Abstract
The purpose of this study was to identify the genes coding for resistance to ceftazidime and imipenem and describe the molecular epidemiology of A. baumannii strains isolated from a clinical center in Colombia. Twenty isolates of imipenem-resistant A. baumannii from an equal number of patients with nosocomial infections were obtained. Primers were used to amplify genes blaIMP, blaVIM, blaOXA-23, blaOXA-24, blaOXA-58, blaOXA-51 and blaADC-7. To detect insertion sequences ISAba1/blaOXA-23, ISAba1/blaOXA-51 and ISAba1/blaADC-7, mapping by PCR using combinations of reverse primers ISAba1 and reverse primers of blaOXA-23, blaOXA-51 and blaADC-7 were used. The amplification products were purified and cloned into PCR 2.1-TOPO vector and transformed into chemically competent Escherichia coli TOP10. These amplicons were then sequenced. PFGE was performed on DNA of A. baumannii isolates digested with ApaI. Results. The DNA profiles obtained included 9 clusters with, four 2–7 isolates per profile, and 5 single-isolate profiles. Of the 20 isolates resistant to imipenem, 15 carried blaOXA-23 gene, 4 contained ISAba1 upstream of blaOXA-51 gene, and 6 contained ISAba1 upstream of blaOXA-23 gene. Eighteen of these isolates carried the blaADC-7 gene, with 9 of the isolates having ISAba1 located upstream of this gene. This is the first report of the ISAba1/ADC-7 associated with OXAs genes in A. baumannii isolates from Colombia.
Keywords: Nosocomial pathogens, Antimicrobial resistance, PFGE
INTRODUCTION
Acinetobacter baumannii can cause a variety of infections including pneumonia, bacteremia, meningitis, urinary tract infections, peritonitis and infections of skin and soft tissue (2). Mortality is high in association with bacteremia (52%) and pneumonia (23–73% (6, 8). Multidrug resistant A. baumannii have become an important nosocomial pathogen that particularly affects critically ill patients. The multiresistance is common in this species which complicates its elimination and therapy in severe infections, with extremely limited therapeutic alternatives currently available (10, 30).
A. baumannii resistant to carbapenems has been isolated in Europe, Asia, North and South America (15). Although carbapenemase production, modification of penicillin-binding proteins (PBPs), loss of porins, and/or altered efflux pump activity are reported as mechanisms contributing to resistance. It is the production of carbapenemases that are the main mechanism involved. These carbapenemases are mainly metalloenzymes (class B), found in several bacterial species of clinical relevance, including members of the family Enterobacteriaceae, Pseudomonas spp. and other non-fastidious Gram-negative nonfermenters. However, the vast majority of OXA carbapenemases (class D) have been discovered in Acinetobacter baumannii (18, 26).
In addition, the insertion sequence ISAba1 has been found in many isolates of A. baumannii located upstream of the blaOXA-23,-51,-58 carbapenemase genes and cephalosporinase blaampC genes. These ISAba1 elements facilitate increased expression of these genes (4, 12). ISAba1 belongs to the family of IS4 insertion sequences and possesses two imperfect inverted repeats of 16 bp. Its transposase is encoded by two open reading frames encoding 189 and 178 amino acids, leading to a functional protein when a frameshift occurs during the translation process (12). Over a 19-month period, it was observed imipenem-resistant A. baumannii isolates in intensive care units of a tertiary care clinic in Monteria-Colombia. The aim of this study was to describe the presence of the ISAba1/ADC-7, which is a novel class C enzyme, associated with oxa genes in A. baumannii isolates from Colombia.
MATERIALS AND METHODS
Bacterial strains
A. baumannii used in this study were collected from a private, tertiary care clinic in Monteria between August 2005 and February 2007. Study strains included 20 isolates resistant to imipenem obtained from an equal number of patients. The identification of A. baumannii was performed using Microscan Neg Combo Panel Type 44 (Dade Behring, Ca, USA). The isolates were obtained from adult (n = 14) and neonatal intensive care units (n = 6).
Determination of breakpoints
Antimicrobial compounds including imipenem, meropenem, ceftazidime, cefepime, aztreonam, amikacin, gentamicin, ciprofloxacin, moxifloxacin, ampicillin/sulbactam and piperacillin/tazobactam were used to establish the breakpoints. Breakpoints were determined by Microscan Neg Combo Panel Type 44 and interpreted according to CLSI standards (3). Escherichia coli ATCC® 25922 and Pseudomonas aeruginosa ATCC® 27853 were used as controls.
