Abstract
Various cultivation parameters were optimized for the production of extra cellular protease by Brevibacterium linens DSM 20158 grown in solid state fermentation conditions using statistical approach. The cultivation variables were screened by the Plackett–Burman design and four significant variables (soybean meal, wheat bran, (NH4)2SO4 and inoculum size were further optimized via central composite design (CCD) using a response surface methodological approach. Using the optimal factors (soybean meal 12.0g, wheat bran 8.50g, (NH4)2SO4) 0.45g and inoculum size 3.50%), the rate of protease production was found to be twofold higher in the optimized medium as compared to the unoptimized reference medium.
Keywords: Brevibacterium linens, Protease, Solid-state fermentation, Plackett–Burman Design, Central Composite Design (CCD)
INTRODUCTION
Brevibacterium linens influences the surface ripening and final characteristics of smear of cheeses due to its enzymatic and biochemical properties. The greatest numbers of studies have focused on the extracellular proteases of the Brevibacterium linens, largely because of their high activity and importance with respect to cheese ripening (23). Proteases, also known as peptidyl-peptide hydrolases, are important industrial enzymes, which are responsible for approximately 60% of all enzyme sales (25), and are utilized extensively in a variety of industries, including laundry detergents, meat tenderization, cheese-making, dehairing, brewery, baking, the production of digestive aids, and the recovery of silver from photographic film. The use of proteases as detergent additives stimulated their commercial development and resulted in a significant expansion of fundamental research into these enzymes (8). Bacterial neutral proteases work within a narrow pH range and have low thermal tolerance. Due to their intermediate rate of reaction, neutral proteases generate less bitterness in hydrolyzed food proteins than do the animal proteases and hence are important for use in the food industry (36).
Solid state fermentation has gained attention due to the several advantages it has over submerged fermentation (18). The protease production in submerged fermentation was found to be 2.57- 3.5 times lower than the highest enzyme production in solid state fermentation using wheat bran as solid support with 50% moisture content as reported by various researchers (30, 32). Solid-state fermentation is also finding increased application in the production of enzymes, antibiotics, biocides surfactants from wastes such as wheat bran, soybean meal used by various researchers (27, 32). Ferracini and Sato (4) have reported an increased level of protease production by using MgS04 and NH4SO4 salts in solid state cultivation medium. There have been significant additions to the science and engineering knowledge of solid-state fermentations recently (4, 32, 34). In the future, solid-state fermentation may well be developed to the same level as submerged fermentation, if validation and standardization continues according to the current trend (31). From the viewpoint of Pandey et al. (19) the production of proteolytic enzymes in solid-state fermentation systems has greater economic feasibility than the submerged method. Optimization of medium by the traditional method, involving changing one independent variable while fixing all others, is extremely time-consuming and expensive for a large number of variables. To overcome this difficulty, experimental factorial design and response methodology can be used to optimize medium components (1). The Plackett–Burman factorial designs allow screening of significant factors from a large number of fermentation variables, and these designs are thus useful for selecting variables for further optimization processes (25).
Response surface methodology (RSM) is effectively used for designing experiments with optimum conditions for factors affecting the various bioprocesses (36). A statistically designed optimization study is helpful in confirming previous effects and interactions of fermentation variables and in determining the optimum values of the critical factors (10). The application of statistical experimental design techniques in fermentation process development can result in an improvement of product yield, reduce process variability, give closer confirmation of the output response to the experimental values and reduce overall costs. RSM can be used to evaluate the relative significance of several factors even in the presence of complex interactions (14, 20).
Keeping in view the green environment, as well as the contribution to meet the increased demand of protease production at industrial level, the present study has been designed to improve the protease yield by using agro-industrial wastes as solid substrates because many researchers have reported the enhanced production of proteases as compared to the submerged fermentation processes. Moreover, statistical approaches offer ideal ways for process optimization studies in solid-state fermentation and have advantages because of the fundamental principles of statistics, randomization and replication (4, 17, 20). There are also some reports on optimization of submerged fermentation using B.linens (24, 33). However, there is no literature found on the production of protease by SSF using B.linens through a statistical approach.
MATERIALS AND METHODS
Microorganism, culture conditions and inoculum preparation
Brevibacterium linens DSM 20158 was used in the present study. The bacterial culture was maintained on a growth medium (M1) containing (g/L): casein peptone (10.0), yeast extract (5.0), glucose (5.0), NaCl (5.0) and agar, for 24 hours at 37°C then stored at 4°C. Inoculum was prepared by transferring a loop full of cells from culture slant to the 50 ml of sterilized growth medium (M1) without agar in an Erlenmeyer flask (250mL). The culture was grown at 37°C for 24 hours at 250 rpm (11.87 ×g).
Cultivation medium
Cultivation medium was prepared as reported by Divakar et al. (3) at 37 °C for 24 hours. At the end of cultivation period protease was extracted with sterile 0.1 M phosphate buffer (pH 7.2). The content was filtered through filter paper, filtrate was centrifuged at 1254 rcf for 15 min and the clear brown supernatant was analyzed for enzyme activity. All experiments were performed in triplicate and mean values were reported.
