Abstract
Direct sensing of intracellular metabolite concentrations by riboswitch RNAs provides an economical and rapid means to maintain metabolic homeostasis. Since many organisms employ the same class of riboswitch to control different genes or transcription units, it is likely that functional variation exists in riboswitches such that activity is tuned to meet cellular needs. Using a bioinformatic approach, we have identified a region of the purine riboswitch aptamer domain that displays conservation patterns linked to riboswitch activity. Aptamer domain compositions within this region can be divided into nine classes that display a spectrum of activities. Naturally occurring compositions in this region favor rapid association rate constants and slow dissociation rate constants for ligand binding. Using X-ray crystallography and chemical probing, we demonstrate that both the free and bound states are influenced by the composition of this region and that modest sequence alterations have a dramatic impact on activity. The introduction of non-natural compositions result in the inability to regulate gene expression in vivo, suggesting that aptamer domain activity is highly plastic and thus readily tunable to meet cellular needs.
Keywords: riboswitch, RNA structure, mutual information, transcriptional regulation, activity tuning
Introduction
Riboswitches are structured regulatory elements contained in the 5′-leader region in many bacterial messenger RNAs. These riboregulators use a secondary structural switch to control gene expression in response to binding an effector molecule, thus providing a rapid and efficient genetic response to changing cellular or environmental conditions (reviewed in Refs. 1 and 2). Currently, over 20 riboswitch classes that control gene expression in response to a structurally diverse range of effector molecules including nucleobases, amino acids, co-factors, metal ions, and second messengers have been identified.3 Although riboswitches are broadly distributed in bacteria, they are most prevalent in the Firmicutes and Fusobacteria.4 In some of these organisms, the same class of riboswitch controls a number of different genes or operons, employing essentially the same aptamer domain for each. For example, in Bacillus subtilis, there are at least 5 purine-binding riboswitches5,6 and 11 S-adenosylmethionine (SAM)-binding riboswitches,7–10 each controlling its own transcriptional unit. Presumably, riboswitches controlling unique transcriptional units will control gene expression at a particular effector concentration dictated by the composition of the transcriptional unit. Furthermore, the diverse genetic backgrounds and environmental niches of bacteria likely necessitate adaptation by individual riboswitches for optimal regulatory control.
It has been hypothesized that the effector binding domains (called the aptamer domain) of riboswitches are “tuned” to elicit the appropriate regulatory response for each transcriptional unit.6,9 A study comparing the properties of the 11 B. subtilis SAM-I riboswitches established a link between functional characteristics of the riboswitch with the gene product being regulated.9 Functional diversity was manifested as a large range in binding affinity (Kd), absolute level of transcript induction, the dynamic range of the expression response, and the propensity to read through a transcriptional terminator for each SAM-I riboswitch variant. It was concluded that SAM-I riboswitches involved in biosynthetic pathways have different regulatory properties than those controlling transport.9 Specifically, riboswitches controlling biosynthetic genes tend to switch gene expression off at lower concentrations of SAM than those controlling genes encoding metabolite transporters. This is consistent with the observation that isolated aptamer domains of some of these SAM-I riboswitches have over a 1000-fold range in their binding affinities for SAM.10 This is interesting since crystal structures of several aptamer domains in complex with SAM show that the core of the RNA is nearly invariant.11,12 Similarly, purine-binding riboswitches exhibit a diversity of effector binding affinities and regulatory properties6 despite nearly invariant secondary structure and nucleotide compositions in the effector-binding site (Fig. 1a). The structural basis for tuning the affinity and therefore, presumably, the regulatory response of a specific riboswitch is unknown.
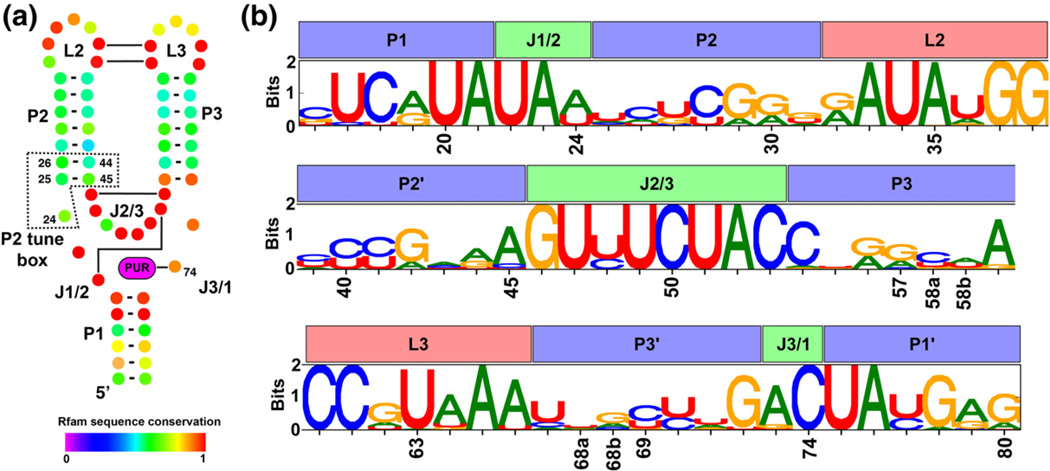
Fig. 1.
