.58’0”E | 9.5 | 7.62 | 31.8 | 27.05 | 412.4 | 4.72 | MBRI 6 |
| MBRI 7 | |||||||||
| 3 | 15°33.01”6”N | 73°56.58’0”E | 14 | 7.75 | 31.5 | 37.64 | 557.6 | 5.32 | MBRI 8 |
| MBRI 11 | |||||||||
| 4 | 18°48”52”N | 72 ° 46’30”E | 13 | 8.02 | 30.4 | 71.61 | 974.7 | 974.7 | MBRI 9 |
| MBRI 10 |
Isolation and characterization of microorganisms
10 % of each individual sample was inoculated in Marine salt medium (MSM) (Composition per liter: 81.0 g NaCl, 10.0 g yeast extract, 9.6 g MgSO4, 7.0 g MgCl2, 5.0 g protease peptone no.3, 2.0 g KCl, 1.0 g glucose, 0.36 g CaCl2, 0.06 g NaHCO3 and 0.026 g NaBr) with pH adjusted to 7.0 + 0.2 and incubated at 30 °C, 120 rpm for 48 h. After 48 h, 0.1 ml of each sample was spread on MSM agar plates and incubated for another 48 h. The isolated colonies were maintained on MSM agar slants. The isolates were characterized morphologically and physiologically. They were studied for their salt-tolerance (8 % and above) and also examined for temperature tolerance by incubating at temperatures ranging from 15 °C to 45 °C for 24 h. Various biochemical tests were performed (8).
Screening of isolates for extracellular protease
Pure cultures isolated from samples were spot inoculated on skim milk agar plates (3) (Composition per liter: 5.0 g peptone, 3.0 g yeast extract, 1.0 g skim milk powder) and incubated at 30 °C for 24 h. A clear zone of hydrolysis appeared after 24 h incubation indicative of protease synthesis. The isolate showing extracellular protease activity was used for further studies.
16S rDNA sequencing
The genomic DNA was isolated as described by Ausubel et al. (4).The PCR assay was performed using Applied Biosystems, model 9800 with 1.5 μL of DNA extract in a total volume of 25 μL. The PCR master mixture contained 2.5 μL of 10X PCR reaction buffer (with 1.5 M MgCl2), 2.5 μL of 2 mM dNTPs, 1.25 μL of 10 pm μL-1 of each oligonucleotide primers 8F and 1392R, 0.2 μL of 3U μL-1 Taq DNA polymerase and 15.76 μL of glass-distilled PCR water.
Initially denaturation accomplished at 94 °C for 3 min. Thirty-two cycles of amplification consisted of denaturation at 94 °C for 30 sec, annealing at 55 °C for 30 sec and extension at 72 °C for 1.30 min. A final extension phase at 72 °C for 10 min was performed. The PCR product was purified by PEG-NaCl method. The sample was mixed with 0.6 times volume of PEG-NaCl, 20 % [PEG (MW 6000) and 2.5 M NaCl] and incubated for 20 min at 37 °C. The precipitate was collected by centrifugation at 3,800 rpm for 20 min. The pellet was washed with 70 % ethanol, air dried and dissolved in 12 μL sterile distilled water.
The sample was sequenced using a 96-well Applied Biosystems sequencing plate as per the manufacturer’s instructions. The thermocycling for the sequencing reactions began with an initial denaturation at 94 °C for 2 min, followed by 35 cycles of PCR consisting of denaturation at 94°C for 10 s, annealing at 50 °C for 10 s, and extension at 60 °C for 4 min using primers 704F (5’GTAGCGGTGAAATGCG TAGA 3’) and 907R(5’ CCGTCAATTCMTTTGAGTTT 3’). The samples were purified using standard protocols described by Applied Biosystems, Foster City, USA. To this, 10 μL of Hi-Di formamide was added and vortexed briefly.
The DNA was denatured by incubating at 95 °C for 3 min, kept on ice for 5–10 min, and was sequenced in a 3730 DNA analyzer (Applied Biosystems) following the manufacturer’s instructions. The obtained sequences of bacterial 16SrDNA (using 704F and 907R primers) were analysed using Sequence Scanner (Applied Biosystems) software. The 16S rDNA sequence contigs were generated using Chromas Pro and then analysed using online databases viz. NCBI-BLAST and Ribosomal Database Project (RDP) to find the closest match of the contig sequence.