Protease assay
Protease activity was determined by incubating 1 mL of the crude extract with 2 mL of 1% (w/v) casein (Sigma) in 50 mM Tris–HCl (pH 7.2) at 37 °C for 60 min by a modified method of Folin and Ciocalteu (6). The reaction was stopped by adding 5 mL of 5% trichloroacetic acid (TCA), followed by centrifugation at 7840 rcf for 10 min. After the addition of 5 mL of Alkaline Copper reagent to 1 mL of the supernatant, 0.5 mL of Folin reagent was added to the mixture and left for 30 minutes, after that the tyrosine liberated was measured at 700 nm. One unit of protease activity was defined as the amount of enzyme that released 1 μg tyrosine per min. A control was run in the same way, except the protease was added after the addition of 5% (w/v) TCA.
Selection of Carbon and nitrogen sources for protease production
In the previous work by Divakar et al, several carbon and nitrogen sources were tested (data not shown) by using one factor optimization method (3). In present study, different solid substrates such as wheat bran, rice husk, and rice bran and soybean meal were used as carbon source for the better production of protease as previously reported (3). Similarly, the different nitrogen sources like corn steep liquor, yeast extract, beef extract and casein were added as organic nitrogen sources. The salts (%): (NH4)2SO4 (0.5), NH4Cl (0.5), NaNO3 (0.5) and KNO3 (0.5) were added as inorganic nitrogen sources to the cultivation medium. The effect of various standard salt solutions (MgSO4.7H2O, 0.5%; KH2PO4, 0.5%; FeSO4.7 H2O, 0.01%) on protease production was also studied.
Screening of significant variables by Plackett- Burman design
This design was used for screening the various physical factors and selected nutritional variables which had positive effects on protease production. Nine variables were screened consisting of solid substrate wheat bran and soybean meal to optimize surface to volume ratio in batch flask culture, yeast extract as organic nitrogen source, (NH4)2SO4 as inorganic nitrogen source, MgSO4 as mineral source, moisture level, incubation period, temperature and inoculum size (v/w). Each factor of this experimental design was examined at three levels as shown by Table 1. Table 2 shows the design with five replications at the central points with seventeen Plackett- Burman runs along the levels and response of each of the nine factors. This statistical design does not involve the interactions between the selected nutrient variables and follows a linear approach for screening of factors (21). A first-order polynomial equation explained the model variables as follows:
Table 1.
Experimental variables at different levels used for the production of protease by Bravibacterium linens using Plackett-Burman design.
| Variables | Codes | Units | Experimental values | ||
|---|---|---|---|---|---|
| Lower (-1.000) | Centre (0.000) | Higher (1.000) | |||
| Incubation Period | A | hrs | 24 | 48 | 72 |
| Moisture content | B | % | 75 | 85 | 95 |
| Inoculum Size | C | % (v/w) | 0.1 | 2.55 | 5 |
| Wheat bran | D | g | 0 | 7.5 | 15 |
| Soybean meal | E | g | 0 | 7.5 | 15 |
| Yeast extract | F | g | 0.01 | 0.51 | 1.0 |
| MgSO4.7H2O | G | g | 0.1 | 0.3 | 0.5 |
| (NH4)2SO4 | H | g | 0.01 | 0.51 | 1.0 |
| Temperature | I | 0C | 30 | 37 | 44 |
Table 2.
Plackett-Burman design used for screening of nine variables with observed and predicted protease activity.
| Run No | Experimental Values | Protease (U/g) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | I | Observed | Predicted | |
| 1 | 24.00 | 75.00 | 5.00 | 0.00 | 15.00 | 1.00 | 0.05 | 1.00 | 44.00 | 2501±218 | 2486 |
