Abstract
In the present study, the efficacy of polymerase chain reaction (PCR) based on mapA gene of C. jejuni was tested for detection of Campylobacter jejuni in naturally infected as well as spiked faecal and food samples of human and animal origin. Simultaneously, all the samples were subjected to the cultural isolation of organism and biochemical characterization. The positive samples resulted in the amplification of a DNA fragment of size ~589 bp in PCR assay whereas the absence of such amplicon in DNA extracted from E. coli, Listeria, Salmonella and Staphylococcus confirmed the specificity of the primers. Of randomly collected 143 faecal samples comprising human diarrheic stools (43), cattle diarrheic faeces (48) and poultry faecal swabs (52) only 4, 3 and 8, respectively, could be detected by isolation whereas 6, 3 and 10, respectively, were found positive by PCR. However, among food samples viz. beef (30), milk (35), cheese (30), only one beef sample was detected both by culture as well as PCR. Additionally, PCR was found to be more sensitive for C. jejuni detection in spiked faecal and food samples (96.1% each) as relative to culture isolation which could detect the organism in 86.7% and 80% samples, respectively. The results depicted the superior efficacy of PCR for rapid screening of samples owing to its high sensitivity, specificity and automation potential.
Keywords: Campylobacter jejuni, isolation, PCR, spiking
INTRODUCTION
Different species within the genus Campylobacter have emerged over the last three decades as important clinical pathogens of human and veterinary concern. The majority of acute bacterial intestinal infections in human beings in the western countries are caused by these organisms, particularly due to thermotolerant campylobacters (11). Among these, C. jejuni and C. coli are the most common pathogens responsible for the majority of human enteritis cases (2, 15). C. jejuni subsp. jejuni has also been reported to cause abortion and mastitis in bovines. Besides, zoonotic campylobacters have been found associated with potentially life threatening complications like Guillain-Barre syndrome, reactive arthritis, hemolytic uraemic syndrome and meningitis etc. (7, 16, 17). The prime cause of campylobacter infections is considered to be contaminated food as the organism is a part of normal flora in various animal species such as poultry, pigs, and cattle.
In the recent past, number of enteritis cases in humans due to campylobacters has exceeded to those caused by Salmonella and Shigella, especially in developed world. However, in developing countries, the true incidence of campylobacteriosis is often underestimated because of lack of adequate laboratory infrastructure. The conventional methods of detection of campylobacters are based on cultural isolation followed by various genus and species specific biochemical tests which are cumbersome and time consuming. The hippurate hydrolysis, a routinely performed test for identification of C. jejuni has its own limitations as false negative as well as false positive results with this test have been reported due to emergence of some hippurate negative C. jejuni strains and some hippurate positive non-C. jejuni strains, respectively (5, 12). Additionally, Campylobacter spp. can survive as viable but non culturable (VBNC) forms which may not grow on selective media. Subsequently, the refrigerated storage under reduced oxygenated conditions that occur in modified atmospheric packaging or vacuum packaging of food products may allow resuscitation of injured or VBNC Campylobacter spp, hereby rendering a potential threat to human health.
Polymerase chain reaction (PCR) assays have been widely employed for identification of the pathogens owing to their sensitivity and cost effectiveness (8). A number of PCR assays have been described for the detection of campylobacters from food and faecal samples (1, 4, 6, 13, 14). The present study was carried to access the prevalence of C. jejuni in various food and faecal samples and to compare the efficacy of cultural and biochemical tests with PCR for detection of the organism.
MATERIALS AND METHODS
Sample collection
A total of 238 faecal and food samples of human and animal origin belonging to Uttar Pradesh state of India were included in the study. Of these, faecal samples comprised human diarrhoeic stools (43), cattle diarrhoeic faeces (48) and poultry faecal swabs (52) where as the food samples included beef (30), milk (35) and cheese (30). The samples were collected over ice maintaining all the sterility measures and brought to the laboratory in enrichment medium.