Secondary structure and positional entropy plot of the aptamer domain of purine riboswitches. (a) The conserved secondary structure of the aptamer domain is depicted such that each nucleotide position is represented as a colored dot with Watson–Crick base-pairing interactions shown as a bar/line connecting the interacting nucleotides. Nucleotide coloring corresponds to their degree of conservation, where a value of 1 denotes universal conservation (adapted from the Rfam database, accession number RF00167). (b) A sequence logo representing the nucleotide conservation as well as information content at each position in our structure-based alignment of 302 sequences. For each position, the height of each nucleotide symbol represents its frequency at that position, while the overall height represents the information content at that position.
To explore the mechanisms of tuning, we examined purine riboswitches that sense intracellular guanine/hypoxanthine (HX) or adenine and generally modulate expression of proteins associated with purine biosynthesis and transport.5–13,14 The structural basis for ligand binding by the aptamer domain is well established.15–19 All guanine/adenine riboswitches contain the same secondary and tertiary architecture, a three-way junction of three helices (P1, P2, and P3) supported by a distal loop–loop interaction (L2–L3). Both the junction and the terminal loops exhibit significant sequence conservation (Fig. 1a). Guanine or adenine recognition is achieved through formation of a Watson-Crick base pair with either a cytosine or a uridine (position 74 in the RNAs of this study), respectively.5,14,15,19 Specificity for either nucleobase is almost entirely determined by the identity of nucleotide 74, as guanine and adenine riboswitches can be inter-converted through a single change to the identity of pyrimidine 74.17 Extensive mutagenesis of several purine riboswitches has further refined our understanding of the nucleotides that directly contribute to the affinity of this RNA for purine nucleobases.6,20,21 The majority of these bases localize to non-helical elements in the junction and loops, with the exception of two Watson-Crick pairs in P1 that are proximal to the ligand-binding site. A broad set of data on this RNA from spectroscopic, single-molecule and chemical probing studies makes the purine riboswitch an ideal model system to study the relationship between sequence variation and activity tuning.22
While, in principle, regulatory tuning can occur in either the aptamer domain or the downstream secondary structural switch, termed the expression platform, a reasonable hypothesis is that tuning primarily occurs within the aptamer domain by altering ligand-binding properties. The principal difficulty in relating sequence variation in the aptamer domain to regulatory tuning is that conservation patterns are dominated by tertiary interactions and secondary structure outside the ligand-binding site. The large number of neutral changes that preserve Watson-Crick pairing frustrates attempts to look for more subtle patterns of variation that may influence the active site.
Rather than test a large set of phylogenetic sequence variants, we sought to identify regions of the RNA that are functionally coupled using a statistical coupling analysis in conjunction with a structurally backed sequence alignment.23 Using this approach, a region adjacent to the three-way junction was identified as containing patterns of covariation with a nucleotide within the junction that does not involve direct pairing of nucleotides. To assess whether this region of the aptamer domain, which we call the “P2 tune box”, influences ligand binding and regulatory activity, we took a reductionist approach by introducing sequence variation into the well-characterized B. subtilis xpt-pbuX guanine riboswitch. We have previously used this approach to understand the relationship between guanine and 2′-deoxyguanosine-sensing RNAs24 whose conclusions were supported by a subsequent crystal structure.25 In this work, we demonstrate that regions flanking the ligand-binding pocket exhibit patterns of covariation that influence riboswitch regulatory properties.
Results
Identification of functional covariation between P2 and the ligand-binding pocket
To identify evolutionarily conserved covariation patterns in the aptamer domain between nucleotides that do not physically interact (i.e., base pair), it is essential to generate a high-quality multiple sequence alignment.26–28 The statistical methods used to infer functional relationships between residues in a sequence work under the assumption that the columns in a multiple sequence alignment are analogous.29 Starting with a high-quality alignment eliminates uncertainty in the position of each base within the RNA prior to performing a statistical analysis, thus decreasing the frequency of false positives and allowing the detection of subtle functional covariation patterns.
A common feature of alignments of RNA sequences is the presence of base-pair insertions or deletions within the middle of helices. In the case of the purine riboswitch aptamer domain, P2 and P3 can vary from five to seven base pairs. The issue that arises is the arbitrary placement of the insertion/ deletion within a helix. An incorrect placement of the insertion/deletion in P2 or P3 would potentially confound potential non-Watson–Crick covariation between nucleotides in a stem. To circumvent this problem, we limited our analysis to 302 of 484 sequences available in Rfam version 9.0.30 These sequences were chosen because they represent the most common secondary structure within the family. Each sequence must contain seven base pairs in both P2 and P3 while P1 was limited to seven base pairs proximal to the three-way junction. P1 lengths of at least four base pairs are observed in all purine riboswitches and the effect of additional base pairs has been determined to have a negligible effect on ligand binding.31 The Shannon Entropy Plot WebLogo for the alignment of these sequences is shown in Fig. 1b and the full alignment is available in Table S1.
