Abstract
Isolation and genetic characterization of an environmental Vibrio cholerae O1 from the Amazon is reported. This strain lacks two major virulence factors - CTX and TCP - but carries other genes related to virulence. Genetic similarity with epidemic strains is evaluated and the importance of V. cholerae surveillance in the Amazon is emphasized.
Keywords: Vibrio cholerae O1, environment, Amazon, virulence genes, PFGE
Vibrio cholerae is a ubiquitous bacterium of aquatic ecosystems and some of its strains are the etiological agents of the Asian cholera disease. It is classified into 206 serogroups (O1-O206) on the basis of an outer-membrane O antigen and, further, into biotypes - e.g. El Tor and Classical - considering several biochemical and genomic markers (1, 2). The ongoing cholera seventh pandemic, as well the previous one, and most of the cholera epidemics up to date, is due to V. cholerae from the O1 serogroup.
Cholera emerged in South America in 1991, reaching Brazil at the same year through the Amazon region. V. cholerae O1 strains from the El Tor biotype - the lineage determinant of the seventh cholera pandemic - were isolated and characterized from clinical cases since the beginning of the epidemic (3, 4). Two distinct V. cholerae O1 El Tor strains were also identified from cholera-like diarrhea in the Brazilian Amazon during the 1991 - 1996 epidemic: a non-toxigenic strain called the Amazonia variant (5) and a sucrose late fermenting variant (6). The Amazonia strain was responsible for outbreaks during 1991-1992, but remained restricted to small villages in the westernmost Amazon. While the sucrose late fermenting variant spread quickly all over the region accounting for most of the cholera cases registered during 1994 to 1995.
The last case of cholera recorded in the state of Pará was in 1995, since then no other clinical or environmental V. cholerae O1 strain was isolated in the Brazilian Amazon. By September 2007, in the context of environmental monitoring (8), a V. cholerae O1 strain was isolated from a superficial water sample. For this isolation, 10 L of superficial water collected in the Tucunduba Stream (Terra Firme district, Belém - PA) was concentrated. All the culture and identification procedures were done as previously described (8). Briefly, the LMA 3984-4 isolate was obtained from the second alkaline peptone water enrichment (6h at 35°C) plated on TCBS agar (18-24h at 35°C). This strain was submitted to several biochemical tests: Kliger Iron Agar (KIA) - MERCK -K/A-, Lysine Iron Agar (LIA) - DIFCO® -K/K-, oxidase-positive, sensitivity to the vibriostatic agent O:129 (10 μg-R) e (150μg-S), glucose fermentation (+), lactose (-), lysine/ornithine decarboxylase (+), arginine dehidrolase (-), indole (+), H2S (-), urease (-), triptophan-deaminase (-), and motility (+), besides positive serology through fast agglutination screening test employing the somatic O1 antiserum and the Ogawa serotype from DIFCO®.
Polymerase chain reactions (PCR) was performed for a set of specific virulence genes associated with the choleragenic El Tor lineage: ctx, zot, ace, orfU, tcpA, pTLC, toxR, hlyA, hlyC, hlyB, rtxA, rtxC and stn/sto (9). Positive results were achieved for toxR, hlyA, hlyC, hlyB, rtxA e rtxC and confirmed by sequencing. Although it doesn´t harbor the major V. cholerae virulence determinants - cholera toxin (CT) and the toxin-corregulated pilus (TCP) - LMA 3984-4 strain could potentially express several toxins with well described biological functions and identified in several clinical V. cholerae strains, as those coded by the hly and rtx genes.
