Abstract
The common bean is one of the most important legumes in the human diet, but little is known about the endophytic bacteria associated with the leaves of this plant. The objective of this study was to characterize the culturable endophytic bacteria of common bean (Phaseolus vulgaris) leaves from three different cultivars (Vermelhinho, Talismã, and Ouro Negro) grown under the same field conditions. The density of endophytic populations varied from 4.5 x 102 to 2.8 x 103 CFU g-1 of fresh weight. Of the 158 total isolates, 36.7% belonged to the Proteobacteria, 32.9% to Firmicutes, 29.7% to Actinobacteria, and 0.6% to Bacteroidetes. The three P. vulgaris cultivars showed class distribution differences among Actinobacteria, Alphaproteobacteria and Bacilli. Based on 16S rDNA sequences, 23 different genera were isolated comprising bacteria commonly associated with soil and plants. The genera Bacillus, Delftia, Methylobacterium, Microbacterium, Paenibacillus, Staphylococcus and Stenotrophomonas were isolated from all three cultivars. To access and compare the community structure, diversity indices were calculated. The isolates from the Talismã cultivar were less diverse than the isolates derived from the other two cultivars. The results of this work indicate that the cultivar of the plant may contribute to the structure of the endophytic community associated with the common bean. This is the first report of endophytic bacteria from the leaves of P. vulgaris cultivars. Future studies will determine the potential application of these isolates in biological control, growth promotion and enzyme production for biotechnology.
Keywords: Endophytic bacteria, common bean, Phaseolus vulgaris, 16S rDNA, diversity indices.
INTRODUCTION
The phyllosphere is the habitat for a large diversity of microorganisms. Although bacteria are the predominant microorganisms present in phyllospheres, others such as filamentous fungi are also important members. Phyllosphere bacteria may be found on the surface of plants (epiphytes) as well as in the interior of plant tissues (endophytes) (3, 32, 40). Endophytic bacteria can be defined as those that can be isolated from healthy, superficially disinfected plant tissues and do not cause any damage to the host plant (15, 17). The population density of endophytic bacteria can vary from 102 to 109 (6, 12, 25, 39, 44) and depends on many factors, including the plant being studied, the part under analysis (31, 46), the developmental stage of the plant (17, 44), the plant cultivar (genotype) (15, 44) and the interaction with other organisms, as well as other environmental-related factors (17).
The interaction between endophytic bacteria and their host plants is not completely understood. However, many isolates seem to have beneficial effects on their hosts (58). These beneficial effects include promoting host growth and biological control of phytopathogens (17, 21).
The common bean (Phaseolus vulgaris) is one of the most important legumes in the human diet and serves as a significant source of proteins (10). The relationship between Rhizobium and other nitrogen-fixing bacteria in the root nodules of beans has been extensively studied (13, 37, 38). Recently, López-López et al. (34) reported the isolation of endophytic bacteria from the seeds and roots of the common bean. However, little is known about endophytic bacteria inhabiting the aerial tissues of the common bean. Therefore, the objective of this study was to isolate the culturable, endophytic bacteria from the leaves of three different common bean cultivars growing in field conditions and characterize the community of culturable bacteria. To our knowledge, this is the first report on endophytic bacteria from the leaves of different cultivars of the common bean.
MATERIALS AND METHODS
Plant materials
Samples were collected from three common bean cultivars during the winter of 2007: Talismã (TAL), Ouro Negro (ONG), and Vermelhinho (VER). The cultivars were planted in an experimental field in the town of Coimbra – MG (altitude: 690 m; latitude: 20º 45’ S; longitude 42º 51’ W). During sowing, 350 kg ha-1 of the 8–28–16 NPK (percentage of nitrogen, phosphorus and potassium) formula was applied, and 25 days after their emergence, the plants were covered in 150 kg ha-1 of ammonium sulfate. The leaves of the superior portion of the plant (20 cm above the soil) were collected in the vegetative phase 45 days after sowing.
Sample preparation and bacterial isolation
The collected leaves were washed in running water and those with superficial injury that was visible to the naked eye were excluded. Each isolation procedure was done in triplicate for each cultivar. Each triplicate was composed of approximately 2 g of leaves belonging to two different plants being evaluated, totaling six plants per cultivar. The disinfection and isolation were performed according to Araujo et al. (1) with minor modifications. Briefly, the leaves were disinfected superficially through the following protocol: 70% alcohol for 1 min, sodium hypochlorite (2.5% Cl-) for 4 min, ethanol for 30 s, and finally 3 rinses in sterile, distilled water. To confirm the disinfection protocol, aliquots of the sterile water used in the final rinse were plated in 10% TSA (1.5 g/L of triptone, 0.5 g/L of soy peptone, 1.5 g/L of NaCl, 15 g/L of agar, pH 7.3) at 28 ºC for 15 days and the plates are examined for the presence or absence of microorganismal growth colony.
Initially, the leaves were ground with 6 mL of aqueous solution (0.9 % NaCl) using a sterile mortar and pestle. The tissue extract was subsequently incubated at 28 ºC for 3 hours to allow the complete release of endophytic microorganisms from the host tissue. For the isolation of endophytic bacteria, the tissue extract was diluted in an aqueous solution (0.9 % NaCl) and plated on five 10% TSA plates for each dilution (10–1 and 10–2). The plates were incubated for up to 15 days at 28ºC. Colonies were selected on days 2, 5, 10, and 15 of incubation and purified in 10% TSA. For each petri dish evaluated, the colonies were selected according to their time of growth and morphology (color, size, shape). After 15 days of incubation, all of the colonies were counted and expressed as CFU per gram of fresh tissue.
Identification and phylogenetic analysis of endophytic bacteria
DNA from each isolate was extracted using the following protocol: 1.5 ml of a 48-hour bacterial culture was centrifuged for 5 minutes at 14000 g and resuspended in 1 ml of TE buffer (mM Tris-HCl, 1 mM EDTA, pH 8.0), centrifuged, resuspended in 500 µl of TE buffer and finally adding 0.5 g of glass pearls (0.1 mm in diameter) (Sigma-Aldrich, USA) and 15 µl of 20% SDS. The cells were then homogenized for 30 s in a vortex mixer (AP56 – Phoenix), 500 µl of buffered phenol was added, and the solution was mixed and centrifuged for 5 min at 14000 g. The aqueous phase was extracted once with phenol-chloroform (1:1) and once more with chloroform. Following the extraction of the aqueous phase, 20 µl of 5M NaCl was added, the DNA was precipitated with isopropanol (5 min at room temperature) and collected by centrifugation for 10 min at 14000 g. The DNA pellet was washed with 70% ethanol, air dried and resuspended in 30 µl of autoclaved, ultrapure water.
The amplification of 16S rDNA was carried out in a reaction with a final volume of 25 µl containing 1 µl (0.5–10 ng) of total DNA, 2.5 µl (0.2 µM) of the P027F primer (5’-GAGAGTTTGATCCTGGCTAG-3’), 2.5 µl (0.2 µM) of the 1378R primer (5’-CGGTGTGTACSSGGCCCGGGAACG-3’), 1.6 µl (200 µM) of each dNTP, 2.5 µl of 5x IB buffer (Phoneutria; Belo Horizonte, Brazil); 1µl (1U) of Taq DNA polymerase (Phoneutria; Belo Horizonte, Brazil), and 2.5 µl (25 µg) of BSA (Promega). A negative control (PCR mix without DNA) was included in all PCR experiments. The PCR reaction conditions were as follows: 94ºC for 4 min, followed by 30 cycles of denaturation at 94ºC for 30 s, annealing at 63ºC for 1 min and extension at 72ºC for 1 min, before a final extension at 72ºC for 7 min. The PCR products were purified and sequenced by Macrogen Inc. (Seoul, South Korea) using an ABI3730 XL automatic DNA sequencer and the primers P027F and 1378R.