PCR, cloning and DNA sequencing
Screening of carbapenems and cephalosporins-resistant A. baumannii isolates was performed by PCR assay for genes blaOXA-51, blaOXA-23, blaOXA-24, blaOXA-58, blaIMP, blaVIM, and blaADC-7 using the primers described elsewhere (13, 14, 17, 19, 33, 34). The presence of the ISAba1 insertion upstream of OXA-23, OXA-51, and ADC-7, were determined by mapping using PCR with cloning and sequencing of products. Combinations of primers reverse ISAba1 and reverse of blaOXA-23, blaOXA-51 and blaADC-7 to detect ISAba1/blaOXA-23,ISAba1/blaOXA-51 and ISAba1/blaADC-7 were used for this purpose. Amplicons were purified with Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI, USA) according to manufacturer’s instructions and cloned in PCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA). These clones were subsequently transformed into chemically competent Escherichia coli TOP10 (Invitrogen, Carlsbad, CA) by heat shock and selected on MacConkey agar containing ampicillin 50 mg/L. Plasmid DNA from transconjugantes was purified using Pure link™ quick plasmid miniprep Kit (Invitrogen, Carlsbad, CA, USA), and both strands were sequenced using an automatic DNA sequencer (MegaBACE 750, Amersham, Biosciences, Piscataway, NJ, USA). BLASTX search engine was used for the initial analysis of sequences (1) and alignments were created using Clustal W version 2.0.8 (29).
PFGE
Purified DNA from all isolates was prepared in agarose blocks and digested with 30U ApaI (Promega, Madison, WI) as described previously (23). Agarose gel electrophoresis was performed in 1% gels (Seakem Gold, Cambrex, USA) in 0.5 X TBE buffer in an orthogonal-field alternating gel electrophoresis Gene Navigator (Pharmacia LKB Biotechnology, Uppsala, Sweden) for 20 hours at 12°C. The run conditions were 200 V with a pulse angle of 120° and pulse times of three phases as follows: 20s for 8h, 10s for 8h and 5s for 4h. A λ ladder (New England Biolabs) was used to provide molecular size markers, and the gels were stained with ethidium bromide (0.5 mg/L). Restricted DNA bands were visualized using a GE Healthcare imager (ImageQuant 100, Uppsala, Sweden). PFGE profiles were interpreted according to the criteria of Tenover et al. (28).
RESULTS
Phenotypic resistance
Breakpoint determinations and molecular detection of carbapenemases and cephalosporinase genes are shown in Table 1. All 20 A. baumannii isolates were resistant to imipenem and 18 to meropenem. Twelve and 17 isolates were resistant to fluoroquinolones and aminoglycosides, respectively, and all were resistant to the piperacillin/tazobactam and cephalosporins tested with the exception of ampicillin/ sulbactam, with 13 resistant isolates.
Table 1.
Carbapenem resistance in relation to presence and location of ISAba1, and the PFGE profile.
| MIC (μg/ml) | ISAba1 | Genes OXA, ADC-7 | ISR/OXA51R | ISR/OXA23R | ISR/ADC-7R | PFGE profile | |||
|---|---|---|---|---|---|---|---|---|---|
| CAZ | FEP | IPM | MER | ||||||
| > 16 | > 16 | > 8 | > 8 | Negative | 23,51, ADC-7 | Negative | Negative | Negative | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Positive | Positive | I |
| > 16 | > 16 | 8 | < 4 | Positive | 51, ADC-7* | Negative | Negative | Negative | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Positive | Positive | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Negative | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Positive | Positive | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Negative | I |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Positive | II |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Positive | II |
| > 16 | > 16 | > 8 | > 8 | Positive | 51, ADC-7 | Positive | Negative | Positive | III |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Positive | III |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51 ADC-7 | Negative | Negative | Negative | IV |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Positive | Negative | IV |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Positive | Negative | IV |
| > 16 | > 16 | > 8 | > 8 | Positive | 51* | Positive | Negative | Negative | IV |
| > 16 | > 16 | > 8 | > 8 | Positive | 23, 51, ADC-7 | Positive | Negative | Positive | V |
| > 16 | > 16 | > 8 | > 8 | Positive | 23, 51, ADC-7 | Negative | Positive | Negative | VI |
| > 16 | > 16 | > 8 | > 8 | Positive | 23,51, ADC-7 | Negative | Negative | Negative | VII |
| > 16 | > 16 | > 8 | > 8 | Positive | 51, ADC-7 | Positive | Negative | Positive | VIII |
| > 16 | > 16 | 8 | < 4 | Positive | 51* | Negative | Negative | Negative | IX |
CAZ: Ceftazidime; FEP: Cefepime; IMP: Imipenem; MER: Meropenem; ISR: ISAba1R;
Imipenem-resistant isolates were negative for blaOXA-23, -24, -58, and blaIMP, VIM.
