Abstract
In this paper we carried out a study about prevalence of the clinically significant coagulase negative staphylococcal (CNS) isolates found in an university hospital. Two hundred four CNS isolates from 191 patients obtained between the period of 1998 to 2002, were studied. About 27% (52/191) of the infection cases studied were confirmed as CNS-associated diseases. Blood stream infection (BSI) was the most frequent CNS associated-disease (25%; 13/52). The great majority of the BSI was verified in the Neonatal Intensive Care Unit (NICU). The analysis of the 52 patients medical history showed that 85% of the BSI was acquired in hospital. Most of the CNS nosocomial infections were associated with the use of indwelling medical devices. The incidence of methicillin-resistance among significant CNS isolates was 38%. In this study, a high percentage of exogenous contaminant was verified (60%), indicating that contamination of clinical specimens during sample collection is critical.
Keywords: Coagulase negative staphylococci, Nosocomial infection, Antimicrobial resistance
INTRODUCTION
Coagulase-negative staphylococci (CNS) colonize skin and mucous membrane of humans and animals (13, 18, 27). Previously, these bacteria were mostly regarded as contaminants of clinical specimens from humans. During the last decades CNS have emerged as important nosocomial pathogens (16, 18, 27, 32). This increase was paralleled by the development of novel therapeutic and surgical interventions, which have promoted an advanced medical care. Unfortunately, these procedures are frequently followed by complications that may result in infections (32, 36). Risk factors for CNS infections include breaches in the natural mucocutaneous barrier, prior exposure to antibiotics, immunosuppression, extreme ages, and most frequently, the presence of venous catheter or indwelling prosthetic device, among others (19, 20, 36).
The recognition of CNS as nosocomial pathogens has stimulated researches involving the development of more accurate methods for its identification and molecular characterization, as well as the study of virulence factors and also the establishment of stringent criteria to discriminate cases of infection from bacterial flora contamination. It has been suggested that the major virulence factor of CNS is the ability to form biofilm on the surface of indwelling medical devices (6). In the biofilm, the bacteria seem to be protected against antibiotics and the immune system (11). Although mechanisms involved in the production of biofilm are not totally clarified, bacterial products that contribute for biofilm formation are among the best-studied CNS virulence determinants, including products involved in the bacterial adhesion to solid surface and cell accumulation (2, 4, 11, 35). Many CNS also produce lipases, proteases, and other exoenzymes, which are possibly implicated with the bacterial persistence and dissemination in the host tissue (34).
Although numerous species of CNS have been recognized recently, the majority is rarely pathogenic (34). Clinical studies have indicated Staphylococcus epidermidis, Staphylococcus haemolyticus, Staphylococcus warneri and Staphylococcus hominis as the most prevalent CNS in hospital infections (1, 31, 34). The emergence of antimicrobial resistant among CNS isolates is also an important factor in nosocomial infections. Furthermore, methicillin-resistant CNS (MRCNS) are cross-resistant to all other ß-lactam antibiotics (28). Multiresistance has been detected in more than 80% of MRCNS hospital isolates (23). In the opposite, approximately 70% of MRCNS obtained from healthy volunteers from the community displayed higher susceptible to non-ß-lactam antibiotics (30).
The aim of this study was to assess the incidence and aetiology of inpatient and outpatient infections caused by CNS in a teaching hospital and to determine the antimicrobial susceptibility of these CNS isolates.
MATERIALS AND METHODS
Hospital
The Antônio Pedro University Hospital (HUAP) is located in Niterói, RJ, Brazil. It is a 291-bed general hospital with 11 medical and 12 surgical units, besides 3 intensive care units (ICU), excluding burns. The hospital provides assistance for adults, children or neonates. In the period studied (1998-2002) there was a total of 25,520 inpatient admittances (annual average of 5,704), 1,041,922 ambulatory visits (annual average of 208,384) and 218,407 emergency medical assistances (annual average 43,681).