Culture Conditions
The isolate showing extracellular protease activity (MBRI 7) was cultivated in MSM and incubated at 30 °C for 24 h. 10 % inoculum (107cells/ml) of this was used for all experiments.
For protease production skim milk broth (3) (Composition per liter : 5.0 g casein hydrolysate, 2.5 g yeast extract, 1.0 g glucose, 1.0 g skim milk powder) was inoculated with 10 % inoculum and incubated at 30 °C for 24 h. The sample was centrifuged at 10000 rpm for 10 minutes at 4 °C. The clear supernatant was used as crude enzyme for further experimental studies.
Enzyme Assay
Protease activity was measured using caseinolytic assay (16) with some modifications. One unit is defined as the amount of enzyme that hydrolyzes casein to produce colour equivalent to 1.0 µmol of tyrosine per minute under standard conditions.
Protein Assay
Total protein of the cell free filtrate was determined by the method of Lowry et al (18). Bovine serum albumin (1000 µg mL-1) was used as a standard.
Growth curve studies
Growth curve characteristics of the isolate were studied by inoculating overnight grown culture in skim milk broth and incubating at 30 °C with shaking at 120 rpm. Growth was determined at the intervals of 4 h for 48 h by measuring absorbance at 660 nm. Each sample was used to determine protease activity.
Effect of temperature and pH on enzyme activity
Protease assay was performed under standard conditions at various temperatures ranging from 30–70 °C to study the effect of temperature on enzyme activity. Optimum pH for protease activity was determined by carrying out the protease assay at various pH between 5–10. Protein assay was carried out as described above.
Effect of temperature and pH on enzyme stability
Cell free extract was exposed to various temperatures ranging from 30–80 °C to study the effect of increase in temperature on enzyme activity. The samples were removed at different time intervals ranging from 30 minutes to 8 h and protease activity was determined. Effect of change in pH on the activity of enzyme was studied as per the method described by Adinarayana et al (1). Different buffers (0.2M)) used were acetate (pH 5.0), potassium phosphate (pH 6.0–7.0), tris/HCl (pH 8.0) and carbonate (pH 9.0) (30). Enzyme samples were exposed to different pH (5–9) for 30 minutes at 40 °C and the assay was performed as described earlier.
Statistical analysis
All experiments were performed in triplicate. For statistical analysis, the standard error of the mean values was calculated and the means were tested according to Students t-test for significant differences among the samples. A statistical significance was accepted at P< 0.05.
RESULTS AND DISCUSSION
In the present study, four different marine samples (Table 1) from the Indian Ocean were used to isolate the bacterial cultures. Total nine cultures were isolated from the samples. They were characterized morphologically and physiologically (Table 2). They were found to be motile, non spore forming, catalase and amylase positive. They i) produced acid for glucose, fructose and maltose ii) showed optimum temperature 35 °C and pH 7.0–8.0 for growth iii) tolerated NaCl up to 10 %. None of them could grow above 45 °C and below 15 °C. Only MBRI 3, MBRI 4, and MBRI 5 tolerated alkaline pH (10.0). MBRI 3 and 5 tolerated NaCl upto 25 % (Table 2).
Table 2.