The identification of the isolates was performed using the Ribosomal Database Project (14, 61) and BLAST (http://blast.ncbi.nlm.nih.gov/blast/Blast.cgi) in NCBI. We used the Sequence Match application and BLAST to verify the similarity of experimental sequences with the reference sequences in the databases (14) and classified them at the genus level.
The DNA sequences of 34 reference strains (“type strain”), 2 strains obtained from the Ribosomal Database Project, and 34 representative strains from experimental isolates were aligned using the Ribosomal Database Project. Phylogenetic trees were constructed using the Neighbor-Joining (NJ) algorithm in MEGA version 4 (56), the Maximum Parsimony (MP) and Maximum Likelihood (ML) algorithm in Paup* (52), and the Bayesian Analysis (BA) algorithm in MrBayes 3.1 (23). The Neighbor Joining method was corrected by the Tamura-Nei multiple base substitution model (55) and by the GAMA distribution (0.4899) established by Modeltest 3.7. The parameters for Maximum Likelihood (GTR+I+G) were selected by AIC in Modeltest 3.7 (45). The Bayesian parameters (GTR+I+G) were selected by AIC in MrModeltest 2.3 (42). A total of 1000 replications were used for the bootstrap tests of the NJ and MP methods, while the ML test had 100 replications. The MB was performed in two independent runs with four Markov chain Monte Carlo (MCMC). A total of 10,000,000 generations were run, with trees being sampled every 1000 generations and the first 1,000,000 trees being discarded. Non-rooted trees were calculated using the 16S rDNA sequence of Methanocaldococcus jannaschii DSM 2661 as an outgroup. The 16S rDNA sequences of each isolate were deposited in the NCBI GENBANK database under the accession numbers HM355592 to HM355749.
Diversity indices
The diversity indices were calculated in the PAST program version 2.01(20), and the expected number of genotypes in the R program version 2.11.1 (47) using the Vegan library (43).
RESULTS
Endophytic bacteria isolation and identification
The density of endophytic populations recovered in 10% Endophytic bacteria from bean TSA medium varied from 4.5 x 102 to 2.8 x 103 CFU g-1 per fresh weight. A total of 158 (about 40 % of the total counted) isolates was obtained, of which 31.01% (49) were isolated from the Talismã cultivar, 37.34% (59) from the Ouro Negro cultivar and 31.65% (50) from the Vermelhinho cultivar (Table 1).
Table 1.
Endophytic isolates obtained from three Phaseolus vulgariscultivars.
| Identified taxum | Cultivar* | ||
|---|---|---|---|
| TAL | ONG | VER | |
| Actinobacteria | 25 | 13 | 9 |
| Actinobacteria (class) | 25 | 13 | 9 |
| Agromyces (A. mediolanus; Agromycessp.) | 0 | 2 | 3 |
| Dietzia (D. cinnamea) | 0 | 0 | 1 |
| Frigoribacterium(F. faeni) | 0 | 3 | 0 |
| Kocuria(K. palustris) | 1 | 0 | 0 |
| Microbacterium(M. foliorum; M. phyllosphaerae; Microbacterium sp.; M. testaceum) | 20 | 7 | 5 |
| Micrococcus(M luteus) | 4 | 0 | 0 |
| Rhodococcus(R. erythropolis) | 0 | 1 | 0 |
| Bacteroidetes | 0 | 0 | 1 |
| Sphingobacteria | 0 | 0 | 1 |
| Sphingobacterium(S. multivorum) Firmicutes | 0 | 0 | 1 |
| Firmicutes | 11 | 23 | 18 |
| Bacilli | 11 | 22 | 18 |
| Bacillus(B. amyloliquefaciens; B. bataviensis; B. muralis; B. niacini, Bacillus sp.; B. subtilis; B. thuringiensis) | 7 | 3 | 5 |
| Brevibacillus(B. agri) | 0 | 1 | 1 |
| Lysinibacillus (Lysinibacillus sphaericus) | 0 | 0 | 1 |
| Paenibacillus(P. cineris; P. lautus; Paenibacillus sp.) | 1 | 2 | 1 |
| Sporosarcina(S. aquimarina; Sporosarcina sp.) | 0 | 1 | 1 |
| Staphylococcus(S. caprae; S. epidermidis; S. kloosii; S. saprophyticus; Staphylococcus sp.; S. warneri;) | 3 | 16 | 9 |
| Proteobacteria | 13 | 23 | 22 |
| Alphaproteobacteria | 5 | 14 | 7 |
| Brevundimonas(B. vesicularis) | 1 | 0 | 0 |
| Methylobacterium(M. populi) | 3 | 8 | 7 |
| Rhizobium(R. larrymoorei) | 1 | 4 | 0 |
| Sphingomonas(S. dokdonensis; S. sanguinis) | 0 | 2 | 0 |
| Betaproteobacteria | 1 | 2 | 1 |
| Delftia(D. tsuruhatensis) | 1 | 2 | 1 |
| Gammaproteobacteria | 7 | 7 | 14 |
| Acinetobacter(A. radioresistens; Acinetobacter sp.) | 0 | 0 | 2 |
| Enterobacter(E. asburiae; E. hormaechei) | 4 | 0 | 0 |
| Stenotrophomonas(S. maltophilia; Stenotrophomonas sp.) | 2 | 7 | 10 |
| Pseudomonas(P. aeruginosa) | 1 | 0 | 2 |
| Total | 49 | 59 | 50 |
TAL = Talismã; ONG = Ouro Negro; VER = Vermelhinho.
Identification and phylogenetic analyses of endophytic bacteria
Sequencing of 16S rDNA was performed in all 158 isolates. Based on the nucleotide sequences each of the isolates was assigned to 23 different genera (Table 1). In terms of phylum, most isolates belonged to Proteobacteria (36.7% of the total number of isolates), followed by Firmicutes (32.9%) and lastly Actinobacteria (29.7%). Isolates from phylum Bacteroidetes comprised only 0.6% of the total and only a single isolate was found from the Sphingobacteria (Sphingobacteriaceae) of the genus Sphingobacterium. The highest number of isolates belonged to the Bacilli class (32.9%), comprised of bacteria from the families Staphylococcaceae (17.7%), Bacillaceae (10.1%), Paenibacillaceae (3.8%) and Planococcaceae (1.3%). The second most prevalent class in isolates was Actinobacteria (29.7%), which includes Microbacteriaceae (24.7%), Micrococcaceae (3.1%), Nocardiaceae (0.6%) and Dietziaceae (0.6%). Among the isolates identified as Proteobacteria, the dominant class in the isolate collection was Gammaproteobacteria (17.71%), with isolates belonging to the families Xanthomonadaceae (12.0%), Enterobacteriaceae (2.5%), Pseudomonadaceae (1.9%) and two (1.3%) isolates from the family Moraxellaceae. Isolates from the Alphaproteobacteria (16.5%) comprised representatives from the families Methylobacteriaceae (11.4%), Rhizobiaceae (3.2%), Sphingomonadaceae (1.3%) and one isolate from the family Caulobacteraceae. Betaproteobacteria (2.5%) contains only members from the family Comamonadaceae (2.5%).
The relative composition of the bacterial isolates by cultivars is shown in Figure 1 according to class. Differences in the proportions of the classes Actinobacteria, Alphaproteobacteria and Bacilli were observed between the three P. vulgaris cultivars. Sphingobacteria were isolated only from the Vermelhinho cultivar, which also exhibited differences in the proportion of isolates belonging to Gammaproteobacteria compared to isolates from the other two cultivars. The proportion of Betaproteobacteria was the same in all cultivars and all isolates of this class belonged to the genus Delftia.