Clinical specimen and bacterial isolate
A total of 204 isolates of CNS were obtained from 191 patients from January 1998 to December 2002. For eleven patients 2 or 3 CNS isolates were obtained in different isolation dates. From these 191 patients, 28 (15%) were assisted in the hospital ambulatories and 163 (85%) were admitted in the hospital wards. Regarding to the source of CNS isolation from the 191 patients, 9% corresponded to respiratory tract secretions, 30% blood and liquor, 18% secretions from surgical and skin infections, 12% urine and urethral secretions, 16% catheter tips and 14% to miscellaneous clinical materials. The Staphylococcus species were identified by Vitek Systems Gram-Positive Identification test (GPI) card (bioMerrieux Vitec, Inc,) according to the manufacturer recommendations. The antibiotic susceptibility was determined by VITEK automatic system with GPS-105.
Clinical relevance
Many factors were considered for the determination of the clinical significance of CNS diseases. All these factors were based on the Center for Disease Control and Prevention criteria for definitions of nosocomial infections (12) and on the criteria described by Sewell et al. (29). Briefly, the clinical relevance was inferred when the following general criteria were observed: i. risk factors for infection, such as invasive procedures (vascular catheter, orotracheal and nasotracheal intubations, urologic manipulation, surgical procedures); ii. classical clinical manifestations (including the presence of fever, local or generalized inflammation, fluctuant wounds or drained purulent material); iii. associated laboratory findings (such as elevated total leukocyte count with granulocyte predominance and other indicator of infections as the presence of leukocytes in cerebrospinal fluid or in the urine); iv. patient response to proper antibiotic therapy; v. CNS isolate classified in the criteria i, ii and iii obtained from patient who has died without the administration of the appropriated antibiotic therapy was also considered clinically relevant and vi. always when CNS isolate, obtained from internal fluids or foreign bodies, was not recovered in pure primary culture, it was considered contaminant.
In addition to the general criteria above, due to the fact that CNS primary pneumoniae is frequently rare, a very stringent criteria was included to assign relevant cases and it was based on the simultaneous presence of i. compatible clinical presentation; ii. chest X-ray evidence; iii. isolation of microorganism from hemocultures or from bronchial or tracheal washing cultures and iv. patient using invasive medical devices, such as intravascular lines or indwelling urinary catheter, was excluded for primary CNS pneumoniae.
Concerning to bloodstream infection, in addition the general criteria above: i. only one positive blood culture was considered enough to assign relevance when infection occurred among neonates; ii. for other patients, at least two blood cultures were obtained on separate occasions and iii. primary bacteremia was only considered for patients using intravascular catheters.
For the purpose of the analysis presented here, infectious diseases associated to CNS that were acquired in the Antônio Pedro University Hospital was called HUAP-acquired infections and the diseases which did not occur in this hospital were all grouped as outside acquired infections.
Statistical analysis
Data related to the clinical significance of the isolates and bacterial resistance were analyzed by chi-square test or Yates correction for continuity when n<20. Fisher test was chosen when n<5. Significance was considered for p<0.05.
RESULTS
CNS isolates
From January 1998 to December 2002 a total of 8,342 positive bacterial cultures were processed in the clinical laboratory of the HUAP. Gram-positive cocci accounted for 27% of this total. Among them, the most frequently represented bacterial genus was the Staphylococcus, accounting for 1,999 isolates (24%); CNS corresponding to 22% of the staphylococcal isolates. Escherichia coli, Klebsiella sp, Pseudomonas sp, Enterobacter sp and Proteus sp, corresponded to 22%, 13%, 9%, 7% and 7%, respectively. Other bacterial genus accounted for a total of 18%. A population of 204 CNS was chosen to be analyzed concerning to its clinical relevance based on the precision of the medical reports. These 204 isolates were obtained from 191 patients. From these 191, 28 (15%) were outpatients and 163 (85%) inpatients. The most frequent species of CNS isolates were Staphylococcus epidermidis (70; 34%), followed by Staphylococcus warneri (35; 17%), Staphylococcus haemolyticus (33; 16%), Staphylococcus simulans (21; 10%), Staphylococcus sciuri (15; 7%), Staphylococcus saprophyticus (5; 3%), Staphylococcus hominis (4; 2%) and Staphylococcus capitis (4; 2%), Staphylococcus xylosus (3; 2%) and Staphylococcus auricularis (1; 0.5%). The 13 isolates whose species could not be determined by the VITEK system were refered here as not determined CNS species (Table 1).