Cultural and biochemical examination
For cultural isolation of the organism, modified selective media (3) was employed. Briefly, the human and cattle diarrhoeic stool samples and faecal swabs of poultry were inoculated into modified enrichment broth and incubated at 37oC for 48 hr under microaerophilic conditions (5% O2, 10% CO2 and 85% N2) using CampyPak (BD, Oxoid) gas generating packs. The grown cultures from broth were streaked onto respective agar plates, incubated as above and were regularly observed for 5-7 days for any bacterial growth. Characteristic Campylobacter colonies were tested for genus specific phenotypic and biochemical characters i.e., Gram’s staining, motility, oxidase, catalase and nitrate reduction tests followed by species specific characters i.e., hippurate hydrolysis, growth in 1% glycine, H2S production on triple sugar iron agar, growth at 25oC and 42oC and sensitivity to nalidixic acid.
Preparation of samples for PCR
Faecal samples, five gram each from cattle, poultry and human were mixed with 50 ml of enrichment broth so as to make a homogeneous suspension. The mixture was incubated under microaerophillic conditions at 37oC for 3 h and then for 18 h at 42oC. Subsequently, it was centrifuged passively to remove the debris and 1 ml of supernatant obtained was further centrifuged at 10000xg for 10 min. The resulting pellet was resuspended in 100 μl of TE buffer (10mM Tris-HCl, 1mM EDTA, pH 8.0), boiled for 10 min followed by immediate chilling on ice. After its centrifugation at 10000xg for 10 min, a five μl of the supernatant was directly used as template in 25 μl PCR reaction. For preparation of template DNA from beef and cheese, five gram minced sample from either of these was diluted ten times (w/v) in enrichment broth and later on processed in same way as that of stools. As regards milk samples, 10 ml milk was added to 90 ml enrichment broth and incubated as above. After incubation, 1 ml suspension was centrifuged at 10000xg for 10 min and the resulting pellet was resuspended in 100 μl of TE buffer (pH 8.0). Subsequent processing was carried out in similar manner as that of faecal samples.
PCR assay
The primers based on mapA gene of C. jejuni (5) were got custom synthesized. The sequences of forward and reverse oligonucleotide primers were as follows:
Forward 5’-CTATTTTATTTTTGAGTGCTTGTG-3’
Reverse 5’-GCTTTATTTGCCATTTGTTTTATTA-3’
The cyclic conditions for PCR were same as those described by Denis et al. (5) which were as follows: initial denaturation at 94oC for 2 min followed by 30 cycles of denaturation at 94oC for 40 sec, annealing at 54oC for 40 sec and extension at 72oC for 1 min and a final extension at 72oC for 5 min. The reaction mixture comprised of 1x PCR buffer [50 mM Tris-HCl, 10 mM KCl, 5 mM (NH4)2SO4, pH8.3], 1.0 mM MgCl2 (MBI fermentas, USA), 0.2 mM dNTP mix (MBI fermentas, USA), 16 p mol of each of the primers (Integrated DNA Technologies, Inc, IA, USA), 1 U of Taq DNA polymerase enzyme (MBI fermentas, USA), 5 μl of template DNA in 25 μl of reaction mixture. The PCR products were analyzed by 1.5% agarose gel (Amersham Pharmacia Biotech AB, Uppsala, Sweden) electrophoresis and photographed using a gel documentation system (Alpha Imager, Germany).
Specificity and sensitivity of PCR
To test the specificity of primers, the PCR assay was also applied on E. coli, Salmonella, Listeria and Staphylococcus organisms. PCR reaction mixture and cyclic conditions were kept same as described above.
For testing the sensitivity of PCR, freshly grown pure culture of Campylobacter was taken and the concentration of cells in liquid culture was estimated to be 109 cells/ml. From this culture, 10 fold serial dilutions were made from 109 to 104 cells/ml. Each of these dilutions was further diluted 1:10 in TE buffer (pH 8.0), boiled for 10 min followed by chilling over ice and centrifugation at 10000xg for 10 min. A 5 μl of the supernatant was used as template in the PCR, resulting in a final concentration ranging from 5x105 to 51 cells per PCR.