Characteristies differentiating marine organisms
| Characteristics | MBRI 3 | MBRI 4 | MBRI 5 | MBRI 6 | MBRI 7 | MBRI 8 | MBRI 9 | MBRI 10 | MBRI 11 |
|---|---|---|---|---|---|---|---|---|---|
| Gram character | + | + | - | + | - | - | - | - | - |
| Temperature ºC | |||||||||
| 15 | - | - | + | - | - | - | + | ++ | - |
| 28 | + | + | + | + | + | + | ++ | + | ++ |
| 35 | ++ | ++ | +++ | +++ | +++ | +++ | +++ | ++ | +++ |
| 45 | +++ | +++ | + | ++ | + | + | + | + | + |
| pH | |||||||||
| 4 | - | - | - | + | - | - | - | - | - |
| 5 | - | - | - | ++ | + | + | - | - | - |
| 6 | +++ | + | + | ++ | ++ | + | - | - | + |
| 7 | + | + | + | +++ | +++ | +++ | +++ | +++ | +++ |
| 8 | + | +++ | +++ | ++ | ++ | ++ | + | + | ++ |
| 9 | + | + | + | - | - | - | - | + | - |
| 10 | + | + | + | - | - | - | + | - | - |
| Salt tolerance (%) | |||||||||
| 2 | + | + | + | + | -- | - | - | - | - |
| 5 | + | + | + | + | + | + | - | + | + |
| 8 | + | +++ | + | + | + | + | + | + | + |
| 10 | + | + | + | + | + | + | + | + | + |
| 15 | +++ | - | +++ | - | + | + | + | + | + |
| 20 | + | - | + | - | - | - | - | - | - |
| 25 | + | - | + | - | - | - | - | - | - |
| Acid production from | |||||||||
| Lactose | + | - | + | - | + | + | - | - | + |
| Sucrose | + | + | + | + | + | - | + | - | - |
| Mannitol | + | - | + | + | + | - | + | + | - |
| Biochemical characterization | |||||||||
| Citrate hydrolysis | + | + | + | - | - | - | - | - | + |
| Urease | + | + | - | - | + | + | + | + | + |
| Gelatin hydrolysis | - | + | + | + | + | + | - | - | - |
| Protease | - | - | - | - | + | - | - | - | - |
Keywords: + = moderate Growth, ++=heavy growth, -=no growth.
Only MBRI 7 exhibited extracellular protease activity on skim milk agar. It was identified using 16S rDNA sequencing. The sequence was deposited in EMBL+Genbank under accession number HQ199216. BLAST analysis of partial 16S rDNA sequence revealed that the strain belonged to Gammaproteobacteria and was closely related to Marinobacter sp (98 % sequence similarity, 1201 bp). Ming-Yang et al. (24) have reported that predominant cultivated protease producing bacteria are Gammaproteobacteria affiliated with the genera Pseudoalteromonas, Marinobacter, Alteromonas, Halomonas, Shewanella etc. Our results are in line with this observation. Marinobacter lipolyticus sp. nov., growing optimally at 7.5 % NaCl showed lipolytic activity along with other hydrolytic enzymes including protease (21).
The growth curve studies revealed that the culture entered late log phase after 16 h (Fig. 1) and entered stationary phase after 28 h. Maximum protease activity was obtained after 16 h of incubation (Fig 1). Earlier studies on Bacillus sp. producing proteases reported longer incubation periods such as 48, 72, 96 h for B. subtilis PE-11 (27), B. subtilis 3411 and Bacillus sp. K-30 (25, 28) respectively. This indicates that MBRI 7 (present work) exhibited maximum protease activity in lesser fermentation time which is a major prerequisite for commercial production.
Figure 1.
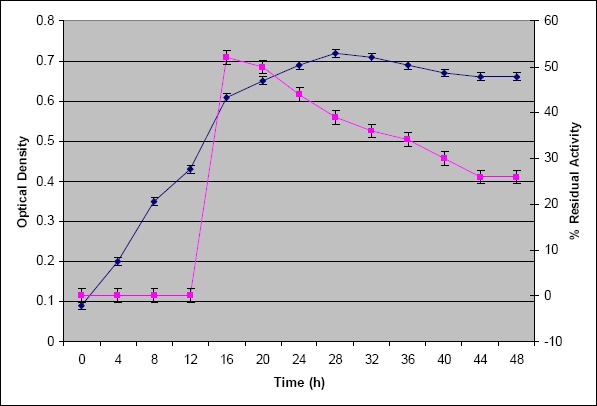
Growth curve of an isolate MBRI 7 (present work). The cells were inoculated in skim milk broth, incubated at 30 °C, 120 rpm and growth was determined by measuring absorbance at 660 nm. Protease activity was estimated at different time intervals and maximum activity was observed after 16 h (♦ optical density, ■ % residual activity).