Figure 1.
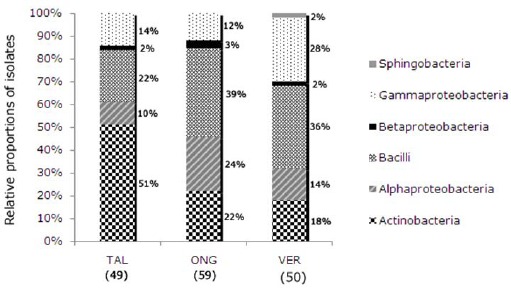
Bacterial class distribution of the culturable endophytic isolates obtained from three Phaseolus vulgaris cultivars: Talismã cultivar (TAL); Ouro Negro cultivar (ONG); Vermelhinho cultivar (VER)
Partial 16S rDNA gene sequences (approximately 1200 bp) from the isolates were used together with sequences taken from the Ribosomal Database Project for construction of phylogenetic trees using four different methods (Neighbor-Joining, Maximum Parsimony, Maximum Likelihood and Bayesian). The tree obtained by the Bayesian method is shown in Figure 2.
Figure 2.
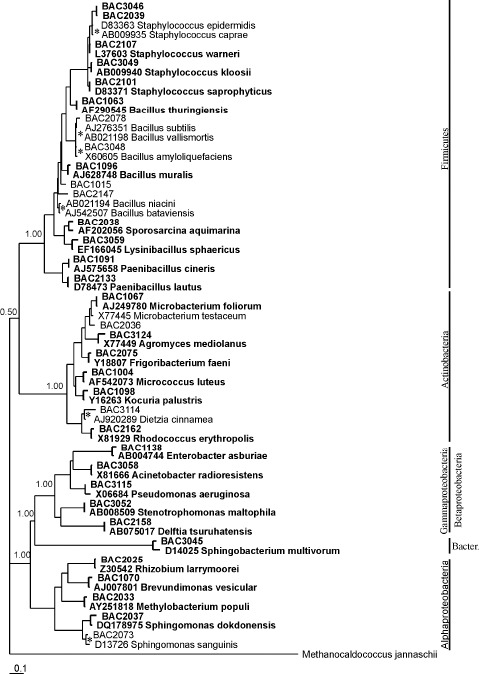
Phylogenetic tree showing the relationship between the 16S rDNA gene sequences from representative isolates of endophytic bacteria from three P. vulgaris cultivars. Terminal nodes in bold have bootstrap values greater than or equal to 94 in the three methods used (NJ, MP, ML) and presented a posteriori probabilities greater than or equal to 0.99. Terminal nodes with a posteriori probabilities equal to 1.00 and with bootstrap values under 90 in any of the other methods are marked with an *. Bacter. = Bacteroidetes.
Two major clades were formed with a posterior probability of 0.50: the first is comprised of Gram positive bacteria and the second of Gram-negative bacteria. The terminal nodes containing isolates BAC2078 and BAC3048 had bootstrap values below 90 as determined by the ML method; the terminal node that contained the isolate BAC3114 BAC2073 had bootstrap values below 90 for the methods MP, had bootstrap values below 90 for both the ML and NJ ML and NJ. The phylum Bacteroidetes aligned with bacteria methods. The terminal node that contained the isolate from the phylum Proteobacteria.
Diversity indices
The diversity index that was calculated in the PAST program and the expected number of genotypes for each cultivar estimated in the R program can be visualized in Table 2.
Table 2.
Number of taxa, individuals, diversity index and expected number of genotypes for each cultivar.
| Diversity indices/Parameters | Formula‡ | Cultivar | |||
|---|---|---|---|---|---|
| TAL | ONG | VER | |||
| Taxa (S) | - | 13 | 14 | 15 | |
| Individuals (n) | - | 49 | 59 | 50 | |
| Dominance (D) | D = Sum(ni/n)2 | 0.212 | 0.135 | 0.122 | |
| Shannon (H) | H = Sum((ni/n)ln(ni/n)) | 2.002 | 2.282 | 2.340 | |
| Simpson (1-D) | 1 - D = 1 - Sum(ni/n)2 | 0.788 | 0.865 | 0.878 | |
| Evenness (E) | E = eH/S | 0.570 | 0.700 | 0.692 | |
| Menhinick (db) | 1.857 | 1.823 | 2.121 | ||
| Margalef (Ma) | Ma = (S-l)ln(n) | 3.083 | 3.188 | 3.579 | |
| Equitability (J) | J = H/Hmax | 0.781 | 0.865 | 0.864 | |
| Fisher alpha (FA) | S = a*ln(l+n/a) | 5.781 | 5.801 | 7.265 | |
| Berger-Parker (d) | d = n/nT | 0.408 | 0.271 | 0.200 | |
| Expected number of genotypes† | - | 13 | 13.38 | 14.86 | |
Calculated in R program version 2.11.1
n = number of individuals; ni = number of individuals of taxon i; S = number of taxa; Nt = number of individuals in the dominant taxon; Hmax = log S.
Fisher’s alpha.
DISCUSSION
Isolation and identification of endophytic bacteria was performed from the leaves of three common bean (P. vulgaris) cultivars grown under the same field conditions in Minas Gerais during the winter season. The population densities of culturable bacteria in this study were similar to the population density of isolates obtained from soybean leaves growing in herbicide-free soil by Kuklinsky-Sobral et al. (30).
All identified isolates corresponded to genera commonly isolated from either the rhizosphere or bacteria associated with plants. Species from the genera Agromyces, Bacillus, Brevibacillus, Delftia, Dietzia, Enterobacter, Methylobacterium, Microbacterium, Micrococcus, Paenibacillus, Pseudomonas, Rhizobium, Rhodococcus, Sphingobacterium and Stenotrophomonas have already been isolated from rhizospheric soil and as endophytic bacteria in many previous studies (4, 5, 8, 18, 19, 24, 27–30, 36, 48–51, 54, 57–60). Additionally, species from the genera Acinetobacter, Brevundimonas, Frigoribacterium, Kocuria, Sphingomonas, Sporosarcina and Staphylococcus have been isolated or reported in studies of culturable and non-culturable endophytic bacteria (5, 8, 27, 29, 30, 48, 50, 51, 58).
Many of the bacterial genera encountered in this work were previously reported by Lopez-Lopez et al., (34), and many species of genera Bacillus were found by Walker et al. (63) in bean seeds. However, some of the species are not the same. The presence of certain genera in different bean cultivars suggest that they are better adapted to live as endophytic bacteria in P. vulgaris than other genera. The genera isolated in this work that have not been previously reported for P. vulgaris are as follows: Agromyces, Brevibacillus, Brevundinomonas, Delftia, Dietzia, Frigoribacterium, Lysinibacillus, Sphingobacterium, Sporosarcina and Stenotrophomonas.
Differences in the composition of the endophytic population according to cultivar or clone of plant have been documented for citrus plants, poplar trees, potato, salix and soybean (2, 11, 29, 41, 44, 58). The results of this study suggest that the cultivar of the plant contributes to the structure of the endophytic community associated with common bean plants or that the observed differences between common bean cultivars could be due to the use of only one sample collected in the Winter of 2007. First, some specific genera were only isolated from a single studied cultivar (Table 1). Second, isolate analyses also indicated that the cultivar of the plant may contribute to the determination of associated bacteria. Some of the genera had been isolated with greater frequency from a particular cultivar, for example, the genus Microbacterium from TAL, the genus Staphylococcus from ONG and Stenotrophomonas from VER. The differences between the number and type of isolates in each cultivar may suggest distinct endophytic communities in each cultivar. The differences in diversity of the endophytic communities of the cultivars may also be observed by the comparison of the relative class percentages presented in Figure 1.