| 2 | 48.00 | 85.00 | 2.55 | 7.50 | 7.50 | 0.51 | 0.10 | 0.51 | 37.00 | 2497±116 | 1607 |
| 3 | 72.00 | 95.00 | 0.10 | 15.00 | 15.00 | 1.00 | 0.05 | 0.01 | 30.00 | 2055±90 | 2048 |
| 4 | 24.00 | 95.00 | 0.10 | 15.00 | 15.00 | 0.01 | 0.15 | 1.00 | 44.00 | 1842±112 | 1817 |
| 5 | 48.00 | 85.00 | 2.55 | 7.50 | 7.50 | 0.51 | 0.10 | 0.51 | 37.00 | 2509±131 | 1607 |
| 6 | 24.00 | 95.00 | 5.00 | 15.00 | 0.00 | 0.01 | 0.05 | 1.00 | 30.00 | 2074±43 | 2029 |
| 7 | 72.00 | 95.00 | 5.00 | 0.00 | 0.00 | 0.01 | 0.15 | 0.01 | 44.00 | 600±180 | 615 |
| 8 | 72.00 | 95.00 | 0.10 | 0.00 | 0.00 | 1.00 | 0.05 | 1.00 | 44.00 | 1058±171 | 1011 |
| 9 | 24.00 | 75.00 | 0.10 | 0.00 | 0.00 | 0.01 | 0.05 | 0.01 | 30.00 | 189±176 | 191 |
| 10 | 24.00 | 95.00 | 5.00 | 0.00 | 15.00 | 1.00 | 0.15 | 0.01 | 30.00 | 1819±248 | 1757 |
| 11 | 48.00 | 85.00 | 2.55 | 7.50 | 7.50 | 0.51 | 0.10 | 0.51 | 37.00 | 1526±298 | 1607 |
| 12 | 72.00 | 75.00 | 5.00 | 15.00 | 0.00 | 1.00 | 0.15 | 1.00 | 30.00 | 2445±25 | 2484 |
| 13 | 48.00 | 85.00 | 2.55 | 7.50 | 7.50 | 0.51 | 0.10 | 0.51 | 37.00 | 2470±34 | 1607 |
| 14 | 72.00 | 75.00 | 5.00 | 15.00 | 15.00 | 0.01 | 0.05 | 0.01 | 44.00 | 2375±2 | 2381 |
| 15 | 48.00 | 85.00 | 2.55 | 7.50 | 7.50 | 0.51 | 0.10 | 0.51 | 37.00 | 2455±170 | 1607 |
| 16 | 24.00 | 75.00 | 0.10 | 15.00 | 0.00 | 1.00 | 0.15 | 0.01 | 44.00 | 888±161 | 896 |
| 17 | 72.00 | 75.00 | 0.10 | 0.00 | 15.00 | 0.01 | 0.15 | 1.00 | 30.00 | 1550±168 | 1567 |
Where as Y is the response, βo is the interception coefficient, βi is the linear coefficient. From the ANOVA as given in Table 3, variables (P< 0.05) considered to have significant effect on response, were optimized by response surface methodology (RSM), a multiple regression analysis technique.
Table 3.
ANOVA (analysis of variance) for the experimental parameters of Plackett Burman design affecting protease production.
| Variable | Coefficient | Standard Error | F-value | p-value |
|---|---|---|---|---|
| Model | - | - | 5.85 | 0.0218* |
| Lack of Fit | - | - | 0.011 | 0.9952** |
| Incubation Period | 64.17 | 101.10 | 0.40 | 0.5490** |
| Moisture Level | -41.67 | 101.10 | 0.17 | 0.6946** |
| Inoculum Size | 352.67 | 101.10 | 12.17 | 0.0130* |
| Wheat Bran | 330.17 | 101.10 | 10.67 | 0.0171* |
| Soybean meal | 407.33 | 101.10 | 16.23 | 0.0069* |
| Yeast Extract | 178.00 | 101.10 | 3.10 | 0.1288** |
| MgSO4 | -92.33 | 101.10 | 0.83 | 0.3963** |
| (NH4)2SO4 | 295.33 | 101.10 | 8.53 | 0.0266* |
| Temperature | -72.33 | 101.10 | 0.51 | 0.5012** |
Significant values
Non-significant values at p< 0.05
Optimization by Central composite design (CCD) & statistical analysis
After screening the significant variables, a CCD was used for optimization of four variables; inoculum size (C), wheat bran (D), soybean meal (E) and (NH4)2SO4 (H), each were studied at five coded levels i.e. (-2, -1, 0, +1, +2) as shown by Table 4 and the model was explained with twenty one runs, having six replicates at the centre value as given in Table 5.
Table 4.
Experimental variables, codes, units, ranges and levels of the independent variables for response surface methodological experiments.
| Variables | Symbol code | Units | Levels | ||||
|---|---|---|---|---|---|---|---|
| -2.0 | -1 | 0 | +1 | +2.0 | |||
| Inoculum size | C | % | -2.35 | 0.1 | 2.55 | 5 | 7.45 |
| Wheat bran | D | g | -7.50 | 0 | 7.5 | 15 | 22.50 |
| Soybean meal | E | g | -7.50 | 0 | 7.5 | 15 | 22.50 |
| (NH4)2SO4 | H | g | -0.48 | 0.01 | 0.51 | 1.0 | 1.50 |
Table 5.
Central Composite Design of the nutrients and physical parameters in real units for the response of protease along with its predicted & observed values.