Table 1.
Distribution of clinical relevance of the 204 isolates of coagulase negative staphylococci (CNS) in the Antônio Pedro University Hospital.
| CNS species a | Geral (%) | CR b | NR b | ||
|---|---|---|---|---|---|
| Total | Outpatient | NI c | |||
| S. epidermidis | 70 ( 34) | 17 | 2 | 15 | 53 |
| S. warneri | 35 ( 17) | 9 | 3 | 6 | 26 |
| S. haemolyticus | 33 ( 16) | 15 | 11 | 4 | 18 |
| S. simulans | 21 ( 10) | 5 | 2 | 3 | 16 |
| S. sciuri | 15 ( 7) | 4 | 3 | 1 | 11 |
| S. saprophyticus | 5 ( 3) | 0 | 0 | 0 | 5 |
| S. hominis | 4 ( 2) | 0 | 0 | 0 | 4 |
| S. capitis | 4 ( 2) | 3 | 0 | 3 | 1 |
| S. xylosus | 3 ( 2) | 1 | 1 | 0 | 2 |
| S. auricularis | 1 ( 0) | 0 | 0 | 0 | 1 |
| NDd | 13 ( 6) | 2 | 1 | 1 | 11 |
| Total isolates | 204 (100) | 56 | 23 | 33 | 148 |
CNS species were identified by VITEK System Gram positive identification test (GPI) card;
Clinically relevant (CR) and non-relevant (NR) CNS isolates were classified on the basis of the criteria described in Materials and Methods;
Nosocomial infection (NI);
ND CNS which species were not identified.
CNS associated infections
Using the criteria described in this paper, of the 191 patients from which CNS isolates were obtained, only 27% (52 patients) had CNS associated diseases (clinical significance). From these 52 patients, 56 CNS isolates were recovered. Thus, the majority of cases from which CNS were isolated was classified as clinically non-relevant (73%; 139/191 patients), corresponding to a total of 148 CNS isolates. When we analyzed the reports from the microbial laboratory, sources of clinical materials, sites of infections and the techniques used for the collection of specimens, among other factors related to clinical relevance, it was verified that the most probable source of contamination (60%) was not related to the patient flora but rather to exogenous contaminants (Fig. 1).
Figure 1.
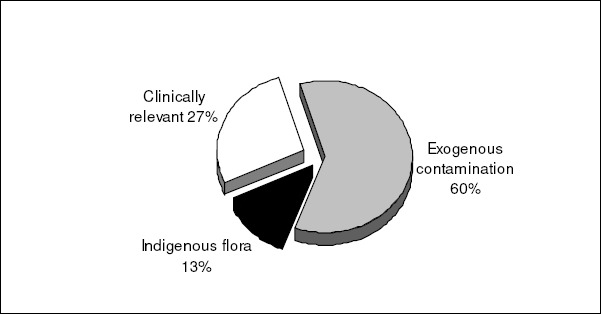
Classification of the 204 coagulase-negative staphylococcal isolates in clinically significant, indigenous flora and exogenous contaminants following the criteria described in Materials and Methods.
Blood stream infection (BSI) was the most frequent CNS associated infection (13/52; 25%). The majority of the sepsis was verified in the Neonatal Intensive Care Unit (NICU; 8/13; 62%). S. epidermidis was defined as the etiologic agent in 57% of the cases, followed by S. warneri and S. capitis (21% each). Two bacteraemia were classified as outside acquired infection (2/52; 4%). Eleven of the 13 were classified as HUAP-acquired bacteraemia (92%), nine from this 11, were associated with the use of intravenous catheter. The remaining 2 had other risk factors as leukemia or prematurity associated to low birth weight and aspiration pneumonia.