To better visualize the community structure of the three common bean cultivars studied, diversity indices (Table 2) were calculated. The diversity indices obtained show that the diversity of bacterial isolates from cultivar Talismã was lower than the diversity of isolates obtained from the other two cultivars while the diversity of bacterial isolates from the cultivar Vermelhinho was the highest. Moreover, the indices Dominance_D and Berger-Parker clearly show that a single taxa of the cultivar Talismã is more abundant in the community, and the number of isolates shown in Table 1 reveal that this is the genus Microbacterium.
Bacteria usually associated with common bean leaf diseases belong to the genera Curtobacterium (22), Pseudomonas (33) and Xanthomonas (62). None of the isolates belong to Curtobacterium or Xanthomonas, while all the isolates belonging to Pseudomonas aligned with different strains of Pseudomonas aeruginosa with scores of 0.999.
The levels of NPK and ammonium sulfate applied to the plants were in accordance with the recommendations for producers in Brazil. However, this high level of nitrogen probably inhibited the nodulation of the bean roots and the association with other nitrogen-fixing bacteria. A few Rhizobium, Pseudomonas, Methylobacterium and Enterobacter species have already been described in the literature as nitrogen-fixing and nodule-forming organisms in the roots of many Leguminosae (7, 26, 37, 53). The five Rhizobium isolates aligned with sequences of Rhizobium larrymoorei, which was originally isolated from tumors affecting aerial parts of Ficus benjamina (9). Some bacterial species considered pathogenic for certain plant species have been isolated as endophytic in other species; from the polar tree, Ulrich et al. (58) isolated endophytes with high similarity to known plant pathogens, such as Clavibacter michiganensis, Pseudomonas syringae and Xanthomonas populi. Maes et. al. (35) also showed that Brenneria salicis could be isolated as an endophyte from poplar (Populus) and alder (Alnus). It is unclear whether these endophytic bacterial species confer some benefit to the host plant or if they merely use the host as a survival strategy in the environment to reach plants on which they can develop disease. The study of endophytic microorganisms is important to comprehend their interaction with their host plants. Additionally, endophytic microorganisms may have biotechnological applications. The potential of the isolated endophytic bacteria to promote bean plant growth and their biocontrol potential in diseases that affect the aerial parts of this important legume for the human diet will be addressed in future studies.
Table S1.
Identity of the 16S rDNA gene sequences of the isolates with the sequences deposited in the database.
| Isolate | Ribossomal Database Project | NCBI | ||
|---|---|---|---|---|
| Similarity score | Sequence name | Sequence name | % identity | |
| BAC1001 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain ODPY 16S; HM770098.1 | 100 |
| BAC1002 | 1.00 | Micrococcus luteus(T); ATCC 4698; AF542073 | Micrococcus luteus strain EHFS1_S04Ha 16S; EU071593.1 | 100 |
| BAC1003 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; AP012052.1 | 99 |
| BAC1004 | 1.00 | Micrococcus luteus(T); ATCC 4698; AF542073 | Micrococcus luteus strain SV21 16S; GU143803.1 | 99 |
| BAC1005 | 1.00 | Micrococcus luteus(T); ATCC 4698; AF542073 | Micrococcus sp. 185 16S ribosomal RNA gene; EU714334.1 | 99 |
| BAC1006 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; AP012052.1 | 99 |
| BAC1007 | 1.00 | Enterobacter asburiae(T); JCM6051; AB004744 | Enterobacter asburiae strain E53; HQ407230.1 | 99 |
| BAC1008 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain ESS21; EF602568.1 | 99 |
| BAC1009 | 0.98 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain PCSB7 16S; HM449703.1 | 98 |
| BAC1O1O | 1.00 | Enterobacter hormaechei(T); CIP 103441; AJ508302 | Enterobacter hormaechei strain Ni-1 16S; HM446004.1 | 99 |
| BAC1011 | 1.00 | Rhizobium larrymoorei(T); 3-10; Z30542 | Agrobacterium larrymoorei strain 13638E 16S; EU741094.1 | 100 |
| BAC1012 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC1013 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; APO 12052.1 | 99 |
| BAC1014 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC1015 | 0.98 | Bacillus niacini(T); IFO15566; AB021194 | Bacillus sp. DL006 16S; GQ355276.1 | 98 |
| BAC1016 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain LCR40 16S; FJ976549.1 | 100 |
| BAC1017 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain ODPY 16S; HM770098.1 | 100 |
| BAC1018 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; APO 12052.1 | 99 |
| BAC1019 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain ESS21 16S; EF602568.1 | 99 |
| BAC1020 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; APO 12052.1 | 99 |
| BAC2021 | 1.00 | Rhizobium larrymoorei(T); 3-10; Z30542 | Agrobacterium larrymoorei strain 13638E 16S; EU741094.1 | 99 |
| BAC2022 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; APO 12052.1 | 99 |
| BAC2023 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 100 |
| BAC2024 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC2025 | 1.00 | Rhizobium larrymoorei(T); 3-10; Z30542 | Agrobacterium larrymoorei strain 13638E 16S; EU741094.1 | 100 |
| BAC2026 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain F71028 16S; HQ908659.1 | 100 |
| BAC2027 | 1.00 | Brevibacillus agri(T); NRRL NRS-1219; D78454 | Brevibacillus agri partial 16S strain R-20121; AJ586388.1 | 99 |
| BAC2028 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium populi BJ001 16S; CP001029.1 | 99 |
| BAC2029 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; AP012052.1 | 99 |
| BAC2030 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16 S; HQ694734.1 | 99 |
| BAC2031 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 99 |
| BAC2032 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16 S; HQ694734.1 | 99 |
| BAC2033 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium populi BJ001; CP001029.1 | 99 |
| BAC2034 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis gene for 16S; AB617573.1 | 100 |
| BAC2035 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain NM62-4 16S; HM218280.1 | 100 |
| BAC2036 | 0.98 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium hominis strain 1P10AE; EU977655.1 | 99 |
| BAC2037 | 0.98 | Sphingomonas dokdonensis(T); DS-4; DQ178975 | Sphingomonas dokdonensis strain 2P01AE; EU977661.1 | 99 |
| BAC2038 | 0.98 | Sporosarcina aquimarina(T); SW28(T); AF202056 | Sporosarcina luteola gene for 16S; AB473560.1 | 99 |