Artificial inoculation/Spiking studies
The experimental inoculation studies were carried out to assess the efficacy of the standardized PCR method for the detection of C. jejuni in spiked faecal and food samples. Faecal samples from cattle, poultry and human were inoculated with overnight grown culture of C. jejuni so as to make a final concentration of 106 bacterial cells/ml of stools and the resulting mixture was centrifuged passively to remove the debris. The resulting supernatant was diluted ten times in TE buffer (pH 8.0) and boiled for 10 min followed by immediate chilling on ice. Afterwards, it was centrifuged at 10000xg for 10 min and a five μl of the supernatant was used as template in PCR. For spiking of milk, overnight grown culture of C. jejuni was added to the pasteurized whole milk in order to achieve a final concentration of 106 bacterial cells/ml of milk. One ml of this spiked milk was diluted ten times in TE buffer (pH 8.0) and later on processed in a way same as that of stools for preparation of template DNA. As regards beef and cheese samples, one gram of minced beef or cheese was mixed with 10 ml of enrichment broth and C. jejuni cells were added to it making a final concentration of 106 cells/ml of suspension. Subsequently, the debris was removed by passive centrifugation and the supernatant was processed for the preparation of template DNA as described elsewhere. Simultaneously, the spiked samples were also streaked onto the modified selective solid media and plates were incubated at 42oC for 3-5 days under microaerophilic conditions for cultural isolation of C. jejuni.
RESULTS AND DISCUSSION
Of 143 faecal samples, 15 were found positive for C. jejuni by cultural and biochemical examination, out of which 4 belonged to human, 3 to cattle and 8 to poultry (Table 1). As regards food samples, only one (beef) was found positive whereas all the milk and cheese samples were found negative. Biochemically, the organisms were positive for oxidase, catalase, nitrate reduction tests as well as species specific tests like hippurate hydrolysis, growth in 1% glycine and were sensitive to nalidixic acid.
Table 1.
Results of cultural isolation and PCR for detection of C. jejuni in randomly collected faecal and food samples
| Type of sample | No. of positive samples | |
|---|---|---|
| Culture | PCR | |
| Faecal samples | ||
| Human diarrhoeic stools (43) | 4 | 6 |
| Cattle diarrhoeic faeces (48) | 3 | 3 |
| Poultry faecal swabs (52) | 8 | 10 |
| Total (143) | 15 (10.5%) | 19 (13.3%) |
| Food samples | ||
| Beef (30) | 1 | 1 |
| Milk (35) | 0 | 0 |
| Cheese (30) | 0 | 0 |
| Total (95) | 1 (1.05%) | 1(1.05%) |
PCR detected all those faecal samples found positive by cultural examination as a fragment of size ~589 bp (Fig. 1) was amplified from these samples. Additionally, four more samples were found positive which were culturally negative; 2 each from human diarrhoeic stools and poultry faecal swabs, respectively. Hence, PCR was found more efficient for detecting C. jejuni from faecal samples (10.5% by culture versus 13.3% by PCR). Earlier workers have also demonstrated the superior efficacy of PCR in detection of the campylobacters from faecal samples declared negative by selective cultural and biochemical tests (9, 10).
Figure 1.
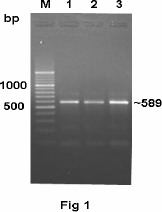
PCR amplification of mapA gene from representative samples for detection of Campylobacter jejuni. Lane M: DNA ladder. Lanes 1 to 3: human diarrhoeic stool, cattle diarrhoeic faeces and poultry faecal swab samples, respectively.
Regarding food samples, no difference was observed in PCR and culture isolation methods for detection of organism as only one beef sample was detected by PCR that was found positive by selective culture method also. Low incidence of C. jejuni in raw beef (3.2%) and raw bulk tank milk samples (1.6%) has been reported earlier also (19).
As isolation and identification of the campylobacters based on selective culture and biochemical differentiation upto species level is tedious, time consuming and has been proved time and again not very reliable. Hence, on the basis of our study, we can say that PCR based methods are more rapid and reliable, particularly while processing a large number of samples.