Surgical site infections accounted to 10% (5/52) of the total CNS diseases and a higher percentage of 19% (10/52) were verified for urinary tract infections and skin/soft tissue infections (10/52; Fig. 2). The use of long-term vesical catheter was linked with 3 of the 5 HUAP-associated UTI. The other 5 UTI were outpatient-acquired and no invasive procedure could be connected to these diseases. Other CSN associated infections were represented by conjunctivitis (8%), meningitis (6%), peritonitis (6%), septic arthritis-osteomyelitis (4%) and finally, endocarditis and pneumonia (2%; Fig. 2). The most frequent species associated with infections was S. epidermidis, S. haemolyticus and S. warneri, corresponding to 72% of the clinically relevant isolates.
Figure 2.
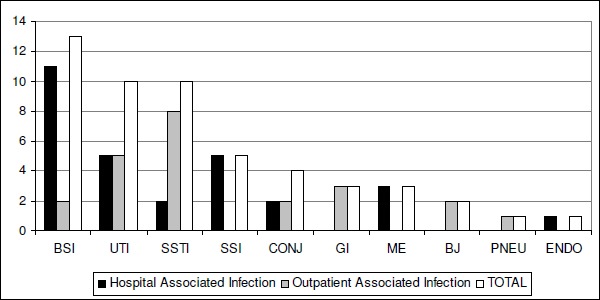
Percentage of Antônio Pedro University Hospital associated infections and outside acquired infections. BSI, blood stream infection; UTI, urinary tract infection; SSTI, skin and soft tissue infections; SSI, surgical site infection; CONJ, conjunctivitis; GI, peritonitis; ME, meningitis; BJ, Septic arthritis and osteomyelitis; PNEU, pneumoniae; and En, endocarditis.
Antimicrobial susceptibility
Among the 56 isolates obtained from the 52 clinically relevant cases, 2 (4%) did not show any resistance trait. The incidence of methicillin-resistance among these isolates was 21/56 (38%). The percentage of MRCNS did not vary significantly among isolates associated with infection in this university hospital (62%), when compared to the outside-acquired (38%). Similarly, the resistance profiles for isolates acquired or not in the HUAP were also quite similar for the other antimicrobial drugs tested. We observed a significant difference in the antimicrobial resistance profiles when contaminant and clinically relevant CNS isolates were compared (p=0.02). The majority of the MRCNS isolates (86%) were resistant to more than 2 antimicrobial classes, beside?ß-lactam antibiotics. Eight isolates of MRCNS (8/56; 14%) were only susceptible to three of the antimicrobials tested, including vancomycin. It was remarkable that one isolate of S. haemolyticus displayed resistance for all antimicrobial drugs tested excepted vancomycin. Table 2 shows the distribution of antimicrobial resistance among the clinically relevant CSN isolates.
Table 2.
Percentage of antimicrobial resistance of 56 clinically relevant CNS isolates in Antônio Pedro University Hospital.
| Antimicrobials | Resistance profile (%) | |||||
|---|---|---|---|---|---|---|
| S. epidermidis | S. haemolyticus | S. warneri | S. simulans | S. sciuri | Other CNS a | |
| Penicillin | 70 | 80 | 78 | 80 | 100 | 50 |
| Oxacillin | 59 | 27 | 22 | 0 | 100 | 17 |
| Erythomycin | 41 | 40 | 22 | 0 | 75 | 17 |
| Gentamicin | 47 | 27 | 22 | 0 | 50 | 0 |
| Trimethoprim-sulfamethoxazole | 53 | 20 | 22 | 0 | 0 | 0 |
| Clindamycin | 24 | 33 | 33 | 20 | 100 | 17 |
| Ciprofloxacin | 27 | 27 | 11 | 20 | 25 | 0 |
| Tetracyclin | 12 | 27 | 33 | 20 | 50 | 0 |
| Rifampin | 12 | 7 | 0 | 0 | 75 | 0 |
| Total isolates | 17 | 15 | 9 | 5 | 4 | 6 |
The other CNS isolates were: S. capitis (3), S. xylosus (1) and CNS not determinated species (2) DISCUSSION
DISCUSSION
Although CNS isolates have been associated with different nosocomial infectious diseases, the clinical relevance of CNS isolates is frequently questionable. In the paper presented here, we evaluated 204 CNS cultures obtained from 191 patients in a university hospital in Brazil. A low percentage of clinical significance (52/191; 27%) was verified among the cases analyzed. This impasse may be mainly due to the surveillance method for classified nosocomial infections. In our study to diagnose nosocomial infections, we use criterium based on the clinical and culture-proven evidence according to CDC definition; however, others authors used culture-proven evidence only (8). In addition, CNS is a group of bacteria belongs to the skin and mucosal human indigenous flora, thus it frequently contaminates directly or indirectly clinical specimens. We classified 12% of the isolates