| BAC2039 | 1.00 | Staphylococcus caprae(T); ATCC 35538T; AB009935 | Staphylococcus capitis strain EHFS2_AU1Hc 16S; EU071603.1 | 100 |
| BAC2040 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens AM1; CP001510.1 | 99 |
| BAC3041 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain NM62-4 16S; HM218280.1 | 100 |
| BAC3042 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis serovar colmeri 16S; EU429660.1 | 100 |
| BAC3043 | 1.00 | Microbacterium foliorum(T); DSM 12966; P 333/02; AJ249780 | Microbacterium foliorum strain 720 16S; EU714376.1 | 99 |
| BAC3044 | 0.97 | Bacillus bataviensis(T); type strain: LMG 21832; AJ542507 | Bacillus sp. R-30632 partial 16S; AM910246.1 | 99 |
| BAC3045 | 0.98 | Sphingobacterium multivorum(T); IFO 14947; D14025 | Sphingobacterium sp. G-2-27-2 16S; EF102865.1 | 99 |
| BAC3046 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain F71028 16S; HQ908659.1 | 100 |
| BAC3047 | 1.00 | Microbacterium testaceum (T); DSM 20166; X77445 | Microbacterium sp. Fek04 16S; EU741023.1 | 99 |
| BAC3048 | 1.00 | Bacillus amyloliquefaciens (T); CR-502; AY603658 | Bacillus amyloliquefaciens LL3; CP002634.1 | 100 |
| BAC3049 | 1.00 | Staphylococcus kloosii (T); ATCC 43959T; AB009940 | Staphylococcus kloosii strain FR2_36con 16S; EU934080.1 | 100 |
| BAC3050 | 0.99 | Microbacterium phyllosphaerae (T); DSM 13468; P 369/06; AJ277840 | Microbacterium foliorum strain 720 16S; EU714376.1 | 99 |
| BAC3051 | 0.99 | Methylobacterium populi (T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 99 |
| BAC3052 | 1.00 | Stenotrophomonas maltophilia (T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC3053 | 0.97 | Bacillus bataviensis(T); type strain: LMG 21832; AJ542507 | Bacillus sp. R-30632 partial 16S; AM910246.1 | 98 |
| BAC3054 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 100 |
| BAC3055 | 0.99 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 98 |
| BAC3056 | 0.92 | Sporosarcina koreensis F73; DQ073393 | Sporosarcina ginsengisoli strain CR5 16S; HQ331532.1 | 90 |
| BAC3057 | 0.82 | Acinetobacter radioresistensINBS1; AM495259 | Acinetobacter radioresistens strain TY37SsD 16S; HQ406757.1 | 81 |
| BAC3058 | 1.00 | Acinetobacter radioresistens(T); DSM 6976; X81666 | Acinetobacter radioresistens strain S13 16S; GU145275.1 | 99 |
| BAC3059 | 0.98 | Lysinibacillus sphaericus;KNUC228; EF166045 | Lysinibacillus sphaericus strain IMAU80223 16S; GU125639.1 | 97 |
| BAC1061 | 0.92 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium sp. CSBd gene for 16S; AB552874.1 | 91 |
| BAC1062 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain NM62-4 16S; HM218280.1 | 100 |
| BAC1063 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain ODPY 16S; HM770098.1 | 100 |
| BAC1064 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens DM4 str. DM4; FP103042.2 | 100 |
| BAC1065 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain DSM 20166 16S; NR_026163.1 | 99 |
| BAC1066 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum StLB037; AP012052.1 | 99 |
| BAC1067 | 1.00 | Microbacterium foliorum(T); DSM 12966; P 333/02; AJ249780 | Microbacterium foliorum strain 720 16S; EU714376.1 | 99 |
| BAC1068 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens AM1; CP001510.1 | 99 |
| BAC1069 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens AM1; CP001510.1 | 100 |
| BAC1070 | 0.99 | Brevundimonas vesicularis(T); ATCC 11426 (T); AJ007801 | Brevundimonas vesicularis DNA for 16S strain LMG 11141; AJ227781.1 | 99 |
| BAC2071 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain LCR40 16S; FJ976549.1 | 100 |
| BAC2072 | 1.00 | Frigoribacterium faeni(T); 801; Y18807 | Frigoribacterium sp. PDD-24b-20 16S; HQ256793.1 | 99 |
| BAC2073 | 0.99 | Sphingomonas sanguinis(T); IFO 13937; D13726 | Sphingomonas pseudosanguinis partial 16S; AM412238.1 | 99 |
| BAC2074 | 0.98 | Staphylococcus warneri(T); L37603 | Staphylococcus pasteuri partial 16S strain PSM NO.15; FR846535.1 | 98 |
| BAC2075 | 0.99 | Frigoribacterium faeni(T); 801; Y18807 | Frigoribacterium sp. 301 16S; AF157479.1 | 99 |
| BAC2076 | 1.00 | Rhizobium larrymoorei(T); 3-10; Z30542 | Agrobacterium larrymoorei strain 2R46 16S; EF178437.1 | 100 |
| BAC2077 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain F71028 16S; HQ908659.1 | 100 |
| BAC2078 | 1.00 | Bacillus subtilissubsp. subtilis (T); DSM10; AJ276351 | Bacillus subtilis strain M-15 16S; HQ401271.1 | 100 |
| BAC2079 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium sp. CSBd gene for 16S; AB552874.1 | 100 |
| BAC2080 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium sp. Fek04 16S; EU741023.1 | 99 |
| BAC3081 | 1.00 | Brevibacillus agri(T); NRRL NRS-1219; D78454 | Brevibacillus agri strain PLIV 16S; HQ166189.1 | 100 |
| BAC3082 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 99 |
| BAC3083 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 99 |
| BAC3084 | 1.00 | Paenibacillus cineris(T); type strain:LMG 18439; AJ575658 | Paenibacillus sp. 3492BRRJ 16S; JF309261.1 | 100 |
| BAC3085 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis strain NM62-4 16S; HM218280.1 | 100 |
| BAC3087 | 1.00 | Microbacterium foliorum(T); DSM 12966; P 333/02; AJ249780 | Microbacterium foliorum strain 720 16S; EU714376.1 | 99 |
| BAC3088 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. DC2c-19 gene for 16S; AB552870.1 | 99 |
| BAC3089 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 99 |
| BAC3090 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium sp. SuP10 16S; EU912450.1 | 99 |
| BAC1091 | 1.00 | Paenibacillus cineris(T); type strain:LMG 18439; AJ575658 | Paenibacillus cineris partial 16S; AJ575658.1 | 99 |
| BAC1092 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain 4CAJ3 16S; GQ383916.1 | 99 |
| BAC1093 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium sp. CSBd gene for 16S; AB552874.1 | 99 |
| BAC1094 | 0.99 | Microbacterium phyllosphaerae(T); DSM 13468; P 369/06; AJ277840 | Microbacterium sp. CSBd gene for 16S | 99 |
| BAC1095 | 1.00 | Micrococcus luteus(T); ATCC 4698; AF542073 | Micrococcus luteus strain EHFS1_S04Ha 16S; EU071593.1 | 100 |
| BAC1096 | 1.00 | Bacillus muralis(T); type strain: LMG 20238; AJ628748 | Bacillus muralis strain REG126 16S; GQ844961.1 | 100 |
| BAC1097 | 0.93 | Microbacteriumsp. S15-M4; AM234160 | Microbacterium sp. HY14(2010) 16S; HM579805.1 | 92 |
| BAC1098 | 1.00 | Kocuria palustris(T); TAGA27 (DSM 11925, type strain); Y16263 | Kocuria palustris strain cT220 16S; JF303036.1 | 99 |
| BAC1099 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 100 |
| BAC1100 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Staphylococcus saprophyticus strain OTUC3 16S; FJ210844.1 | 100 |