Detection of blaOXA-23, blaOXA-51, and ISAba1/OXA-23–51
Amplification of purified DNA yielded a PCR product with specific primers for blaOXA-23,-51 genes (Figure 1); the blaOXA-24 and blaOXA-58 genes were not detected by this method. The complete sequence of the fragment 507 bp was 100% identical to that described for the blaOXA-23 gene (Accession number GU253894). The 641 bp fragment was identical to the blaOXA-51 gene (Accession number EU255296).
Figure 1.
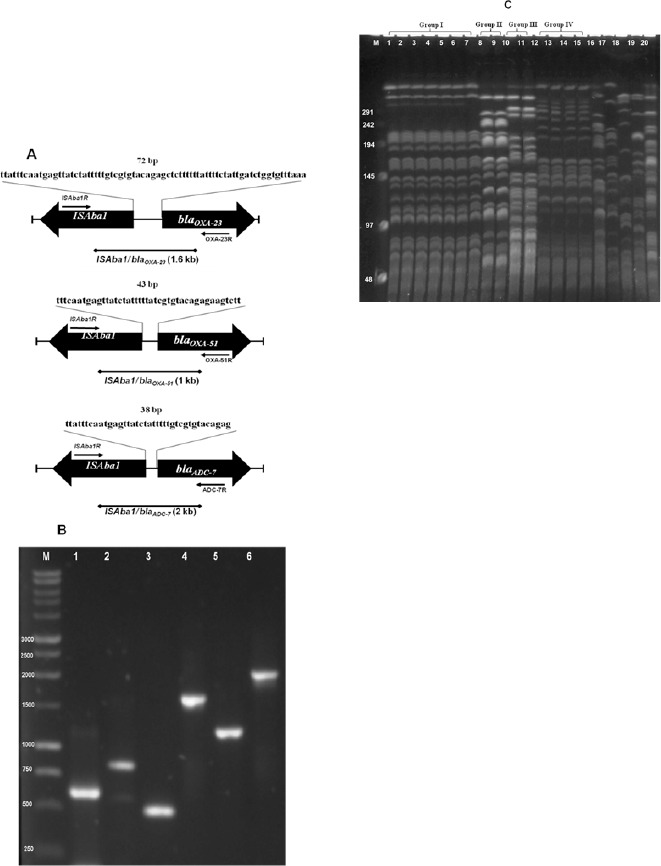
A. Location of the primers used in this work to amplify ISAba1 located upstream of the genes blaOXA-23, 51 and blaADC-7. B. PCR amplification of genes blaOXA-23, 51 and blaADC-7. Line 1 OXA-51, line 2 ISAba1/OXA-51, Line 3 OXA-23, Line 4 ISAba1/OXA-23, Line 5 ADC-7, Line 6; ISAba1/ADC-7. C. PFGE profiles of 20 A. baumannii isolates digested with ApaI in lanes 1–20. Lane M, lambda DNA markers with molecular weights in kb shown.
Fifteen of the isolates were positive for the blaOXA-23 gene, 4 others were producers of carbapenemase OXA-51 with the insertion of ISAba1 in the promoter region of the blaOXA-51gene, and one of the 15 isolates carrying blaOXA-23 also contained the ISAba1/OXA-51 gene. Two isolates were negative for carbapenemase genes (Table 1).