as contaminants originated from patient flora and 60% as exogenous contaminants. Besides that, the CNS isolates are one of the most important agents of nosocomial bacteremia, worldwide. This problem is complicated by the high level of methicillin resistance frequently observed among these isolates (22). In the paper presented here, the incidence of methicillin-resistance among CNS isolates was about 38% (21/56). The majority of the MRCNS (86%) was resistant to more than 2 antimicrobial classes besides ß-lactam antibiotic. In hospitals, the frequency of methicillin resistance among CNS isolates range from 30%-80% (22, 25, 30).
Searching on the PubMed (http://www.ncbi.nlm.nih.gov/entrez) we verified that the great majority of the studies involving clinical significance of CNS focused on blood isolates. Recently, 234 of 411 (57%) episodes of CNS bacteraemia in the presence of signs of sepsis were considered clinically relevant in a large university teaching hospital in Geneva, Switzerland (9). Data obtained from 49 USA hospitals showed that, in 1999, CNS isolates accounted for nearly 1/3 of the total BSI (7). Jarlov et al. (15) studying two small hospitals in Denmark observed that the majority of the 124 blood cultures of CNS lacks clinical significance. A study involving 160 patients with at least one positive blood culture for non-epidermidis CNS was performed by Ruhe et al. (26) and they diagnosed true bacteraemia in only 32 (20%) of the patients. Similarly, in another work, it was determined a rate of 30% of true bacteraemia in a total of 137 episodes of positive CNS blood cultures (10). In a study, carried out in a NICU of a Brazilian hospital, it was verified a percentage of 51% of clinical significance for a total of 117 CNS cultures of blood, urine and secretions (5). The higher percentage of clinical significance observed by those authors might be associated with the fact that most of the patients presented with very low birth weight (<1,500 g) and were submitted to two or more invasive procedures, including the use of catheter, parenteral nutrition and mechanical ventilation.
In the study presented here, 33% of the infections were associated to an invasive medical device (17/52). It is well known that infections caused by CNS isolates are commonly associated to the use of indwelling medical devices (6, 33). The production of biofilm is considered the most important virulence factor in infections caused by S. epidermidis and others CNS isolates associated to indwelling medical devices (6, 35). Thus, some researches have investigated the production of biofilm by clinically relevant CNS isolates. It was reported that the identification of biofilm producer S. epidermidis predicted the clinical significance of blood isolates of CNS with an overall accuracy of 89% (14). Conversely, in their studies, Jones et al. (17) concluded that biofilm production had a limited role on the differentiation between clinically significant and contaminant CNS isolates obtained from blood cultures. However, they observed that absence of biofilm in isolates of S. epidermidis suggests a lack of pathogenicity. In a recent study from our group it was verified that isolates of S. epidermidis obtained from nasal colonization had a lower probability to produce biofilm (about 60%) when compared with S. epidermidis isolates obtained from infections diseases at HUAP (about 80%). However, beside the difference observed, these results indicate that S. epidermidis from human flora or from infectious diseases can be both equipped with this virulence factor (3).
In agreement with other studies, this paper showed S. epidermidis was the most common CNS causing sepsis in the NICU (4, 5, 24, 25). Nevertheless, analysing the total of CNS-associated diseases, in the study presented here only 30% of the total 56 clinically relevant CNS isolates detected were S. epidermidis. On the other hand, recently, S. capitis is also point to the possible emergence of this species in NICUs, being the second most common species (4). This finding was also confirmed by our results.