| BAC2101 | 1.00 | Staphylococcus saprophyticussubsp. saprophyticus (T); ATCC 15305 (= MAFF 911473); D83371 | Staphylococcus saprophyticus strain OTUC3 16S; FJ210844.1 | 100 |
| BAC2102 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 100 |
| BAC2103 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens gene for 16S rRNA; AB298401.1 | 99 |
| BAC2104 | 1.00 | Delftia tsuruhatensis(T); T7; AB075017 | Delftia tsuruhatensis strain IPPBC R15 16S; HQ436355.1 | 100 |
| BAC2105 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens gene for 16S; AB298401.1 | 99 |
| BAC2106 | 0.86 | Bacillus cereusme-5; EU652058 | Bacillus cereus partial 16S; FR749846.1 | 85 |
| BAC2107 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 99 |
| BAC2108 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium extorquens gene for 16S rRNA; AB298401.1 | 99 |
| BAC2109 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 99 |
| BAC2110 | 1.00 | Frigoribacterium faeni(T); 801; Y18807 | Frigoribacterium faeni partial 16S; AM410686.1 | 99 |
| BAC3111 | 0.99 | Pseudomonas aeruginosa(T); DSM50071; X06684 | Pseudomonas aeruginosa strain CRC5 16S; HQ995502.1 | 100 |
| BAC3112 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 100 |
| BAC3113 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium chloromethanicum gene for 16S; AB175630.1 | 99 |
| BAC3114 | 0.97 | Dietzia cinnamea(T); type strain:IMMIB RIV-399; AJ920289 | Dietzia timorensis gene for 16S; AB377289.1 | 100 |
| BAC3115 | 0.99 | Pseudomonas aeruginosa(T); DSM50071; X06684 | Pseudomonas aeruginosa strain MTH8 16S; HQ202541.1 | 100 |
| BAC3116 | 0.95 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 94 |
| BAC3117 | 1.00 | Agromyces mediolanus(T); DSM 20152; X77449 | Agromyces mediolanus gene for 16S; D45054.1 | 99 |
| BAC3118 | 1.00 | Agromyces mediolanus(T); DSM 20152; X77449 | Agromyces mediolanus strain c18 16S; FJ950540.1 | 100 |
| BAC3119 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA 3088 16S; GQ222399.1 | 99 |
| BAC3120 | 0.88 | Stenotrophomonas maltophilia;AY484506 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 87 |
| BAC3121 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC3122 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4; HM143858.1 | 100 |
| BAC3123 | 0.99 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 99 |
| BAC3124 | 0.98 | Agromyces mediolanus(T); DSM 20152; X77449 | Agromyces mediolanus strain c18 16S; FJ950540.1 | 97 |
| BAC3125 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain JKR32b 16S; HQ671069.1 | 100 |
| BAC2126 | 0.99 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium trichotecenolyticum strain 3370 16S; EU714362.1 | 99 |
| BAC2127 | 0.99 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 99 |
| BAC2128 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC2129 | 1.00 | Agromyces mediolanus(T); DSM 20152; X77449 | Agromyces mediolanus strain c18 16S; FJ950540.1 | 100 |
| BAC2130 | 0.92 | Agromyces mediolanusDSM 20152; X77449 | Agromyces mediolanus strain c70 16S; FJ950561.1 | 91 |
| BAC2131 | 1.00 | Staphylococcus epidermidis(T); ATCC 14990; D83363 | Staphylococcus epidermidis gene for 16S; AB617573.1 | 100 |
| BAC2132 | 0.99 | Methylobacterium populi(T); BJ001; ATCC BAA-705; NCIMB 13946; AY251818 | Methylobacterium populi strain TNAU10 16S; EF116588.1 | 98 |
| BAC2133 | 0.99 | Paenibacillus lautus(T); NRRL NRS-666T; D78473 | Paenibacillus lautus strain DS19 16S; EU834247.1 | 99 |
| BAC2134 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain JKR32b 16S; HQ671069.1 | 99 |
| BAC2135 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC1136 | 0.99 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 98 |
| BAC1137 | 0.99 | Pseudomonas aeruginosa(T); DSM50071; X06684 | Pseudomonas aeruginosa strain MTH8 16S; HQ202541.1 | 100 |
| BAC1138 | 1.00 | Enterobacter asburiae(T); JCM6051; AB004744 | Enterobacter hormaechei strain Ni-1 16S; HM446004.1 | 99 |
| BAC1139 | 1.00 | Enterobacter hormaechei(T); CIP 103441; AJ508302 | Enterobacter cancerogenus strain M119 16S; HQ407292.1 | 99 |
| BAC1140 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain ODPY 16S; HM770098.1 | 100 |
| BAC1141 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain ODPY 16S; HM770098.1 | 100 |
| BAC2142 | 1.00 | Rhizobium larrymoorei(T); 3-10; Z30542 | Agrobacterium larrymoorei strain 13638E 16S; EU741094.1 | 100 |
| BAC2143 | 0.95 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 95 |
| BAC2144 | 0.88 | Staphylococcus saprophyticusATCC 15305; AP008934 | Staphylococcus saprophyticus strain T86 16S; HQ407261.1 | 88 |
| BAC2145 | 0.81 | Paenibacillus lautusJCM 9073; AB073188 | Paenibacillus lactis strain ZYb1 16S; FJ445392.1 | 80 |
| BAC2147 | 0.98 | Bacillus bataviensis(T); type strain: LMG 21832; AJ542507 | Bacillus circulans strain RIGLD BC1 16S; HQ315829.1 | 98 |
| BAC3148 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas sp. 2A9S2 16S; HQ246220.1 | 100 |
| BAC3149 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas sp. 2A9N6 16S; HQ246302.1 | 100 |
| BAC3150 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain AhsB4 16S; HM143858.1 | 100 |
| BAC3151 | 1.00 | Bacillus thuringiensis(T); ATCC10792; AF290545 | Bacillus thuringiensis strain NBB6 16S; HQ256544.1 | 100 |
| BAC1152 | 1.00 | Staphylococcus warneri(T); L37603 | Staphylococcus warneri strain FUA2075 16S; HQ694734.1 | 100 |
| BAC2153 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain BAC2153 16S; HM355741.1 | 100 |
| BAC3154 | 1.00 | Microbacterium testaceum(T); DSM 20166; X77445 | Microbacterium testaceum strain BAC3154 16S; HM355742.1 | 100 |
| BAC3155 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas sp. 2A9N6 16S; HQ246302.1 | 100 |
| BAC1156 | 0.92 | Delftia tsuruhatensis(T); T7; AB075017 | Delftia tsuruhatensis strain BN-HKY6 16S; HQ731453.1 | 92 |
| BAC1157 | 1.00 | Microbacterium foliorum(T); DSM 12966; P 333/02; AJ249780 | Microbacterium foliorum strain DS42 16S; EU834263.1 | 99 |
| BAC2158 | 1.00 | Delftia tsuruhatensis(T); T7; AB075017 | Delftia tsuruhatensis strain BN-HKY6 16S; HQ731453.1 | 100 |
| BAC3159 | 1.00 | Delftia tsuruhatensis(T); T7; AB075017 | Delftia tsuruhatensis strain BN-HKY6 16S; HQ731453.1 | 100 |
| BAC2160 | 1.00 | Stenotrophomonas maltophilia(T); ATCC 13637T; AB008509 | Stenotrophomonas maltophilia strain BAC3148 16S; HM355736.1 | 100 |
| BAC2162 | 1.00 | Rhodococcus erythropolis(T); ATCC 4277T; X81929 | Rhodococcus erythropolis strain BAC2162 16S; HM355749.1 | 100 |
ACKNOWLEDGEMENTS
We would like to thank the Brazilian institutions CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico) and FAPEMIG (Fundação de Amparo à Pesquisa do Estado de Minas Gerais) for financial support.