PCR mapping showed ISAba1 to be located in the promoter region of the blaOXA-23 gene in 6 isolates; these amplicons were 1.5-kb in size. The sequence obtained for the ISAba1-blaOXA-23 amplicon was 97% identical to that described for the insertion sequence ISAba1 transposase gene and the blaOXA-23 gene (Accession number GU292795). Four other isolates also had ISAba1 located upstream of the blaOXA-51gene, and these amplicons were 1-kb in size (Figure 1). The sequence obtained the latter showed 96% identity to that described for the insertion sequence ISAba1 transposase gene and blaOXA-51 gene (Accession number HM545089)
Identification of blaADC-7 and ISAba1/ADC-7
All 20 isolates were resistant to third-and fourth-generation cephalosporins and piperacillin-tazobactam (Table 1). The PCR product for blaADC-7 produced an amplicon of 1.15-kb (Figure 1). Eighteen isolates contained the blaADC-7gene. The nucleotide sequence of the 1.15-kb fragment was identical to the blaADC-7 gene (Accession number AY648950).
Mapping the position of ISAba1 relative to blaADC-7.
Among the 18 isolates with blaADC-7 that were also resistant to carbapenems and cephalosporins, a band of 2-kb was obtained for 9 isolates using the reverse primer for ISAba1 and the reverse primer for blaADC-7 (Figure 1 and Table 1). Sequencing showed 98% identity to that described for the insertion sequence ISAba1 transposase gene and the β-lactamase ADC-7 (Accession number GU292796).
PFGE
Fifteen of the isolates clustered into four groups, while the remaining 5 isolates were unrelated to other strains and were categorized as having unique profiles. Group I contained seven isolates, groups II and III two isolates each, and group IV four isolates (Figure 1).
DISCUSSION
Carbapenems are the antibiotics of choice for the treatment of infections caused by A. baumannii when these bacteria are resistant to other β-lactam antibiotics. However, carbapenem resistance has increased, limiting the use of this class of agent for empiric antibiotic therapy (9). There is mounting evidence that A. baumannii has a natural intrinsic carbapenemase resistance mediated by blaOXA-51 (27, 31). However, carbapenem resistance is only expressed when the insertion sequence ISAba1 is present upstream of the blaOXA-51gene (31).
Of the 20 carbapenem resistant A. baumannii isolates included in our study, 15 expressed the carbapenemase OXA-23 responsible for their carbapenem resistance. PFGE distinguished nine types clustered into four groups and involving 15 isolates. Although many nosocomial outbreaks are caused by a common clone (5, 24), the polymorphism observed mong isolates of this study suggest the existence of different lineages as a result of the mobilization of patients between clinics and hospital institutions.
Furthermore, studies of Villegas et al (32) have found widespread dissemination of OXA-23 among clinical isolates of A. baumannii, both clonal and non-clonal, in hospitals in several Colombian cities. Our data report similar findings and highlight the clinical and epidemiological problems associated with the existence of these resistant strains in the ICUs of hospitals due to the versatility of A. baumannii for nosocomial spread and contamination of the environment. Naturally this clinical picture complicates the choice of empirical antibiotic therapy in severely ill patients, even when using antibiotic alternatives such as polymyxins, minocycline and sulbactam (11).
The presence of the insertion sequence ISAba1 upstream of carbapenemases genes can influence the expression of resistant genes (19). In our isolates containing blaOXA-51, these were resistant to imipenem and meropenem if they possessed ISAba1 upstream of blaOXA-51 (Table 1).
In addition, 6 isolates that were positive for blaOXA-23 gave a band of 1.5-kb using the ISAba1R and OXA-23R primers. Although these isolates were also positive for blaOXA-51, no amplicons were produced with primer combinations using ISAba1F/OXA-51R or ISAba1R/OXA-51R, suggesting that co-expression of these two carbapenemases genes was not occurring and that blaOXA-23 alone was responsible for the carbapenem resistance phenotype of these strains.
Similarly, Poirel et al (16) found that expression of blaPER-1 in A. baumannii, P. aeruginosa and P. stuartii was associated with the promoter sequence ISPa12. Futhermore, multiple copies of ISAba1 are frequently found in A. baumannii isolates (21). These may be associated with several genes, as found in three isolates belonging to the same PFGE profile in this study, where ISAba1 was associated with blaOXA-23, and blaADC-7(Table 1). Moreover, the PCR product amplified with the ISAba1R/ADC-7 reverse primers amplified a fragment of 2-kb in nine of the isolates, seven of these were also positive for blaOXA-23, and 3 carried the promoter sequence ISAba1 upstream of blaOXA-23 gene (Table 1). Similar findings were reported by Héritier et al (12), who studied six isolates of A. baumannii resistant to ceftazidime and here these contained ISAba1 upstream of the blaAmpC gene; however, they did not find ISAba1/ADC in isolates positive for blaOXA-23 and positive for ISAba1/OXA-23. As suggested by Segal et al (21), ISAba1 could play an important role in the acquisition and expression of resistance genes.