Besides the importance of CNS isolates as hospital pathogens, our results showed that many of CNS laboratory isolates are contaminants from the patient flora or from exogenous sources. Similar results were also demonstrated by others (3, 21).
Concluding, until specific markers can be established to distinguish clinically relevant from non-relevant CNS isolates, stringent criteria for classification of these bacterial infections are recommended. Cases of neonatal bacteremia by CNS showed high clinical significance. The use of more effective precautions during the collection of samples should be observed, in order to avoid exogenous contaminations.
ACKNOWLEDGEMENTS
This work was supported by FAPERJ (Fundação Carlos Chagas de Amparo a Pesquisa do Estado do Rio de Janeiro), PRONEX/FAPERJ, CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). We thank Jupira Miron Carballido for providing access in the microbiology laboratory of the Hospital Universitário Antônio Pedro.
All the content of the journal, except where otherwise noted, is licensed under a Creative Commons License
REFERENCES
- 1.Alcaráz, L.E.; Satorres, S.E.; Lucero, R.M.; Puig de Centorbi, O.N. (2003). Species identification, slime production and oxacillin susceptibility in coagulase-negative Staphylococci isolated from nosocomial specimes. Braz. J. Microbiol. 34, 45-51. [Google Scholar]
- 2.Araújo, G.L.; Coelho, L.R.; Carvalho, C.B.; Maciel, R.M.; Coronado, A.Z.; Rozenbaum, R.; Ferreira-Carvalho, B.T.; Figueiredo, M.A.S.; Teixeira, L.A. (2006). Commensal isolates of methicillin-resistant Staphylococcus epidermidis are also well equipped to produce biofilm on polystyrene surfaces. J. Antimicrob. Chemother. 57, 855-864. [DOI] [PubMed] [Google Scholar]
- 3.Bodonaik, N.C.; Moonah, S. (2006). Coagulase negative Staphylococci from blood cultures: contaminants or pathogens? West Indian Med. J. 55, 174-82. [DOI] [PubMed] [Google Scholar]
- 4.Bradford, R.; Abdul Manan, R.; Daley, A.J.; Pearce, C.; Ramalingam, A.; D'Mello, D.; Mueller, Y.; Uahwatanasakul, W.; Qu, Y.; Grando, D.; Garland, S.; Deighton, M. (2006). Coagulase-negative staphylococci in very-low-birth-weight infants: inability of genetic markers to distinguish invasive strains from blood culture contaminants. Eur. J. Clin. Microbiol. Infect. Dis. 25, 283-90. [DOI] [PubMed] [Google Scholar]
- 5.Cunha, M.L.R.S.; Lopes, C.A.M.; Rugolo, L.M.S.S.; Chalita, L.V.A.S. (2002). Clinical significance of coagulase-negative staphylococci isolated from neonates. J. Pediatr. 78, 279-288. [PubMed] [Google Scholar]
- 6.Donlan, R.M. (2001). Biofilms and device-associated infections. Emerg. Infect. Dis. 7, 277-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmond, M.E.; Wallace, S.E.; McClish, D.K.; Pfaller, M.A.; Jones, R.N.; Wenzel, R.P. (1999). Nosocomial bloodstream infections in United States hospitals: a three-year analysis. Clin. Infect. Dis. 29, 239-244. [DOI] [PubMed] [Google Scholar]
- 8.Efird, M.M.; Rojas, M.A.; Lozano, J.M.; Bose, C.L.; Rojas, M.X.; Rondon, M.A.; Ruiz, G.; Pineros, J.G.; Rojas, C.; Robayo, G.; Hoyos, A.; Gosendi, M.E.; Cruz, H.; Leon, A. (2005). Epidemiology of nosocomial infections in selected neonatal intensive care units in Colombia, South America. J. Perinatol. 25, 531-6. [DOI] [PubMed] [Google Scholar]