Quantitative estimation of protease activity using crude cell free extract of MBRI 7 yielded 52 U mL-1 activity. Streptomyces microflavus (31) and Bacillus amyloliquefaciens CMB01 (5) showed 726 U mL-1 and 2110 U mL-1 activity in crude cell free extract respectively. However neutral protease of Bacillus sp. 158 exhibited 30 U mL-1 activity in crude cell free extract (29). Thus MBRI 7 showed moderate protease activity in cell free extract that may be increased by mutations and/or by genetic manipulations.
Our results of temperature and pH optimization showed that the enzyme had maximum activity at 40 °C and at pH 7.0 (Fig. 2a and 2b) indicating that it was a neutral protease. Strain R2 of Marinobacter sp. produced protease optimally at 20 °C, pH 9.0 to 10.0 (36).
Figure 2.
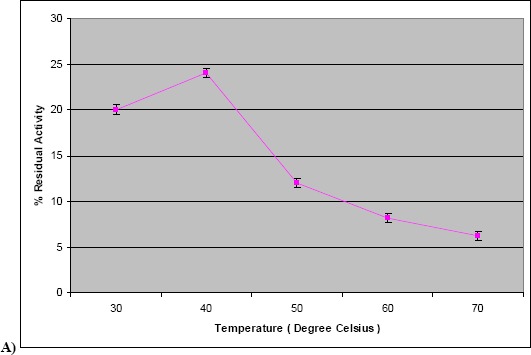
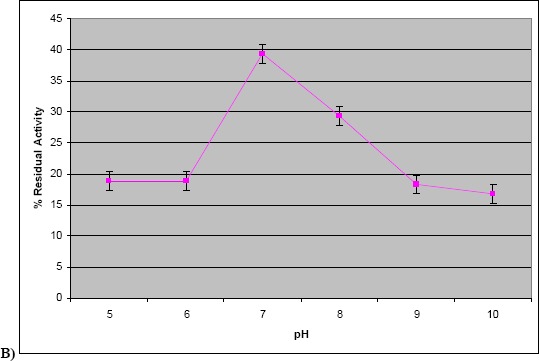
a) Effect of temperature on protease activity. The assay was carried out using crude culture supernatant at different temperatures ranging from 30 to 70 °C. b) Effect of pH on protease activity. The assay was carried out using crude culture supernatant at different pH in the range of 5.0–10.0.
Effect of varying temperature and pH on enzyme stability of protease was determined. Figure 3a depicts kinetics of thermal inactivation of crude enzyme. D values of the enzyme at different temperatures were determined and used to calculate z value. D values were in the range of 342 minutes to 78 minutes for temperatures ranging from 40 °C to 80 °C. Based on D values for the crude enzyme at different temperatures, z value was determined graphically and it was found to be 56 °C (Fig. 3b). The enzyme retained 100 % activity at 40 °C after 4 h. It showed 60 % activity at 80 °C after 4 h and more than 70 % activity was retained at 70 °C after 1 hr. Extracellular alkaline proteases have been reported from halophilic and alkaline bacteria isolated from saline habitat of costal Gujarat in India with optimum pH 9.0 to 9.5 (10). Extracellular protease from thermophilic Bacillus sp SMIA 2 was stable for 2 h at 30 °C while at 40 ºC and 80 ºC, 14 % and 84 % of the original activities were lost, respectively (26). Heat stable protease from P. fluroescences P26 was found to require 15 hrs at 62.8 °C and 9 minutes at 121 °C for complete inactivation (23). The neutral protease of Pseudomonas aeruginosa CCRC 15541 retained 85% activity for 1 hr at 60 °C (19) while the acid protease of Penicillium duponti K1014 was stable at 60 °C for 1 hr and retained more than 65 % of original activity for 1 hr at 70 °C (12). In the light of above, protease produced by Marinobacter sp.MBRI 7 (present work) is a novel extracellular neutral protease with significant thermostability. Besides, the enzyme exhibited 50 % activity at pH 9 after 1 h indicating reasonable stability at alkaline pH.
Figure 3.