However, Segal et al. (22) showed that transcription of the blaADC-like gene is dependent on the promoter sequence within an ISAba1 located upstream region of this gene. In the present study, the blaADC-7 gene was found upstream of ISAba1 in 45% of the isolates, which could be related to the regulation of this gene by activation or repression of resistance to third generation cephalosporins in isolates of A. baumannii. Similar results were reported by Corvec et al (4), who found ISAba1 located upstream of the blaADC gene (no blaADC-7) in 52.4% of isolates of A. baumannii. However, Ruiz et al (19), also found ISAba1 located upstream blaADC gene in 54% of the isolates of A. baumannii. Furthermore, ISAba1 was located upstream from the blaADC gene in only one of the two A. baumannii strains belonging to the same clone, and was located upstream from the blaADC gene in only one of the two strains belonging to the same clone. In our study PFGE showed the presence of 9 profiles, with ISAba1 upstream of the blaADC-7 gene in only 3 of 7 isolates belonging to profile 1. These results show the wide dissemination of ISAba1 related to resistance genes in A. baumannii isolates in the hospital environment.
These findings highlight the emerging problem of carbapenem resistant A. baumannii in this Colombian hospital associated with OXA-23 and OXA-51 carbapenemases, and the links with ISAba1 in some isolates. Two isolates that expressed resistance to imipenem but not meropenem and were negative for blaVIM, IMP, OXA23, OXA24, OXA58, and ADC genes (Table 1), and it is likely they have mutations in outer membrane porins or altered PBPs (7, 25).
In conclusion, this is the first report of the ISAba1/ADC-7 associated with OXAs genes in carbapenem-resistant A. baumannii isolates from Colombia. Furthermore, production of the OXA-23, ISAba1/OXA-23 and ISAba1/OXA-51 carbapenemases presents an emerging threat of carbapenem resistance among A. baumannii isolates which is particularly worrisome due to the difficult choice of empirical antibiotic therapy in seriously ill patients and the possible contribution to increased hospital stay and associated costs.
ACKNOWLEDGEMENTS
We thank to the University of Cordoba by support to this study with grant FMV 04–06 and contract 1120223, and Dr. Seamus Fanning for reviewing and critics the manuscript.
REFERENCES
- 1.Altschul S., Gish W., Miller W., Myers E., Lipman D. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ben Othman A., Zribi M., Masmoudi A., Abdellatif S., Ben Lakhal S., Fendri C. Multiresistance and endemic status of Acinetobacter baumannii associated with nosocomial infections in a tunisian hospital: a critical situation in the intensive care units. Braz. J. Microbiol. 2011;42(2):415–422. doi: 10.1590/S1517-83822011000200001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brenner P., Otaíza F., Bustamante R. Nosocomial infections outbreaks in Chile 1985–2002. Am. J. Infect. Control. 2004;32(3):E49. [Google Scholar]
- 4.Clinical and Laboratory Standards Institute (CLSI). Approved standard M7-A8. Eighth. Wayne, PA: EE.UU. Clinical and Laboratory Standards Institute; 2009. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Informational. [Google Scholar]
- 5.Corvec S., Caroff N., Espaze E., Giraudeau C., Drugeon H., Reynaud A. AmpC cephalosporinase hyperproduction in Acinetobacter baumannii clinical strains. J. Antimicrob. Chemother. 2003;52(4):629–635. doi: 10.1093/jac/dkg407. [DOI] [PubMed] [Google Scholar]
- 6.Dalla Costa L., Coelho J., Souza H., Castro M., Stier C., Bragagnolo K., Rea Neto A., Penteado Filho S., Livermore D., Woodford N. Outbreak of carbapenem-resistant Acinetobacter baumannii producing OXA-23 enzymes in Curitiba, Brazil. J. Clin. Microbiol. 2003;41(7):3403–3406. doi: 10.1128/JCM.41.7.3403-3406.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fagon J., Chastre J., Domart Y., Trouillet J., Gibert C. Mortality due to ventilator-associated pneumonia or colonization with Pseudomonas or Acinetobacter species: assessment by quantitative culture of samples obtained by a protected specimen brush. Clin. Infect. Dis. 1996;23(3):538–542. doi: 10.1093/clinids/23.3.538. [DOI] [PubMed] [Google Scholar]