REFERENCES
- 1.Araújo W. L., Lima A.O.S., Azevedo J.L., Marcon J., Kuklinsky-Sobral J., Lacava P.T. Piracicaba, SP: Departamento de Genética Escola Superior de Agricultura “Luiz de Queiroz”—Universidade de São Paulo; 2002. Manual: Isolamento de microrganismos endofíticos. [Google Scholar]
- 2.Araujo W.L., Marcon J., Maccheroni W., Jr., van Elsas J.D., van Vuurde J.W.L., Azevedo J.L. Diversity of endophytic bacterial populations and their interaction with Xylella fastidiosain citrus plants. Appl. Environ. Microbiol. 2002;68(10):4906–4914. doi: 10.1128/AEM.68.10.4906-4914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold A.E., Maynard Z., Gilbert G.S., Coley P.D., Kursar T.A. Are tropical fungal endophytes hyperdiverse? Ecol. Lett. 2000;3(4):267–274. [Google Scholar]
- 4.Azanza M., Azanza R., Vargas V., Hedreyda C. Bacterial endosymbionts of Pyrodinium bahamense var. compressum. Microb. Ecol. 2006;52(4):756–764. doi: 10.1007/s00248-006-9128-7. [DOI] [PubMed] [Google Scholar]
- 5.Barzanti R., Ozino F., Bazzicalupo M., Gabbrielli R., Galardi F., Gonnelli C., Mengoni A. Isolation and characterization of endophytic bacteria from the nickel hyperaccumulator plant Alyssum bertolonii. Microb. Ecol. 2007;53(2):306–316. doi: 10.1007/s00248-006-9164-3. [DOI] [PubMed] [Google Scholar]
- 6.Bell C.R., Dickie G.A., Harvey W.L.G., Chan J. Endophytic bacteria in grapevine. Can. J. Microbiol. 1995;41(1):46–53. [Google Scholar]
- 7.Benhizia Y., Benhizia H., Benguedouar A., Muresu R., Giacomini A., Squartini A. gamma proteobacteria can nodulate legumes of the genus Hedysarum. Syst. Appl. Microbiol. 2004;27(4):462–468. doi: 10.1078/0723202041438527. [DOI] [PubMed] [Google Scholar]
- 8.Berg G., Krechel A., Ditz M., Sikora R.A., Ulrich A., Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 2005;51(2):215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Bouzar H., Jones J. Agrobacterium larrymoorei sp. nov., a pathogen isolated from aerial tumours of Ficus benjamina. Int. J. Syst. Evol. Microbiol. 2001;51(3):1023–1026. doi: 10.1099/00207713-51-3-1023. [DOI] [PubMed] [Google Scholar]
- 10.Broughton W.J., Hernández G., Blair M., Beebe S., Gepts P., Vanderleyden J. Beans (Phaseolus spp.)—model food legumes. Plant Soil. 2003;252(1):55–128. [Google Scholar]
- 11.Cambours M.A., Nejad P., Granhall U., Ramstedt M. Frost-related dieback of willows. Comparison of epiphytically and endophytically isolated bacteria from different Salix clones, with emphasis on ice nucleation activity, pathogenic properties and seasonal variation. Biomass Bioenergy. 2005;28(1):15–27. [Google Scholar]
- 12.Chi F., Shen S.-H., Cheng H.P., Jing Y.X., Yanni Y.G., Dazzo F.B. Ascending migration of endophytic rhizobia, from roots to leaves, inside Rice plants and assessment of benefits to rice growth physiology. Appl. Environ. Microbiol. 2005;71(11):7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocking E.C. Endophytic colonization of plant roots by nitrogen-fixing bacteria. Plant Soil. 2003;252(1):169–175. [Google Scholar]
- 14.Cole J.R., Wang Q., Cardenas E., Fish J., Chai B., Farris R.J., Kulam-Syed-Mohideen A.S., McGarrell D.M., Marsh T., Garrity G.M., Tiedje J.M. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(S1):D141–145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Compant S., Duffy B., Nowak J., Clement C., Barka E.A. Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects. Appl. Environ. Microbiol. 2005;71(9):4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromin N., Achouak W., Thiéry J.M., Heulin T. The genotypic diversity of Pseudomonas brassicacearumpopulations isolated from roots of Arabidopsis thaliana: influence of plant genotype. FEMS Microbiol. Ecol. 2001;37(1):21–29. [Google Scholar]
- 17.Hallmann J., QuadtHallmann A., Mahaffee W.F., Kloepper J.W. Bacterial endophytes in agricultural crops. Can. J. Microbiol. 1997;43(10):895–914. [Google Scholar]
- 18.Hallmann J., Rodríguez-Kábana R., Kloepper J.W. Chitinmediated changes in bacterial communities of the soil, rhizosphere and within roots of cotton in relation to nematode control. Soil Biol. Biochem. 1999;31(4):551–560. [Google Scholar]
- 19.Han J., Xia D., Li L., Sun L., Yang K., Zhang L. Diversity of culturable bacteria isolated from root domains of Moso Bamboo (Phyllostachys edulis) Microb Ecol. 2009;58:363–373. doi: 10.1007/s00248-009-9491-2. [DOI] [PubMed] [Google Scholar]
- 20.Hammer Ø., Harper D.A.T., Ryan P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electronica. 2001;4(1):1–9. http://palaeo-electronica.org/2001_1/past/issue1_01.htm. [Google Scholar]
- 21.Hardoim P.R., van Overbeek L.S., Elsas J.D.v. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 22.Hedges F. A bacterial wilt of the bean caused by Bacterium flaccumfaciens nov. sp. Science. 1922;55(1425):433–434. doi: 10.1126/science.55.1425.433. [DOI] [PubMed] [Google Scholar]
- 23.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 24.Idris R., Trifonova R., Puschenreiter M., Wenzel W., Sessitsch A. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl. and Environ. Microbiol. 2004;70(5):2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs M.J., Bugbee W.M., Gabrielson D.A. Enumeration, location, and characterization of endophytic bacteria within sugar-beet roots. Can. J. Bot. 1985;63(7):1262–1265. [Google Scholar]
- 26.Jourand P., Giraud E., Bena G., Sy A., Willems A., Gillis M., Dreyfus B., Lajudie P. Methylobacterium nodulans sp. nov., for a group of aerobic, facultatively methylotrophic, legume root-nodule-forming and nitrogen-fixing bacteria. Int. J. Syst. Evol. Microbiol. 2004;54(6):2269–2273. doi: 10.1099/ijs.0.02902-0. [DOI] [PubMed] [Google Scholar]
- 27.Kang S., Cho H., Cheong H., Ryu C., Kim J., Park S. Two bacterial entophytes eliciting both plant growth promotion and plant defense on pepper (Capsicum annuum L.) J. Microbiol. Biotechnol. 2007;17(1):96–103. [PubMed] [Google Scholar]
- 28.Kuffner M., Puschenreiter M., Wieshammer G., Gorfer M., Sessitsch A. Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil. 2008;304(1):35–44. [Google Scholar]
- 29.Kuklinsky-Sobral J., Araújo W.L., Mendes R., Geraldi I.O., Pizzirani-Kleiner A.A., Azevedo J.L. Isolation and characterization of soybean-associated bacteria and their potential for plant growth promotion. Environ. Microbiol. 2004;6(11):1244–1251. doi: 10.1111/j.1462-2920.2004.00658.x. [DOI] [PubMed] [Google Scholar]
- 30.Kuklinsky-Sobral J., Araújo W.L., Mendes R., Pizzirani-Kleiner A.A., Azevedo J.L. Isolation and characterization of endophytic bacteria from soybean (Glycine max) grown in soil treated with glyphosate herbicide. Plant Soil. 2005;273(1):91–99. [Google Scholar]
- 31.Lamb T.G., Tonkyn D.W., Kluepfel D.A. Movement of Pseudomonas aureofaciensfrom the rhizosphere to aerial plant tissue. Can. J. Microbiol. 1996;42(11):1112–1120. [Google Scholar]