- 9.Favre, B.; Hugonnet, S.; Correa, L.; Sax, H.; Rohner, P.; Pittet, D. (2005). Nosocomial bacteremia: clinical significance of a single blood culture positive for coagulase-negative staphylococci. Infect. Control Hosp. Epidemiol 26, 697-702. [DOI] [PubMed] [Google Scholar]
- 10.Finkelstein, R.; Fusman, R.; Oren, I.; Kassis, I.; Hashman, N. (2002). Clinical and epidemiologic significance of coagulase-negative staphylococci bacteremia in a tertiary care university Israeli hospital. Am. J. Infect. Control 30, 21-25. [DOI] [PubMed] [Google Scholar]
- 11.Götz, F. (2002). Staphylococcus and biofilms. Mol. Microbiol. 43, 1367-1378. [DOI] [PubMed] [Google Scholar]
- 12.Horan, T.C.; Gaynes, R.P. (2004). Surveillance of nosocomial infections. In: Mayhall, CG (ed). Hospital Epidemiology and Infection Control Willians & Wilkins, Philadelphia: Lippincott, E.U.A., p.1695-1702. [Google Scholar]
- 13.Hu, L.; Umeda, A.; Amako, K. (1995). Typing of Staphylococcus epidermidis colonizing human nares by pulsed-field gel electrophoresis. Microbiol. Immunol. 39, 315-319. [DOI] [PubMed] [Google Scholar]
- 14.Ishak, M.A.; Groschel, D.H.; Mandell, G.L.; Wenzel, R.P. (1985). Association of slime with pathogenicity of coagulase-negative staphylococci causing nosocomial septicemia. J. Clin. Microbiol. 22, 1025-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarlov, J.O.; Prag, J.; Rosdahl, V.T.; Espersen, F. (1995). Evaluation of staphylococci isolated from a blood culture system (Colorbact). APMIS 103, 383-387. [DOI] [PubMed] [Google Scholar]
- 16.Jeong, I.S.; Jeong, J.S.; Choi, E.O. (2006). Nosocomial infection in a newborn intensive care unit (NICU), South Korea. BMC Infect. Dis. 23, 103-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, J.W.; Scott, R.J.; Morgan, J.; Pether, J.V. (1992). A study of coagulase-negative staphylococci with reference to slime production, adherence, antibiotic resistance patterns and clinical significance. J. Hosp. Infect. 22, 217-227. [DOI] [PubMed] [Google Scholar]
- 18.Kloos, W.E.; Bannerman, T.L. (1994). Update on clinical significance of coagulase-negative staphylococci. Clin. Microbiol. Rev. 7, 117-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marra, A.R.; Opilla, M.; Edmond, M.B.; Kirby, D.F. (2007). Epidemiology of bloodstream infections in patients receiving long-term total parenteral nutrition. J. Clin. Gastroenterol 41, 19-28. [DOI] [PubMed] [Google Scholar]
- 20.Mattos, E.M.; Teixeira, L.A.; Alves, V.M.; Resende, C.A.F.R.; Coimbra, M.V.S.; Carvalho, M.C.S.; Ferreira-Carvalho, B.T.; Figueiredo, A.M.S. (2003). Isolation of methicillin-resistant coagulase-negative staphylococci from patients undergoing continuous ambulatory peritoneal dialysis (CAPD) and comparison of different molecular techniques for discriminating isolates of Staphylococcus epidermidis Diagn. Microbiol. Infect. Dis. 45, 13-22. [DOI] [PubMed] [Google Scholar]
- 21.Mirrett, S.; Weinstein, M.P.; Reimer, L.G.; Wilson, M.L.; Reller, L.B. (2001). Relevance of the number of positive bottles in determining clinical significance of coagulase-negative staphylococci in blood culture. J. Clin. Microbiol. 39, 3279-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.NNIS System (2004). National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1992 through June 2004, issue October 2004. Am. J. Infec. Report. 32, 470-485. [DOI] [PubMed] [Google Scholar]