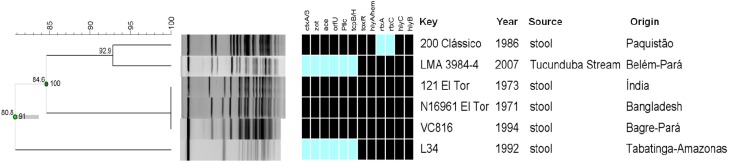
In order to determine the genetic similarity of the LMA 3984-4 strain with other epidemic V. cholerae O1 strains, NotI digested genomic DNA was separated by pulsed field gel electrophoresis and analyzed as previously described (10, 11). Five reference strains were used in this comparison: strain N16961 from Bangladesh and 121 from India, both representatives of the current seventh cholera pandemic El Tor biotype; strain 200, O1 representative of the classical biotype; strain Amazonia L34, described above and; a clinical isolate from the recent Brazilian cholera epidemic (VC816). All strains used in this comparison were obtained from the V. cholerae Collection of the Laboratory of Molecular Genetics of Microorganisms/Oswaldo Cruz Foundation.
Two clusters were defined by the PFGE patterns: one grouping strains from the classical biotype, LMA 3984-4 and the El Tor biotypes and the other with the Amazonia variant (L34) (Figure 1). The LMA 3984-4 differs genetically from the El Tor strains at the approximate rate of 15,4%. The similarity between the Amazonia strain (L34) and the others (N16961, 121, V816, LMA3984-4 and 200) was of 80,8%. When comparing the LMA 3984-4 and the classical biotype, the similarity was of 92,9%. These similarity rates did not reveal a clonal relation in between the LMA 3984-4 strain and other epidemic V. cholerae strains. Concerning the set of virulence associated genes tested, the LMA 3984-4 isolate shares the same profile as the Amazonia strain (Figure 1).
Figure 1.
PFGE profile (NotI) of the V.cholerae O1 LMA3984-4 strain and reference strains. Grouping by similarity was determined by the Dice coefficient and the UPGMA method. The blue/black matrix stands for presence/absence of the virulence genes named in the sidebar. The samples name codes are listed at the right side.
Studies have shown that even without the major virulence determinants some V. cholerae strains are the causing agents of severe diarrhea and are considered a potential threat to human health (12, 13, 14). Some strains could also acquire the TCP and CTX genes - determinants of choleragenic virulence - that are mobile genomic elements that can transit in between susceptible strains (15). Further extended genomic analysis, such as complete genome sequencing, is necessary to determine if the differences in the LMA 3984-4 and the epidemic strains reflect the acquisition, loss, or alteration of mobile genomic elements or, alternatively, if it´s not directly related to known epidemic lineages but acquired the O1 antigen through horizontal gene transfer (HGT).
This isolation reinforces the concept that the O1 serotype label is not absolute in the identification of epidemic V. cholerae strains (15) even though it is largely used as the major routine monitoring element for pathogenic isolates. The identification of an environmental V. cholerae O1 strain, carrying a set of virulence related genes, clustering by PFGE pattern with pandemic isolates and probably adapted to the fresh water ecosystem of the Amazon, emphasizes the importance of the environmental surveillance of this bacterium in the region.
Acknowledgments
We thank Evandro Chagas and Oswaldo Cruz Institute for financial suport, Raimundo Pio Girard, Geralda Amélia Rezende, Ritangela Santos and Rita Felix for technical support and to the program of Biology of Infecctious and Parasite Agents (BAIP) from the Federal University of Pará.