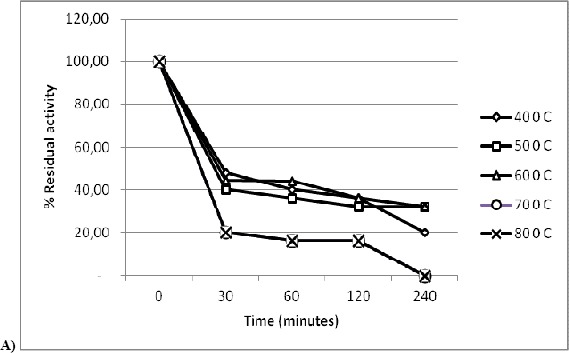
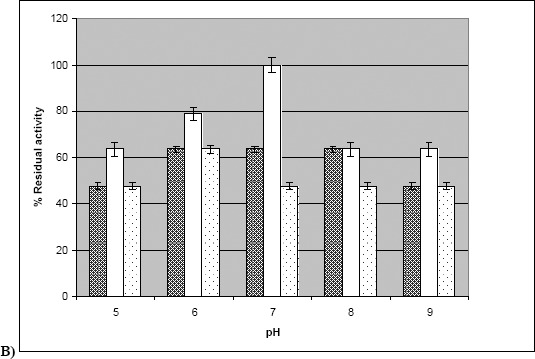
a) Kinetics of thermal inactivation of crude enzyme. Culture supernatant was exposed to different temperatures ranging from 30–80 °C. Enzyme activity was measured at time intervals of 30 minutes, 1 h, 2 h, and 4 h. b) Stability of crude enzyme at various pH. Culture supernatant was exposed to various pH ranging from 5.0–9.0. Enzyme activity was measured at time intervals of 30 minutes and 60 minutes.(100 % activity corresponds to 52 U mL-1) (▓ - 0 minutes, □ - 30 minutes, ░ - 60 minutes).
Increased awareness of the use of biocatalytic capabilities of the enzymes and microorganisms has made possible the development of biologically based processes and products (9). Interest in thermostable enzymes has grown, mainly due to the fact that most of the existing industrial enzyme processes are run at high temperatures using enzymes from mesophilic sources. There are certain advantages in using thermostable enzymes in industrial processes such as high reaction rate, longer half life, lesser risk of contamination, and decrease in viscosity allowing higher concentration of less soluble materials (7). The main application for thermostable enzymes has predominantly been food processing and in detergents industries (38). The results of the present study indicate that the neutral protease from MBRI 7, a novel thermostable enzyme from mesophilic organism, appears to have considerable potential for industrial applications. Advances in the field of genetic and enzyme engineering coupled with the novel properties of this enzyme from halotolerant microorganism will make it possible to move the desired gene coding for this novel enzyme from prokaryote to eukaryote and to amplify its expression.
ACKNOWLEDGEMENTS
The authors are indebted to Dr. S. S. Kadam, Vice Chancellor, Bharati Vidyapeeth University, Pune, Maharashtra, India and Dr. R. M. Kothari, Principal, Rajiv Gandhi Institute of IT and Biotechnology for allowing to conduct this work at Rajiv Gandhi Institute of IT and Biotechnology, Bharati Vidyapeeth University. The authors are also thankful to Dipti Salunkhe and Vipra Jadhav for their frequent help.