| Run No. | Experimental values | Protease (U/g) | ||||
|---|---|---|---|---|---|---|
| Inoculum size | Wheat bran | Soybean meal | (NH4)2SO4 | Obserevd | Predicted | |
| 1 | 2.55 | 7.50 | 7.50 | 0.51 | 6324±224 | 6383 |
| 2 | 0.10 | 0.00 | 15.00 | 0.01 | 3367±119 | 3367 |
| 3 | 2.55 | -7.50 | 7.50 | 0.51 | 2610±92 | 2569 |
| 4 | 7.45 | 7.50 | 7.50 | 0.51 | 3246±115 | 3097 |
| 5 | 2.55 | 22.50 | 7.50 | 0.51 | 3808±135 | 3735 |
| 6 | 0.10 | 0.00 | 0.00 | 0.01 | 1256±44 | 1263 |
| 7 | 5.00 | 0.00 | 15.00 | 1.00 | 5207±185 | 5188 |
| 8 | 2.55 | 7.50 | -7.50 | 0.51 | 2364±175 | 2310 |
| 9 | 0.10 | 15.00 | 0.00 | 1.00 | 2430±179 | 2408 |
| 10 | 2.55 | 7.50 | 7.50 | -0.48 | 5395±248 | 5382 |
| 11 | 5.00 | 15.00 | 15.00 | 0.01 | 6733±298 | 6782 |
| 12 | 0.10 | 15.00 | 15.00 | 1.00 | 2220±28 | 2237 |
| 13 | 5.00 | 0.00 | 0.00 | 1.00 | 1287±37 | 1326 |
| 14 | -2.35 | 7.50 | 7.50 | 0.51 | 55±2 | 41.6 |
| 15 | 2.55 | 7.50 | 7.50 | 0.51 | 6309±185 | 6383 |
| 16 | 2.55 | 7.50 | 7.50 | 1.50 | 5981±176 | 6036 |
| 17 | 2.55 | 7.50 | 7.50 | 0.51 | 6232±183 | 6383 |
| 18 | 2.55 | 7.50 | 7.50 | 0.51 | 6504±207 | 6383 |
| 19 | 2.55 | 7.50 | 22.50 | 0.51 | 5483±175 | 5509 |
| 20 | 2.55 | 7.50 | 7.50 | 0.51 | 6268±200 | 6383 |
| 21 | 5.00 | 15.00 | 0.00 | 0.01 | 6089±194 | 6182 |
The four variables used were coded according to the following equation:
Where “Z” is the coded value of the independent variable, “X” is the corresponding real value, “Xo” is at the real value of an independent variable at the centre point and “ΔX” is the step change of the real value at the variable for the value “Z”.
The relationship between the response and the independent variables was explained by using the second order polynomial equation:
Where Y is the predicted response, βo is the interception coefficient, βi is the linear coefficient, βii is quadratic coefficient and βij is the interaction coefficient. The multiple regression analysis of the model and construction of response surface graphs were performed by using Design-Expert, version 7.1.6 (STATEASE Inc., Minneapolis, MN, USA). The quality of regression equation was determined by the coefficient of determination R2 and its significance was judged by an F-test. The fitted 2nd order polynomial equation was explained in the form of counter plot graphs to show the relationship between the response and experimental variables. The point optimization method was used to optimize the maximum response of each variable.
Statistical model validation
To validate the optimization of medium composition, an experiment with optimized conditions was conducted in triplicate for 48 hours and their mean values were used to confirm the results from an analysis of the RSM.
Characteristics of Protease: The effect of different concentrations of casein substrate (5, 10, 15, 20, 25, 30 and 35 mg/mL) to protease activity was detected to calculate the Vmax and Km value by Lineweaver-Burk (LB) plot kinetics.
RESULTS AND DISCUSSION
Selection of carbon and nitrogen sources for Protease production
Selection of best carbon and nitrogen source was made according to previous study by using “one factor at a time” method (3). In our study, among solid media; maximum protease production was shown by soybean meal (2051 U/g) and wheat bran (1435 U/g) followed by, rice bran (931 U/g) and rice husk (821 U/g). Yeast extract and (NH4)2SO4 were found to be excellent stimulants for the better production of protease among organic yeast extract (2334 U/g), beef extract (2289 U/g), corn steep liquor (2167 U/g), casein (2011 U/g) and inorganic nitrogen sources (NH4)2SO4 (2412U/g); NH4Cl (2314 U/g); NaNO3 (2025 U/g); and KNO3 (2013 U/g) respectively. Among the salt solutions (MgSO4.7H2O (2498 U/g); KH2PO4, (2315 U/g); FeSO4.7 H2O (2143 U/g) ) MgSO4.7H2O was found to be the best for protease production. On the basis of these results, wheat bran, yeast extract, (NH4)2SO4 and MgSO4.7H2O were selected as carbon, organic nitrogen, inorganic nitrogen sources and mineral source respectively for further study by using the Plackett-Burman design and response surface methodology.
Screening of significant variables by Plackett-Burman design
Nine variables were screened by Plackett-Burman design as shown in Table 2. Two of the six significant variables screened, MgSO4 and temperature, exerted a negative effect on protease production whereas all other non significant variables were neglected. The significance of the model was calculated by the P-value. P < 0.05 was considered significant (13). Soybean meal and inoculum size were most significant followed by wheat bran and (NH4)2SO4. The effect of the significant variables was determined by ANOVA (Table 2) by putting the data in the linear equation as follows:
Where C= Inoculum size, D= Wheat bran, E= Soybean meal, H = (NH4)2SO4
Non-significant lack-of-fit (P: 0.4746) showed that model is fit and its significant variables could be used for further optimization by RSM (9).