- 23.Raad, I.; Alrahwan, A.; Rolston, K. (1998). Staphylococcus epidermidis: Emerging resistance and the need for alternative agents. Clin. Infect. Dis. 26, 1182-1187. [DOI] [PubMed] [Google Scholar]
- 24.Raimundo, O.; Heussler, H.; Bruhn, J.B.; Suntrarachun, S.; Kelly, N.; Deighton, M.A.; Garland, S.M. (2002). Molecular epidemiology of coagulase-negative staphylococcal bacteraemia in a newborn intensive care unit. J. Hosp. Infect. 51, 33-42. [DOI] [PubMed] [Google Scholar]
- 25.Refsahl, K.; Andersen, B.M. (1992). Clinically significant coagulase-negative staphylococci: identification and resistance patterns. J. Hosp. Infect. 22, 19-31. [DOI] [PubMed] [Google Scholar]
- 26.Ruhe, J.; Menon, A.; Mushatt, D.; Dejace, P.; Hasbun, R. (2004). Non-epidermidis coagulase-negative staphylococcal bacteremia: clinical predictors of true bacteremia. Eur. J. Clin. Microbiol. Infect. Dis. 23, 495-498. [DOI] [PubMed] [Google Scholar]
- 27.Rupp, M.E.; Archer, G. (1994). Coagulase-negative staphylococci: pathogens associated with medical progress. Clin. Infect. Dis. 19, 231-243. [DOI] [PubMed] [Google Scholar]
- 28.Sader, H.S.; Jones, R.N.; Gales, A.C.; Silva, J.B.; Pignatari A.C. (2004). SENTRY Antimicrobial Surveillance Program Report: Latin American and Brazilian Results for 1997 through 2001. Braz. J. Infect. Dis. 8, 25-79. [DOI] [PubMed] [Google Scholar]
- 29.Sewell, C.M.; Clarridge, J.E.; Young, E.J.; Guthrie, R.K. (1988). Clinical significance of coagulase-negative staphylococci. J. Clin. Microb. 16, 236-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva, F.R.; Mattos, E.M.; Coimbra, M.V.S.; Ferreira-Carvalho, B.T.; Figueiredo, A.M.S. (2001). Isolation and molecular characterization of methicillin-resistant coagulase-negative staphylococcus from nasal flora of healthy humans at three community institutions in Rio de Janeiro City. Epidemiol. Infect. 127, 57-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sivadon, V.; Rottman, M.; Chaverot, S.; Quincampoix, J.C.; Avettand, V.; de Mazancourt, P.; Bernard, L.; Trieu-Cuot, P.; Feron, J.M.; Lortat-Jacob. A.; Piriou, P.; Judet, T.; Gaillard, J.L. (2005). Use of genotypic identification by sodA sequencing in a prospective study to examine the distribution of coagulase-negative Staphylococcus species among strains recovered during septic orthopedic surgery and evaluate their significance. J. Clin. Microbiol. 43, 2952-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spencer, R.C. (1996). Predominant pathogens found in the European prevalence of infections in intensive care study. Eur. J. Clin. Microbiol. Infect. Dis. 15, 281-285. [DOI] [PubMed] [Google Scholar]
- 33.von Eiff, C.; Jansen, B.; Kohnen, W.; Becker, K. (2005). Infections associated with medical devices: pathogenesis, management and prophylaxis. Drugs 65, 179– 214. [DOI] [PubMed] [Google Scholar]
- 34.von Eiff, C.; Peters, G.; Heilmann, C. (2002). Pathogenesis of infections due to coagulase-negative staphylococci. Lancet Infect. Dis. 2, 677-685. [DOI] [PubMed] [Google Scholar]
- 35.Vuong, C.; Otto, M. (2002). Staphylococcus epidermidis infections. Microbes Infect. 4, 481-489. [DOI] [PubMed] [Google Scholar]
- 36.Ziebuhr, W.; Hennig, S.; Eckart, M.; Kranzler, H.; Batzilla, C.; Kozitskaya, S. (2006). Nosocomial infections by Staphylococcus epidermidis: how a commensal bacterium turns into a pathogen. Int. J. Antimicrob. Agents 28 (1), S14-20. [DOI] [PubMed] [Google Scholar]