REFERENCES
- 1.Cameron D.N., Khambaty F.M., Wachsmuth I.K., Tauxe R.V., Barrett T.J. Molecular characterization of Vibrio cholerae O1 strains by pulsed-field gel electrophoresis. J Clin Microbiol. 1994;32:1685–1690. doi: 10.1128/jcm.32.7.1685-1690.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chatterjee S., Ghosh K., Raychoudhuri A., Chowdhury G., Bhattacharya M.K., Mukhopadhyay A.K., Ramamurthy T., Bhattacharya S.K., Klose K.E., Nandy R.K. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2009;47(4):1087–95. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun J., Grim C., Hasan N., Lee J., Choi S., Haley B., Taviani E., Jeon Y., Kim D., Lee J., Brettin T., Bruce D., Challacombe J., Detter J., Han C., Munk A., Chertkov O., Meincke L., Saunders E., Walters R., Huq A., Nair G., Colwell R. Comparative genomics reveals mechanism for short-term and long-term clonal transitions in pandemic Vibrio cholerae. PNAS. 2009;106(36):15442–47. doi: 10.1073/pnas.0907787106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coelho A., Andrade J.R.C., Vicente A.C.P., Salles C.A. New variant of Vibrio choleae O1 from clinical isolates in Amazonia. J Clin Microbiol. 1995;33:114–118. doi: 10.1128/jcm.33.1.114-118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalsgaard A., Albert M.J., Taylor D.N., Shimada T., Meza R., Serichantalergs O., Echeverria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33(10):2715–22. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faruque S. M., Albert M. J., Mekalanos J. J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faruque S., Mekalanos J. Pathogenicity islands and phages in Vibrio cholerae evolution. Trends Microbiol. 11(11):505–10. doi: 10.1016/j.tim.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Gerolomo M., Penna M.L.F. Os primeiros cinco anos da sétima pandemia de cólera no Brasil. Inf. Epidemiol. Sus. 1999;8(3):49–58. [Google Scholar]
- 9.Kaper J.B., Morris J.B.Jr., Levine M.M. Cholera. Clin Microbiol Rev. 1995;8(1):48–86. doi: 10.1128/cmr.8.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pichel M., Rivas M., Chinen I., Martín F., Ibarra C., Binsztein N. Genetic diversity of Vibrio cholerae O1 in Argentina and emergence of a new variant. J Clin Microbiol. 2003;41:124–134. doi: 10.1128/JCM.41.1.124-134.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramos F.L.P., Lainson Z.C.L., Silva E.L., Proiete A.A., Mareco M.L., Lamarão M.L.N. Cholera in North Brasil: on the Occurrence of strains of Vibrio cholerae O1 wich fail to Ferment Sucrose During Routine Plating on Thiosulphate-Citrate-Bile Salt-Sucrose Agar (TCBS). A New Problem in Diagnosis and Control? Rev Latinoam Microbiol. 1997;39:141–144. [PubMed] [Google Scholar]
- 12.Raychoudhuri A., Mukhopadhyay A.K., Ramamurthy T., Nandy R.K., Takeda Y., Oshifumi, Nair G.B. Biotyping of Vibrio cholerae O1: Time to redefine the scheme. Indian J Med Res. 2008;128:695–698. [PubMed] [Google Scholar]
- 13.Sá L.L.C. Diversidade genética de isolados ambientais de Vibrio cholerae da Amazônia brasileira. Belém, Pará, Brasil: 2009. p. 137. (D.Sc. Tesis. Biologia de Agentes Infeciciosos e Parasitários, Instituto de Ciências Biológicas. UFPA) [Google Scholar]
- 14.Sá L.L.C., Vale E.R., Santos E.C.O., Jesus I.M., Mendonça N.M., Theophilo G.N.D., Ferreira F.S.F., Hofer; E., Loureiro E.C.B. Vigilância ambiental para cólera na região metropolitana de Belém, Pará, no período de 1999 a 2006. Cad Saude Colet. 2007;15(4):449–466. [Google Scholar]
- 15.Teixeira L.M., Carvalho M.G.S., Merquior V.L.C., Steigerwalt A.G., Brenner D.J., Facklam R.R. Phenotypic and genotypic characterization of Vagococcus fluvialis, including strains isolated from human sources. J Clin Microbiol. 1997;35:2778–2781. doi: 10.1128/jcm.35.11.2778-2781.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vicente A.C., Coelho A.M. 1990s Vibrio cholerae epidemic, Brazil. Emerg Infect Dis. 2005;11(1):171–2. doi: 10.3201/eid1101.040484. [DOI] [PMC free article] [PubMed] [Google Scholar]