No amplification was observed in PCR using DNA extracted from E. coli, Listeria, Salmonella and Staphylococcus organisms (Fig. 2). The absence of desired amplicon from these organisms confirmed the specificity of the primers. Regarding sensitivity of PCR on DNA extracted from pure culture of C. jejuni by heat lysis method, upto a minimum of 50 cells per PCR reaction (corresponding to 105 cells/ml of culture) were detected. However, the intensity of amplicons gradually improved with increase in concentration of cells (Fig. 3). High sensitivity of PCR assay for detection of C. jejuni from pure culture was in agreement with observation of Perssson and Olsen (13) who detected 10-100 cells per PCR reaction.
Figure 2.
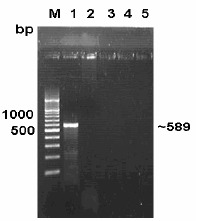
Specificity study of PCR amplification of mapA gene. Lane M: DNA ladder. Lane 1: Campylobacter jejuni. Lane 2: E. coli. Lane 3: Salmonella. Lane 4: Listeria. Lane 5: Staphylococcus.
Figure 3.
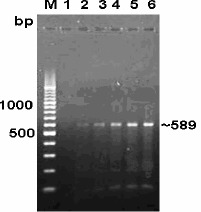
Sensitivity assay of PCR by 10 fold dilution of bacterial DNA derived from Campylobacter jejuni culture. Lane M: DNA ladder. Lanes 1 to 6: 104, 105, 106, 107, 108, 109 cells/ml respectively.
As regards artificial inoculation studies, culture and biochemical identification could detect 26 (86.7%) of 30 faecal samples and 24 (80%) of 30 food samples whereas 29 (96.1%) each of faecal and food samples were found positive in PCR assay (Table 2). Persson and Olsen (13) and Waage et al. (18) have also found PCR to be quite effective in detection of C. jejuni from spiked samples. Our study on spiked samples further underscores the better efficacy of PCR over cultural identification of the organism and bolsters the application of technique in randomly collected faecal and food samples.
Table 2.
Comparison of culture and PCR for detection of C. jejuni in spiked faecal and food samples
| No. of positive samples | ||
|---|---|---|
| Type of Sample | Culture | PCR |
| a) Faecal samples | ||
| Cattle diarrhoeic faeces (10) | 9 | 10 |
| Human diarrhoeal stools (10) | 8 | 9 |
| Poultry faecal swabs (10) | 9 | 10 |
| Total (30) | 26 (86.7%) | 29 (96.1%) |
| b) Food samples | ||
| Beef (10) | 9 | 10 |
| Cheese (10) | 7 | 9 |
| Milk (10) | 8 | 10 |
| Total (30) | 24 (80%) | 29 (96.1%) |
REFERENCES
- 1.Al Amri A., Senok A.C., Ismaeel A.Y., Al-Mahmeed Ali E., Botta Giuseppe A. Multiplex PCR for direct identification of Campylobacter spp. in human and chicken stools. J. Med. Microbiol. 2007;56:1350–1355. doi: 10.1099/jmm.0.47220-0. [DOI] [PubMed] [Google Scholar]
- 2.Allos B.M., Blaser M.J. C. jejuni and expanding spectrum of related infections. Clin. Infect. Dis. 1995;20:1092–1099. doi: 10.1093/clinids/20.5.1092. [DOI] [PubMed] [Google Scholar]
- 3.Barua R., Rathore R.S. Development of modified selective media for the isolation of Campylobacter jejuni from poultry. J. Food Sci. Technol. 2006;43:305–307. [Google Scholar]
- 4.Debruyne L., Samyn E., De Brandt E., Vandenberg O., Heyndrickx M., Vandamme P. Comparative performance of different PCR assays for the identification of Campylobacter jejuni and Campylobacter coli. Res. Microbiol. 2008;159:88–93. doi: 10.1016/j.resmic.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 5.Denis M., Soumet C., Rival K., Ermel G., Salavat G., Colin P. Development of a mPCR assay for simultaneous identification of Campylobacter jejuni and C. coli. Lett. Appl. Microbiol. 1999;29:406–410. doi: 10.1046/j.1472-765x.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 6.Jackson C.J., Fox A.J., Jones D.M. A novel polymerase chain reaction assay for the detection and speciation of thermophilic Campylobacter spp. J. Appl. Bacteriol. 1996;81:467–473. doi: 10.1111/j.1365-2672.1996.tb03534.x. [DOI] [PubMed] [Google Scholar]