Mixture of cheap solid substrates, wheat bran and soybean meal, was used for maximum protease production as reported by Boer and Peralta (2), because it is a good source of carbohydrates, proteinaceous nitrogen and other nutritious substances essential for bacterial growth. Kammoun et al. (12) reported that organic nitrogen sources traditionally have a positive effect since they are also a carbon source and contain trace of minerals and ions that could enhance the enzyme secretion. On the other hand, (NH4)2SO4 resulted in enhanced protease production. The maximum production of protease being supported by the combination of organic and inorganic nitrogen source corroborated the results of Rossa et al. (29). The optimization of inoculum size was quite important, as high inoculum density could attenuate enzyme production due to competition for available nutrients (26). These four variables: soybean meal, wheat bran, (NH4)2SO4, and inoculum size were selected for further optimization for maximum protease production. On the other hand the parameters like incubation period, temperature and moisture content which were not selected for further optimization study but due to their importance in solid state fermentation, were kept at their central values in the present study for the production of high yield of enzyme. However in the present study, incubation period was not significant and supported by the previously reported results of Reddy et al. (26). The temperature was significant but had negative effect on enzyme production. This is in agreement with the results reported by Francis et al. (7), Kammoun et al. (12) and Shabbiri et al. (31) showing a decrease in production at temperatures outside the mesophilic range. However Kunamneni et al. (14) reported a range of initial moisture content (50–110%) with maximum production of enzyme at 90% in solid state fermentation. But in the present study, it was observed that microbial activity was increased up to 85% moisture content. Further increase in the moisture level decreased the enzyme production. This may be associated with decreased porosity and reduced oxygen tension in the substrate.
Optimization by Central composite design (CCD) & statistical analysis
Following screening, CCD with twenty one experiments was used to determine the optimal levels of the four significant factors (soybean meal, wheat bran, (NH4)2SO4, and inoculum size) that affected protease production. According to the regression analysis of the RSM (Table 6) the model terms C, D, E, H, CD, CE, CH, DE, DH, EH, C 2, D2, E2 and H2 were significant (p < 0.05). The linear effects of C and E the interactive effects of CD, CH and DE and the second order effects of C 2, D2,E2 were most significant, all having P-value <0.0001, thus corroborating the results of Wang et al. (37). These results indicated that inoculum size and soybean meal had direct relationship to the protease yield. The regression equation coefficients were studied by putting data in the 2nd order polynomial equation as follows
Table 6.
Regression analysis of the central composite design.
| Variables | Coefficient Estimate | Standard Error | F-value | p-value |
|---|---|---|---|---|
| Intercept | 2464.12 | 18.90 | - | - |
| C | 797.88 | 33.85 | 555.61 | < 0.0001 |
| D | 299.47 | 33.85 | 78.27 | 0.0001 |
| E | 793.85 | 23.94 | 1100.03 | < 0.0001 |
| H | 146.62 | 33.85 | 18.76 | 0.0049 |
| CD | 934.16 | 47.87 | 380.81 | < 0.0001 |
| CE | 332.67 | 33.85 | 96.59 | < 0.0001 |
| CH | -494.89 | 47.87 | 106.88 | < 0.0001 |
| DE | -699.57 | 33.85 | 427.14 | < 0.0001 |
| DH | -457.32 | 47.87 | 91.27 | < 0.0001 |
| EH | 119.23 | 33.85 | 12.41 | 0.0125 |
| C2 | -1174.74 | 19.11 | 3778.61 | < 0.0001 |
| D2 | -785.12 | 19.11 | 1687.80 | < 0.0001 |
| E2 | -606.44 | 19.11 | 1007.00 | < 0.0001 |
| H2 | -165.39 | 19.11 | 74.90 | 0.0001 |
The ANOVA for the second order polynomial model as given in Table 7 showed that R2 was 0.9994 which means that model could explain 99.94% of the variation in response (protease production). The square of the linear correlation is termed as the coefficient of determination (R2), which is the measure of the strength of the linear relationship between the experimental and the predicted values (23). The predicted R2 of 0.9865 was in reasonable agreement with the adjusted R2 of 0.9979 indicating good adjustment between the observed and predicted values. i.e. the regression model could be used to analyze trends of responses. The high model F-value 544.24 implies that protease production by Brevibacterium linens has a good fit with the model and non-significant lack-of-fit (0.6392) showed that the model was significant as previously reported (11). The coefficient of variation % (CV%) is a measure of residual variation of the data relative to the size of the mean. Usually, the higher the value of CV, the lower is the reliability of experiment (15). A low coefficient variation (CV=2.25) showed the reliability of the experiments conducted. Adequate precision 82.899 explained significantly the good fit of the model as this value should be above “4” explaining the signal to noise to navigate the design space excellently (27). The contour plots as shown by Figure 1 provided information about the interaction between two variables and interpreted the optimum experimental conditions. These plots were generated for the pair wise combination of the four variables, while keeping the other two at their centre point levels. Thus the elliptical contour plots clearly indicate that the mutual interaction is prominent among factors (15,37). Figure 1A depicts the contour plot of the calculated response from the interaction between wheat bran and inoculum size while keeping the other two variables ((NH4)2SO4 and soybean meal) at zero level. The result demonstrated that with increase in wheat bran and inoculum size up to 11.21g and 3.85%, respectively, the enzyme production had increased up to 6621 U/g and with further increases, it declined. Fig. 1B shows the effect of soybean meal and inoculum size on enzyme production, keeping other variables at zero level ((NH4)2SO4 and wheat bran). The plot showed that the maximum protease production (6813 U/g) occurred at (13.48g) soybean meal and an inoculum size of 3.56%, which was in agreement with the model. An interaction between (NH4)2SO4 and inoculum size on enzyme production was studied by keeping the wheat bran and soybean meal at central level (Fig. 1C). The graph shows that the maximum protease production (6464 U/g) occurred at inoculum size of 3.46% and of 0.46g (NH4)2SO4, which was in agreement with the model. Whereas the interaction between the two parameters (wheat bran at 6.31g and soybean meal at 12.88g) resulted in protease production of 6597 U/g as shown by Fig. 1D. The interaction between (NH4)2SO4 (0.63g) and wheat bran (8.35g) was represented by Fig. 1E with maximum enzyme production at 6365 U/g. However the interaction between (NH4)2SO4 (0.85g) and soybean meal (12.88g) resulted in the highest enzyme titer which was 6667 U/g as shown by Fig. 1F.