REFERENCES
- 1.Adinarayana K., Ellaih P. Response surface optimization of the critical medium component for the production of alkaline protease by a newly isolated Bacillus sp. J. Pharm. Pharmaceut. Sci. 2002;5:272–278. [PubMed] [Google Scholar]
- 2.Anisworth S.J. Soaps and detergents. Chem. Eng. News. 1994;72:34–59. [Google Scholar]
- 3.Atlas R.M. Handbook of Microbiological Media. Florida, USA: CRC Press, Boca Raton; 1997. [Google Scholar]
- 4.Ausubel F.M., Rrent R. E., Kingston D. D., Moore J. G., Seidman J. A., Struhl Smith and K. Newyork: Wiley; 1987. Current Protocol in Molecular Biology. [Google Scholar]
- 5.Ban O.H., Han S.S., Lee Y.N. Identification of a potent protease-producing bacterial isolate, Bacillus amyloliquefaciens CMB01. Ann. Microbiol. 2003;53:95–103. [Google Scholar]
- 6.Brenner D.J., Krieg N.R., Staley J.T. Bergey’s Manual of Systematic Bacteriology. New York, USA: Springer; 2005. [Google Scholar]
- 7.Rocha da Silva C., Delatorre A., Martins M. Effect of the culture conditions on the production of an extracellular protease by thermophilic bacillus sp and some properties of the enzymatic activity. Braz. J. Microbiol. 2007;38:253–258. [Google Scholar]
- 8.Cappuccino J.G., Sherman N. Microbiology A Laboratory Manual. Singapore: Pearson Education Pte. Ltd; 2005. [Google Scholar]
- 9.Chandrashekharan M, M.M. Industrial enzymes from marine microorganism: The Indian Scenario. J. Mar. Biotechnol. 1997;5:86–94. [Google Scholar]
- 10.Dodia1 M. S., Joshi1 R.H., Patel R.K., Singh S. P. Characterization and stability of extracellular alkaline proteases from halophilic and alkaliphilic bacteria isolated from saline habitat of coastal Gujarat, India Braz. J. Microbiol. 2006;37:276–282. [Google Scholar]
- 11.Gajju H., Bhalla T.C., Agarwal O.H. Thermostable alkaline protease from thermophilic Bacillus coagulans PB-77. Indian. J. Microbiol. 1966;36:153–155. [Google Scholar]
- 12.Hashimoto H., Iwaasa T., Yokotsuka T. Thermostable acid protease produced by Penicillium duponti K1014, a true thermophilic fungus newly isolated from compost. Appl. Environ. Microbiol. 1972;24:986–992. doi: 10.1128/am.24.6.986-992.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jadhav G., Salunkhe D., Nerkar D., Bhadekar R. Novel Staphylococcus sp. isolated from wall scrapings of a historical building in India. Ann. Microbiol. 2010;60:197–201. [Google Scholar]
- 14.Jadhav G.G., Salunkhe D.S., Nerkar D.P., Bhadekar R.K. Isolation and characterization of salt-tolerant nitrogen-fixing microorganisms from food. EurAsia. J. BioSci. 2010;4:33–34. [Google Scholar]
- 15.Jadhav V. V., Jamle M.M., Pawar P.D., Devare M.N., Bhadekar R.K. Fatty acid profiles of PUFA producing Antarctic bacteria: correlation with RAPD analysis. Ann. Microbiol. 2010 (In Press) DOI: 10.1007/s13213–010–0114–4. [Google Scholar]
- 16.Jayaraman J. Laboratory manual in Biochemistry. New Delhi: New Age International Publishers; 2007. [Google Scholar]
- 17.Leiola M., Jokela J., Pastinen O., Turunen O. Industrial use of enzymes. The Netherlands: [Google Scholar]
- 18.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with Folin phenol reagent. J. Biol. Chem. 1915;193:265–275. [PubMed] [Google Scholar]
- 19.Lu S.F., Chang P.P. A thermostable neutral protease from Pseudomonas aeruginosa CCRC 15541. Society. For. Applied. Microbiology. 1996;22:5–9. [Google Scholar]
- 20.Malashetty V., Prakash S., Mahajan V., Shouche Y.S., Sreeramulu1 K. Purification and characterization of an extreme halothermophilic protease from a halophilic bacterium Chromohalobacter sp. TVSP101. Braz. J. Microbiol. 2009;40:12–19. doi: 10.1590/S1517-83822009000100002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martı´n S., Ma´ rquez. M. C., Sa´nchez-Porro C., Mellado E., Arahal D. R., Ventosa A. Marinobacter lipolyticus sp. nov., a novel moderate halophile with lipolytic activity. Int. J. Syst. Evol. Microbiol. 2003;53:1383–1387. doi: 10.1099/ijs.0.02528-0. [DOI] [PubMed] [Google Scholar]