- 8.Fernández Cuenca F., Martínez Martínez L., Conejo M., Ayala J., Perea E., Pascual A. Relationship between β-lactamase production, outer membrane protein and penicillin-binding protein profiles on the activity of carbapenems against clinical isolates of Acinetobacter baumannii. J. Antimicrob. Chemother. 2003;51(3):565–574. doi: 10.1093/jac/dkg097. [DOI] [PubMed] [Google Scholar]
- 9.García J., Ortiz C., Garnacho J., Jiménez Jiménez F., Pérez Paredes C., Barrero Almodóvar A., Miner M. Risk factors for Acinetobacter baumannii nosocomial bacteremia in critically ill patients: a cohort study. Clin. Infect. Dis. 2001;33(7):939–946. doi: 10.1086/322584. [DOI] [PubMed] [Google Scholar]
- 10.Gaspareto P., Martins A., Zavascki A., Barth A. Ocurrence of blaSPM-1 and blaIMP-1 genes of metallo-beta-lactamases in clinical isolates of Pseudomonas aeruginosa from three universitary hospitals in the city of Porto Alegre, Brazil. Braz. J. Microbiol. 2007;38(1):108–109. [Google Scholar]
- 11.Go E., Urban C., Burns J., Kreiswirth B., Eisner W., Mariano N., Mosinka Snipas K., Rahal J. Clinical and molecular epidemiology of Acinetobacter infections sensitive only to polymyxin B and sulbactam. Lancet. 1994;344(8933):1329–1332. doi: 10.1016/s0140-6736(94)90694-7. [DOI] [PubMed] [Google Scholar]
- 12.Guzmán Blanco M., Casellas J., Sader H. Bacterial resistance to antimicrobial agents in Latin America. The patient is awakening. Infect. Dis. Clin. North. Am. 2000;14(1):67–81. doi: 10.1016/s0891-5520(05)70218-x. [DOI] [PubMed] [Google Scholar]
- 13.Héritier C., Poirel L., Nordmann P. Cephalosporinase over-expression resulting from insertion of ISAba1 in Acinetobacter baumannii. Clin. Microbiol. Infect. 2006;12(2):123–130. doi: 10.1111/j.1469-0691.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 14.Hujer K., Hamza N., Hujer A., Perez F., Helfand M., Bethel C., Thomson J., Anderson V., Barlow M., Rice L., Tenover F., Bonomo R. Identification of a new allelic variant of the Acinetobacter baumannii cephalosporinase, ADC-7 b-lactamase: defining a unique family of class C enzymes. Antimicrob. Agents. Chemother. 2005;49(7):2941–2948. doi: 10.1128/AAC.49.7.2941-2948.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyobe S., Minami S., Yamada H. Insertion of a carbapenemase gene cassette into an integron of a Pseudomonas aeruginosa plasmid. J. Antimicrob. Chemother. 1996;38(6):1114–1115. doi: 10.1093/jac/38.6.1114. [DOI] [PubMed] [Google Scholar]
- 16.Livermore D. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs. 2002;3:218–224. [PubMed] [Google Scholar]
- 17.Poirel L., Cabanne L., Vahaboglu H., Nordmann P. Genetic Environment and Expression of the Extended-Spectrum β-Lactamase blaPER-1 Gene in Gram-Negative Bacteria. Antimicrob. Agents. Chemother. 2005;49(5):1708–1713. doi: 10.1128/AAC.49.5.1708-1713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poirel L., Marqué S., Héritier C., Segonds C., Charbanon G., Nordmann P. OXA-58, a Novel Class D β-Lactamase Involved in Resistance to Carbapenems in Acinetobacter baumannii. Antimicrob. Agents. Chemother. 2005;49(1):202–208. doi: 10.1128/AAC.49.1.202-208.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Queenan A., Bush K. Carbapenemases: the Versatile β-Lactamases. Clin. Microbiol. Rev. 2007;20(3):440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ruiz M., Marti S., Fernandez Cuenca F., Pascual A., Vila J. High prevalence of carbapenem-hydrolysing oxacillinases in epidemiologically related and unrelated Acinetobacter baumannii clinical isolates in Spain. Clin. Microbiol. Infect. 2007;13(12):1192–1198. doi: 10.1111/j.1469-0691.2007.01825.x. [DOI] [PubMed] [Google Scholar]