- 32.Lindow S.E., Brandl M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003;69(4):1875–1883. doi: 10.1128/AEM.69.4.1875-1883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindow S.E., Arny D.C., Upper C.D. Distribution of ice nucleation-active bacteria on plants in nature. Appl. Environ. Microbiol. 1978;36(6):831–838. doi: 10.1128/aem.36.6.831-838.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.López-López A., Rogel M.A., Ormeño-Orrillo E., Martínez-Romero J., Martínez-Romero E. Phaseolus vulgaris seed-borne endophytic community with novel bacterial species such as Rhizobium endophyticum sp. nov. Syst. Appl. Microbiol. 2010;33:322–327. doi: 10.1016/j.syapm.2010.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Maes M., Huvenne H., Messens E. Brenneria salicis, the bacterium causing watermark disease in willow, resides as an endophyte in wood. Environ Microbiol. 2009;11:1453–1462. doi: 10.1111/j.1462-2920.2009.01873.x. [DOI] [PubMed] [Google Scholar]
- 36.Mano H., Tanaka F., Nakamura C., Kaga H., Morisaki H. Culturable endophytic bacterial flora of the maturing leaves and roots of rice plants (Oryza sativa) cultivated in a paddy field. Microb Environ. 2007;22:175–185. [Google Scholar]
- 37.Martínez-Romero E. Diversity of Rhizobium-Phaseolus vulgarissymbiosis: overview and perspectives. Plant Soil. 2003;252(1):11–23. [Google Scholar]
- 38.Martínez-Romero E. Coevolution in Rhizobium-legume symbiosis? DNa Cell Biol. 2009;28(8):361–370. doi: 10.1089/dna.2009.0863. [DOI] [PubMed] [Google Scholar]
- 39.Misaghi I.J., Donndelinger C.R. Endophytic bacteria in symptom-free cotton plants. Phytopathology. 1990;80(9):808–811. [Google Scholar]
- 40.Monier J.-M., Lindow S.E. Frequency, size, and localization of bacterial aggregates on bean leaf surfaces. Appl. Environ. Microbiol. 2004;70(1):346–355. doi: 10.1128/AEM.70.1.346-355.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moore F.P., Barac T., Borremans B., Oeyen L., Vangronsveld J., van der Lelie D., Campbell CD., Moore E.R.B. Endophytic bacterial diversity in poplar trees growing on a BTEX-contaminated site: the characterization of isolates with potential to enhance phytoremediation. Syst. Appl. Microbiol. 2006;29(7):539–556. doi: 10.1016/j.syapm.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 42.Nylander J.A.A. Uppsala University: Evolutionary Biology Centre; 2004. MrModeltest 2.3. Program distributed by the author. [Google Scholar]
- 43.Oksanen J., Blanchet F.G., Kindt R., Legendre P., O’Hara R.B., Simpson G.L., Solymos P., Stevens M.H.H., Wagner H. vegan: Community Ecology Package. R package version 1. 2010. pp. 17–3. http://CRAN.R-project.org/package=vegan.
- 44.Overbeek L.v., Elsas J.D.v. Effects of plant genotype and growth stage on the structure of bacterial communities associated with potato (Solanum tuberosumL.) FEMS Microbiol. Ecol. 2008;64(2):283–296. doi: 10.1111/j.1574-6941.2008.00469.x. [DOI] [PubMed] [Google Scholar]
- 45.Posada D., Crandall K. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 46.QuadtHallmann A., Kloepper J.W. Immunological detection and localization of the cotton endophyte Enterobacter asburiaeJM22 in different plant species. Can. J. Microbiol. 1996;42(11):1144–1154. [Google Scholar]
- 47.47. R Development Core Team. Vienna, Austria: R Foundation for Statistical Computing; 2010. [Accesed 08 February 2011]. R: A language and environment for statistical computing. ISBN 3–900051–07–0, Available at http://www.R-project.org. [Google Scholar]
- 48.Rijavec T., Lapanje A., Dermastia M., Rupnik M. Isolation of bacterial endophytes from germinated maize kernels. Can. J. Microbiol. 2007;53(6):802–808. doi: 10.1139/W07-048. [DOI] [PubMed] [Google Scholar]
- 49.Rivas R., Trujillo M., Mateos P., Martinez-Molina E., Velaquez E. Agromyces ulmi sp nov., a xylanolytic bacterium isolated from Ulmus nigrain Spain. Int. J. Syst. Evol. Microbiol. 2004;54(6):1987–1990. doi: 10.1099/ijs.0.63058-0. [DOI] [PubMed] [Google Scholar]
- 50.Sun L., Qiu F., Zhang X., Dai X., Dong X., Song W. Endophytic bacterial diversity in rice (Oryza sativa L.) roots estimated by 16S rDNA Sequence Analysis. Microb. Ecol. 2008;55(3):415–424. doi: 10.1007/s00248-007-9287-1. [DOI] [PubMed] [Google Scholar]
- 51.Surette M., Sturz A., Lada R., Nowak J. Bacterial endophytes in processing carrots (Daucus carota L. var. sativus): their localization, population density, biodiversity and their effects on plant growth. Plant Soil. 2003;253(2):381–390. [Google Scholar]
- 52.Swofford D.L. Sunderland, Massachusetts: Sinauer Associates; 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods) [Google Scholar]
- 53.Sy A., Giraud E., Jourand P., Garcia N., Willems A., de Lajudie P., Prin Y., Neyra M., Gillis M., Boivin-Masson C., Dreyfus B. Methylotrophic Methylobacteriumbacteria nodulate and fix nitrogen in symbiosis with legumes. J. Bacteriol. 2001;183(1):214–220. doi: 10.1128/JB.183.1.214-220.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takeuchi M., Hatano K. Agromyces luteolus sp nov., Agromyces rhizospherae sp nov and Agromyces bracchium sp nov., from the mangrove rhizosphere Int. J. Syst. Evol. Microbiol. 2001;51(4):1529–1537. doi: 10.1099/00207713-51-4-1529. [DOI] [PubMed] [Google Scholar]
- 55.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10(3):512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 56.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular evolutionary genetics analysis (MEGA) Software Version 4.0. Mol. Biol. Evol. 2007;24(8):1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 57.Tian F., Ding Y., Zhu H., Yao L., Du B. Genetic diversity of siderophore-producing bacteria of tobacco rhizosphere. Braz. J. Microbiol. 2009;40(2):276–284. doi: 10.1590/S1517-838220090002000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ulrich K., Ulrich A., Ewald D. Diversity of endophytic bacterial communities in poplar grown under field conditions. FEMS Microbiol. Ecol. 2008;63(2):169–180. doi: 10.1111/j.1574-6941.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- 59.Van Aken B., Peres C.M., Doty S.L., Yoon J.M., Schnoor J.L. Methylobacterium populi sp. nov., a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoidesxnigra DN34) Int. J. Syst. Evol. Microbiol. 2004;54(4):1191–1196. doi: 10.1099/ijs.0.02796-0. [DOI] [PubMed] [Google Scholar]
- 60.Vivas A., Biró B., Ruíz-Lozano J.M., Barea J.M., Azcón R. Two bacterial strains isolated from a Zn-polluted soil enhance plant growth and mycorrhizal efficiency under Zn-toxicity. Chemosphere. 2006;62(9):1523–1533. doi: 10.1016/j.chemosphere.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 61.Wang Q., Garrity G.M., Tiedje J.M., Cole J.R. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. and Environ. Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Webster D.M., Atkin J.D., Cross J.E. Bacterial blights of snap beans and their control. Plant Disease. 1983;67(10):935–940. [Google Scholar]
- 63.Walker R., Powell A.A, Seddon B. Bacillusisolates from the spermosphere of peas and dwarf French beans with antifungal activity against Botrytis cinereaand Pythiumspecies. J. Appl. Microbiol. 1998;84:791–801. doi: 10.1046/j.1365-2672.1998.00411.x. [DOI] [PubMed] [Google Scholar]