- 22.Masse F.W.J.L., Tilburg R.V. The benefit of detergent enzymes under changing washing conditions. J. Am. Oil. Chem. Soc. 1983;60:1672–1675. [Google Scholar]
- 23.Mayerhofer H. J., Marshall R. T., White C. H., Lu M. Characterization of a heat stable protease of Pseudomonas fluroscens P26. Appl. Microbiol. 1973;25:44–48. doi: 10.1128/am.25.1.44-48.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ming-Yang Z., Xiu-Lan C., Hui-Lin Z., Hong-Yue D., Xi-Wu L., Xi-Ying Z, Hai-Lun H, Bai-Cheng Z., Yu-Zhong Z. Diversity of Both the Cultivable Protease-Producing Bacteria and Their Extracellular Proteases in the Sediments of the South China Sea. Microbial ecology. 2009;58:582–590. doi: 10.1007/s00248-009-9506-z. [DOI] [PubMed] [Google Scholar]
- 25.Naidu K.S.B., Devi K.L. Optimization of thermostable alkaline protease production from species of Bacillus using rice bran. Afr. J. Biotechnol. 2005;4:724–726. [Google Scholar]
- 26.Nascimento W., Martins M. Production and properties of an extracellular protease from thermophilic Bacillus sp. Braz. J. Microbiol. 2004;35:91–96. [Google Scholar]
- 27.Oghbady L.T., Okagbue R.N., Ahmed A.A. Glutamic acid production by Bacillus isolates from Nigerian fermented vegetable proteins. World. J. Microbiol. Biotechnol. 1990;6:337–382. doi: 10.1007/BF01202118. [DOI] [PubMed] [Google Scholar]
- 28.Pastor M.D., Lorda G.S., Balatti A. Protease 0btention using Bacillus subtilis 3411 and Amaranth seed meal medium of different aeration rate. Braz. J. Microbiol. 2001;32:6–9. [Google Scholar]
- 29.Pawar R., Zambare V., Barve S., Paratkar G. Application of Protease Isolated from Bacillus sp. 158 in Enzymatic Cleansing of Contact Lenses. Biotechnol. 2009;8:276–278. [Google Scholar]
- 30.Plummer D.T. An Introduction To Practical Biochemistry. New Delhi: Tata Mc Graw Hill Publishing Company Ltd; 1988. [Google Scholar]
- 31.Rifaat H.M., Hassanein S.M., El-Said O.H., Saleh S.A., Selim M.S.M. Purification and characterization of extracellular neutral protease from Streptomyces microflavus. Arab. J. Biotech. 2006;9:51–60. [Google Scholar]
- 32.Salunkhe D.S., Tiwari N., Walunjkar S.A., Bhadekar R.K. Halomonas sp. Nov., an EPA producing mesophilic marine isolate from the Indian Ocean. Pol. J Microbiol. 2010 (In press) [PubMed] [Google Scholar]
- 33.Sangeetha R., Geetha A., Arulpandi I. Concomitant production of protease and lipase by Bacillus licheniformis vsg1: production, purification and characterization. Braz. J. Microbiol. 2010;41:179–185. doi: 10.1590/S1517-838220100001000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tunga R., Banerjee I.T., Bhattarcharya B.C. Optimizing some factors affecting protease production under solid-state fermentation. Bioprocess. Eng. 1988;19:187–190. [Google Scholar]
- 35.Ventosa A., Nieto J.J. Biotechnological applications and potentialities of halophilic microorganism. World. J. Microbial. Biotechnol. 1995;11:85–94. doi: 10.1007/BF00339138. [DOI] [PubMed] [Google Scholar]
- 36.Wei L.N., Rui Z., Jing Z., Ying Y. Z. Cold-active Protease from Antarctic Bacterium Marinobacter sp. Strain R2: Fermentation Condition and Enzyme Properties. Journal of Xiamen University (NATURAL SCIENCE) 2004;43:865–870. [Google Scholar]
- 37.Wolf A.M., Showell M.S., Venegas M.G., Barnett B.L., Wertz W.C. Laundry performance of subtilisin enzyme - Practical protein engineering. New York: Plenum Press; 1993. [Google Scholar]
- 38.Zamost B.L., Nielsen H.K., Starnes R.L. Thermostable enzymes for industrial applications. J. Ind. Microbiol. 1991;8:71–82. [Google Scholar]