Table 7.
Analysis of variance (ANOVA) for the second-order polynomial model.
| Source | SS | DF | MS | F-value | p-value |
|---|---|---|---|---|---|
| Model | 8.88E+007 | 14 | 6.34E+006 | 544.24 | < 0.0001 |
| Residual | 54997 | 6 | 9166.3 | ||
| Lack of Fit | 11026.3 | 2 | 5513.1 | 0.50 | 0.6392 |
| Pure Error | 43971.3 | 4 | 10992.7 | - | - |
| Total | 8.891E+007 | 20 | - | - | - |
Std Dev =, 95.74 Mean = 4246.6, C.V% = 2.25, R2 = 0.9994, Adj R2 =0.9979, Pre R2 = 0.9865, Adeq Precision = 82.899
Figure 1.
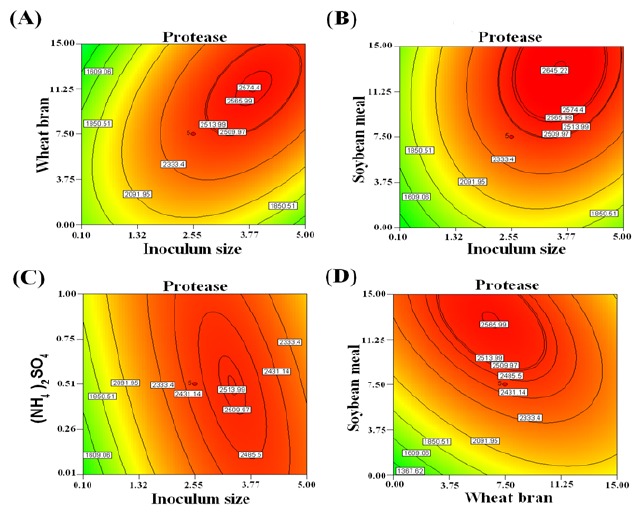
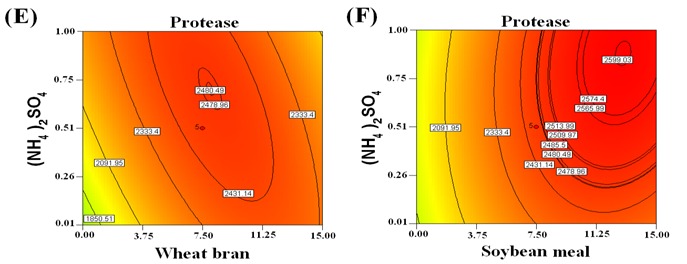
Contour plots for Protease production showing the interactive effects of wheat bran and inoculum size (A), soybean meal and inoculum size (B), (NH4)2SO4 and inoculum size (C), soybean meal and wheat bran (D), (NH4)2SO4 and wheat bran (E), soybean meal and (NH4)2SO4 (F). Other variables were kept at central level.
Statistical model validation
With the help of the special features of RSM, point prediction and contour plot generation the optimum value of the combination of the four fermentation variables was determined for maximum production of protease. Experiments were carried out in shake flasks under predicted conditions to validate the model. There was high degree of similarity between predicted and experimental values, showing the validity of the RSM. The model was validated by further fitting the best optimized values of wheat bran (8.50g), (NH4)2SO4 (0.45g), inoculum size (3.50%), soybean meal (12.0g) in the 2nd order polynomial equation while keeping other variables at their central levels. The closeness of the predicted response for protease (6850 U/g) and the experimental response (6811 U/g) proved the validity of the model.
Characteristics of Protease: The Lineweaver-Burk (LB) plot (Fig. 2) for the proteolytic reaction of casein revealed that the Vmax of the reaction was 8025 μg/min. The Michaelis-Menten constant constant (Km) was 7.5 mg/mL. The reaction rate remained constant beyond a substrate concentration of 30mg/mL of casein (data not shown).
Figure 2.