- 21.Ruiz M., Marti S., Fernandez Cuenca F., Pascual A., Vila J. Prevalence of ISAba1 in epidemiologically unrelated Acinetobacter baumannii clinical isolates. FEMS. Microbiol. Lett. 2007;274(1):63–66. doi: 10.1111/j.1574-6968.2007.00828.x. [DOI] [PubMed] [Google Scholar]
- 22.Segal H., Garny S., Elisha B. Is ISAba-1 customized for Acinetobacter? FEMS Microbiol. Lett. 2005;243:425–429. doi: 10.1016/j.femsle.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 23.Segal H., Nelson E., Elisha B. Genetic environment and transcription and transcription of AmpC in Acinetobacter baumannii clinical isolate. Antimicrob. Agents. Chemother. 2004;48(2):612–614. doi: 10.1128/AAC.48.2.612-614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seifert H., Dolzani L., Bressan R., van der Reijden T., van Strijen B., Stefanik D., Heersma H., Dijkshoorn L. Standardization and Interlaboratory Reproducibility Assessment of Pulsed-Field Gel Electrophoresis-Generated Fingerprints of Acinetobacter baumannii. J. Clin. Microbiol. 2005;43(9):4328–4335. doi: 10.1128/JCM.43.9.4328-4335.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siroy A., Molle V., Lemaitre Guillier C., Vallenet D., Pestel Caron M., Cozzone A., Jouenne T., Dé E. Channel Formation by CarO, the Carbapenem Resistance-Associated Outer Membrane Protein of Acinetobacter baumannii. Antimicrob. Agents. Chemother. 2005;49(12):4876–4883. doi: 10.1128/AAC.49.12.4876-4883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunde M. Prevalence and characterization of class 1 and class 2 integrons in Escherichia coli isolated from meat and meat products of Norwegian origin. J. Antimicrob. Chemother. 2005;56(6):1019–1024. doi: 10.1093/jac/dki377. [DOI] [PubMed] [Google Scholar]
- 27.Takagi E., Lincopan N., Cassettari V., Passadore L., Mamizuka E., Martinez M. Carbapenem-resistant Acinetobacter baumannii outbreak at university hospital. Braz. J. Microbiol. 2009;40(2):339–341. doi: 10.1590/S1517-838220090002000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tenover F., Arbeit R., Goering R., Mickelsen P., Murray B., Pershing D., Swaminathan B. Interpreting Chromosomal DNA Restriction Patterns Produced by Pulsed-Field Gel Electrophoresis: Criteria for Bacterial Strain Typing. J. Clin. Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson J., Higgins D., Gibson T. CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic. Acids. Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turton J., Kaufmann M., Warner M., Coelho J., Dijkshoorn L., van der Reijden T., Pitt T. A prevalent, multiresistant, clone of Acinetobacter baumannii in South East England. J. Hosp. Infect. 2004;58(3):170–179. doi: 10.1016/j.jhin.2004.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Turton J., Ward M., Woodford N., Kaufmann M., Pike R., Livermore D., Pitt T. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS. Microbiol. Lett. 2006;258:72–77. doi: 10.1111/j.1574-6968.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- 32.Villegas M., Kattan J., Correa A., Lolans K., Guzman A., Woodford N., Livermore D., Quinn J. Dissemination of Acinetobacter baumannii Clones with OXA-23 Carbapenemase in Colombian Hospitals. Antimicrob. Agents. Chemother. 2007;51(6):2001–2004. doi: 10.1128/AAC.00226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodford N., Ellington M., Coelho J., Turton J., Ward M., Brown S., Amyes S., Livermore D. Multiplex PCR for genes encoding prevalent OXA carbapenemases in Acinetobacter spp. Internat. J. Antimicrob. Agents. 2006;27:351–353. doi: 10.1016/j.ijantimicag.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Yang J., Hsueh P., Ko W., Luh K., Tsai S., Wu H., Wu J. Metallo-b-Lactamases in Clinical Pseudomonas Isolates in Taiwan and Identification of VIM-3, a Novel Variant of the VIM-2 Enzyme. Antimicrob. Agents. Chemother. 2001;45(8):2224–2228. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]