- 7.Kopyta I., Wardak S. Campylobacter jejuni infection in patient with Guillain Barre syndrome- Clinical case report. Med. Dosw. Mikrobiol. 2008;60:59–63. [PubMed] [Google Scholar]
- 8.Kricka L.J. Prospects for chemiluminescent and bioluminescent immunoassay and nucleic acid assays in food testing and the pharmaceutical industry. J. Biolumin. Chemilumin. 1998;13:189–193. doi: 10.1002/(SICI)1099-1271(199807/08)13:4<189::AID-BIO487>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 9.Kulkarni S.P., Lever S., Logan J.M., Lawson A.J., Stanley J., Shafi M. Detection of Campylobacter species: a comparison of culture and polymerase chain reaction based methods. J. Clin. Pathol. 2002;55:749–753. doi: 10.1136/jcp.55.10.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawson A.J., Shafi M.S., Pathak K., Stanley J. Detection of Campylobacter in gastroenteritis: Comparison of direct PCR assay of faecal samples with selective culture. Epidemiol. Infect. 1998;121:547–553. doi: 10.1017/s0950268898001630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher M., Finnegan C., Collins E., Ward B., Carroll C., Cormican M. Evaluation of culture methods and a DNA probe-based PCR assay for detection of Compylobacter spp. in clinical specimens of faeces. J. Clin. Microbiol. 2003;41:2980–2986. doi: 10.1128/JCM.41.7.2980-2986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morris G.K., El Sherbeeny M.R., Patton C.M., Kodaka H., Lombard G.L., Edmonds P., Hollis D.G., Brenner D.J. Comparison of four hippurate hydrolysis methods for identification of thermophilic Campylobacter spp. J. Clin. Microbiol. 1985;22:714–718. doi: 10.1128/jcm.22.5.714-718.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Persson Søren, Olsen Katharina E.P. Multiplex PCR for identification of Campylobacter coli and Campylobacter jejuni from pure cultures and directly on stool samples. J. Med. Microbiol. 2005;54:1043–1047. doi: 10.1099/jmm.0.46203-0. [DOI] [PubMed] [Google Scholar]
- 14.Sails Andrew D., Fox A.J., Bolton F.J., Wareing D.R.A., Greenway D.L.A. A Real-Time PCR Assay for the detection of Campylobacter jejuni in foods after enrichment culture. Appl. Environ. Microbiol. 2003;69:1383–1390. doi: 10.1128/AEM.69.3.1383-1390.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skirrow M.B. Disease due to Campylobacter, Helicobacter and related bacteria. J. Comp. Pathol. 1994;111:113–149. doi: 10.1016/s0021-9975(05)80046-5. [DOI] [PubMed] [Google Scholar]
- 16.Stanfford T.D., Tenkate R.J. Risk factors for Campylobacter infection in infants and young children: A matched case control study. Epidemiol. Infect. 2001;127:399–404. doi: 10.1017/s0950268801006306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stern N.D., Line J.E. Campylobacter. 2000. pp. 1040–1056. In: The microbiological safety and quality of food.
- 18.Waage A.S., Vardund T., Lund V., Kapperud G. Detection of small numbers of Campylobacter jejuni and Campylobacter coli cells in environmental water, sewage, and food samples by a seminested PCR assay. Appl. Environ. Microbiol. 1999;65:1636–1643. doi: 10.1128/aem.65.4.1636-1643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whyte P., McGill K., Cowley D., Madden R.H., Moran L., Scates P., Cauoll C., O’Leary A., Funning S., Collins J.D., McNamara E., Moore J.E., Cormican M. Occurrence of Campylobacter in retail food in Ireland. Int. J. Food Microbiol. 2004;95:111–118. doi: 10.1016/j.ijfoodmicro.2003.10.018. [DOI] [PubMed] [Google Scholar]