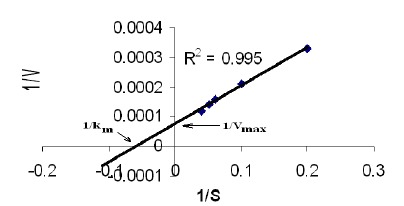
Lineweaver-Burk plot for neutral protease under varying substrate (casein) concentrations (5–35 mg/mL) indicating the Km and Vmax values
CONCLUSIONS
To meet the industrial demand of proteases, the present study was conducted to produce industrially important proteases by Brevibacterium linens DSM 20158 by optimizing various cultivation parameters, including fermentation conditions and low cost medium composition, using statistical approach. The nine variables were screened by using the Plackett–Burman design, and four variables (i.e. wheat bran, soybean meal, (NH4)2SO4 and inoculum size) exerted significant effects on protease production. In order to optimize low-cost fermentation conditions for protease production by B.linens, we conducted RSM experiments using the screened variables. After having the optimum levels of each variable while keeping the other variables at their central values, a successful and significant two fold improvement in the production of protease by B.linens was accomplished using cheap carbon and nitrogen sources. The optimized medium established in this work might result in a significant reduction in the cost of medium constituents.
REFERENCES
- 1.Adinarayana K., Ellaiah P., Srinivasulu B., Devi R.B., Adinarayana G. Response surface methodological approach to optimize the nutritional parameters for neomycin production by Streptomyces marinensis under solid-state fermentation. Process. Biochem. 2003;38:1565–1572. [Google Scholar]
- 2.Boer C.G., Peralta R.M. Production of extra cellular protease by Aspergillus tamari. J. Basic. Microbiol. 2000;40:75–81. [Google Scholar]
- 3.Divakar G., Sunitha M., Vasu P., Shankar P.U., Ellaiah P. Optimization of process parameters for alkaline protease production under solid-state fermentation by Thermoactinomyces thalpophilus PEE 14. Indian. J. Biotechnol. 2006;5:80–83. [Google Scholar]
- 4.Ferracini-Santos L., Sato H.H. Production of alkaline protease from Cellulosimicrobium cellulans. Braz. J. Microbiol. 2009;40:54–60. doi: 10.1590/S1517-83822009000100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fiedler F., Schäffler M.J., Stackebrandt E. Biochemical and nucleic acid hybridization studies on Brevibacterium linens and related strains. Arch. Microbiol. 1981;129:85–93. [Google Scholar]
- 6.Folin O., Ciocalteu V. On tyrosine and tryptophane determinations in proteins. J. Biol. Chem. 1927;73:627–650. [Google Scholar]
- 7.Francis F., Sabu A., Nampoothiri K.M., Ramachandran S., Ghosh S., Szakacs G., Pandey A. Use of response surface methodology for optimizing process parameters for the production of a-amylase by Aspergillus oryzae. Biochem. Eng. J. 2003;15:107–115. [Google Scholar]
- 8.Germano S., Pandey A., Osaku C.A., Rocha S.N., Soccol C.R. Characterization and stability of proteases from Penicillium sp. produced by solid-state fermentation. Enzyme. Microb. Technol. 2003;32:246–251. [Google Scholar]
- 9.González A.G. Two level factorial experimental designs based on multiple linear regression models: a tutorial digest illustrated by case studies. Analytica. Chimica. Acta. 1998;360:227–241. [Google Scholar]
- 10.He G.Q., Kong Q., Dingm L.X. Response surfacemethodology for optimizing the fermentation medium of Clostridium butyricum. Lett. Appl. Microbiol. 2004;39:363–368. doi: 10.1111/j.1472-765X.2004.01595.x. [DOI] [PubMed] [Google Scholar]
- 11.Kahyaoglu T. Optimization of the pistachio nut roasting process using response surface methodology and gene expression programming. Food. Scie. Technol. 2008;41:26–33. [Google Scholar]
- 12.Kammoun R., Naili B., Bejar S. Application of a statistical design to the optimization of parameters and culture medium for a-amylase production by Aspergillus oryzae CBS 819.72 grown on gruel (wheat grinding by-product) Bioresour. Technol. 2008;99:5602–5609. doi: 10.1016/j.biortech.2007.10.045. [DOI] [PubMed] [Google Scholar]
- 13.Khuri I., Cornell A.J. Response Surfaces: Design and Analysis. New York: Marcel Dekker, Inc.; 1987. p. 405. [Google Scholar]
- 14.Kunamneni A, Permaul K., Singh S. Amylase production in solid state fermentation by the thermophilic fungus Thermomyces lanuginosus. J. Biosci Bioeng. 2005;100:168–171. doi: 10.1263/jbb.100.168. [DOI] [PubMed] [Google Scholar]
- 15.Lotfy W.A., Ghanem K.M., El-Helow ER. Citric acid production by a novel Aspergillus niger isolate: II. Optimization of process parameters through statistical experimental designs. Bioresour Technol. 2007;98:3470–3477. doi: 10.1016/j.biortech.2006.11.032. [DOI] [PubMed] [Google Scholar]
- 16.Myers R.H., Montgomery D.C. Response surface methodology: process and optimization using designed experiments. 1st. Wiley Interscience; 1995. [Google Scholar]
- 17.Oskouie S.F.G., Tabandeh F., Yakhchali B., Eftekhar F. Response surface optimization of medium composition for alkaline protease production by Baccillus clausii. Biochem. Engineer. J. 2008;39:37–42. [Google Scholar]
- 18.Pandey A. Solid-state fermentation. Biochem. Eng. J. 2003;13:81–84. doi: 10.1016/s1369-703x(00)00065-6. [DOI] [PubMed] [Google Scholar]
- 19.Pandey A., Selvakumar P., Soccol C.R., Nigam P. Solid state fermentation for the production of industrial enzymes. Curr. Sci. 1999;77:149–162. [Google Scholar]
- 20.Pansuriya R.C., Singhal R.S. Response surface methodology for optimization of production of lovastatin by solid state fermentation. Braz. J. Microbiol. 2010;41:164–172. doi: 10.1590/S1517-838220100001000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plackett R.L., Burman J.P. The design of optimum multifactorial experiments. Biometrika. 1946;33:305–325. [Google Scholar]
- 22.Raghavarao K.S.M.S., Ranganathan T.V., Karanth N.G. Some engineering aspects of solid-state fermentation. Biochem. Eng. J. 2003;13:127–135. [Google Scholar]
- 23.Rajendran A., Thangavelu V. Evaluation of various unstructured kinetic models for the production of protease by Bacillus Sphaericus Mtcc511. Eng.Life Sci. 2008;8:179–185. [Google Scholar]
- 24.Rattray F.P., Fox P.F. Aspects of enzymology and biochemical properties of Brevibacterium linens relevant to cheese ripening. J. Dairy. Sci. 1999;82:891–909. doi: 10.3168/jds.S0022-0302(99)75308-7. [DOI] [PubMed] [Google Scholar]
- 25.Rattray F.P., Fox P.F., Healy A. Specificity of an extracellular proteinase from Brevibacterium linens ATCC 9174 on bovine b-casein. Appl. Environmental. Microbiol. 1997;63:2468–2471. doi: 10.1128/aem.63.6.2468-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy L.V.A., Wee Y.J., Yun J.S., Ryu H.W. Optimization of alkaline protease production by batch culture of Bacillus sp. RKY3 through Plackett–Burman and response surface methodological approaches. Bioresour. Technol. 2008;99:2242–2249. doi: 10.1016/j.biortech.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 27.Ren J.Y., Zhao M.M., Shi J., Wang J.S., Jiang Y.M., Cui C., Kakuda Y., Xue S.J. Optimization of antioxidant peptide production from grass carp sarcoplasmic protein using response surface methodology. Food. Sci. Technol. 2008;41:1624–1632. [Google Scholar]
- 28.Rigo E., Ninow J.L., Luccio M.D., Oliveira J.V., Polloni A.E., Remonatto D., Arbter F., Vardanega R., Oliveira D.D., Treichel H. Lipase production by solid fermentation of soybean meal with different supplements. LWT - Food Sci Technol. 2010;43:1132–1137. [Google Scholar]
- 29.Rossa C.A.A., Yantorno O.M., Arcas J.A., Ertola R.J. Organic and inorganic nitrogen source ratio effects on Bacillus thuringiensis varisraelensis delta-endotoxin production. World. J. Microbiol. Biotechnol. 2005;6:27–31. doi: 10.1007/BF01225351. [DOI] [PubMed] [Google Scholar]
- 30.Sandhyaa C., Sumanthaa A., Szakacsb G., Pandey A. Comparative evaluation of neutral protease production by Aspergillus oryzae in submerged and solid-state fermentation. Proc. Biochem. 2005;40:2689–2694. [Google Scholar]
- 31.Shabbiri K., Adnan A., Noor B., Jamil S. Optimized production, purification and characterization of alpha amylase by Brevibacterium linens DSM 20158 using bio-statistical approach. Ann Microbiol. 2011 doi: 10.1590/S1517-838220120003000031. DOI: 10.1007/s13213-011-0286-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singhania R.R., Patel A.K., Soccol C.R., Pandey A. Recent advances in solid-state fermentation. Biochem. Eng. J. 2009;44:13–18. [Google Scholar]
- 33.Soares F.E.F., Braga F.R., André Geniêr H.L., Victor de Araújo J., Ferreira S.R., Araujo J.M., Tavela A.O., Vilela V.L.R, de Queiróz H.J. Optimization of medium composition for protease production by Paecilomyces marquandii in solid-statefermentation using response surface methodology. Afr. J. Microbiol. Res. 2010;4:2699–2703. [Google Scholar]
- 34.Strauss S., Kopetcky A., Zemanovicand J., Hampel W.A. Production of Proteases from Brevibacterium Linens. Biocatalysis. Biotransformations. 1994;10:317–323. [Google Scholar]
- 35.Thumar J.T., Singh S.P. Secretion of an alkaline protease from a salt- tolerant and alkaliphilic, Streptomyces clavuligerus strain MIT-1. Braz. J. Microbiol. 2007;38:766–772. [Google Scholar]
- 36.Thys R.C.S., Guzzon S.O., Olivera F.C., Brandelli A. Optimization of protease production by Microbacterium sp. in feather using response surface methodology. Process. Biochem. 2006;41:67–73. [Google Scholar]
- 37.Wang Q., Hou Y., Xu Z., Miao J., Li G. Optimization of cold-active protease production by the psychrophilic bacterium Colwellia sp. NJ341 with response surface methodology. Bioresour. Technol. 2008;99:1926–1931. doi: 10.1016/j.biortech.2007.03.028. [DOI] [PubMed] [Google Scholar]