Since the structural framework of these sequences is identical, an unambiguous analysis of functionally coupled regions of the RNA can be performed using mutual information (MI), a statistical technique often used to predict RNA structure.32,33 This method uses a multiple sequence alignment to identify coevolving residues in a sequence, where one nucleotide changes in response to a change at another position in the sequence. Functionally coupled nucleotide positions are expected to yield greater MI scores than expected for a random interaction. In order to correct for the effects that high sequence conservation has on the MI calculation, normalized mutual information (NMI)34,35 was chosen to allow us to detect subtle functional interactions. A statistical method for assessing the significance of NMI scores has been previously shown to successfully predict functional interactions in protein sequences.35
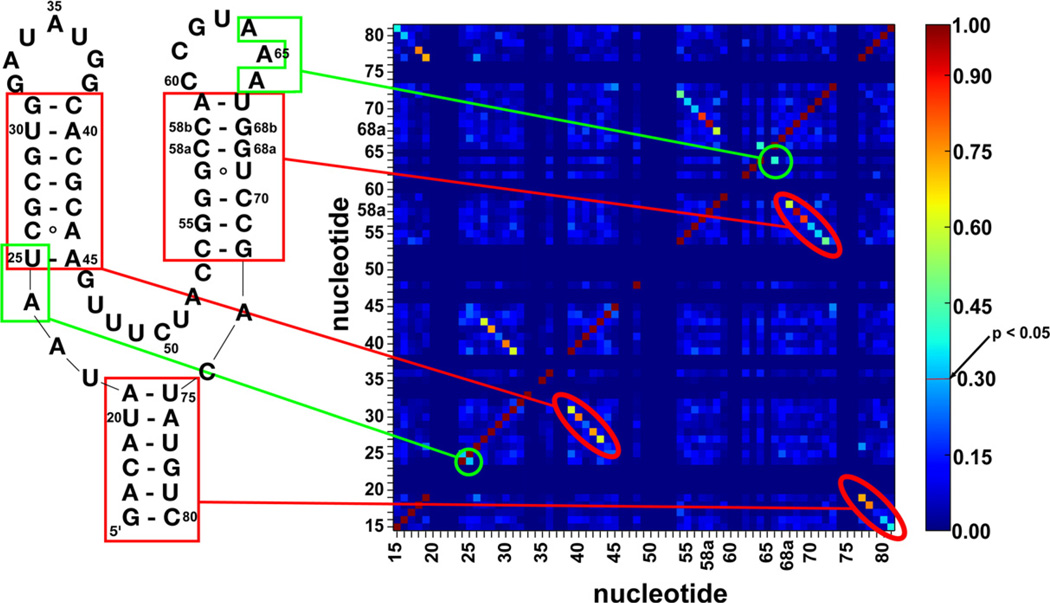
NMI scores were determined for all possible pairwise interactions and three regions demonstrated significant covariation patterns corresponding to paired regions P1, P2, and P3 (Fig. 2, red). In addition to these expected regions of covariation, a significant NMI score (NMI = 0.320, p=0.0197) between positions 24 and 25 is observed (Fig. 2, green). Covariation between positions 24 and 25 is especially interesting as position 24 is located in J1/2 and uniquely anchors J1/2 to J3/1 by stacking between positions 72 and 73. While position 25 shows low conservation, position 24 is either an adenine (66%) or a uracil (34%) (Fig. 1b). The other significant NMI score for non-interacting bases is between nucleotides 64 and 66 (NMI = 0.401, p= 0.00579) in L3. In the majority of sequences (80%), both of these nucleotides are adenosines that form non-canonical base-pairing interactions with L2. In ~ 10% of total sequences, these two nucleotides are uracil (U64/U66); in 8.5%, G64/A66; and in 1.5%, U64/A66. A mutagenic survey of the pbuE aptamer domain revealed that the U64/A66 variant binds 2-aminopurine (2AP) as well as wild type (A64/A66).21 Furthermore, it has been shown that the A35–A64 pair in the L2–L3 is tolerant to changes in the 35 position, suggesting that the L2–L3 interaction can accommodate different pairs at this site. In this study, we have chosen to focus on understanding the 24/25 covariation because of its adjacency to the ligand-binding pocket. Notably, these results were not obvious from sequence alignments using gap alignment in the paired regions,4 illustrating the utility of performing covariation analyses on structurally backed alignments.
Fig. 2.
NMI scores indicate covariation patterns in the aptamer domain. Secondary structural elements (red) and novel covariation interaction (green) are correlated to a two-dimensional heat map with NMI scoring represented on heat map colored by NMI values. The color bar to the right of the heat map indicates that an NMI value of 0.3 or greater is a significant value (p<0.05).
Classification of aptamer domains by P2 composition
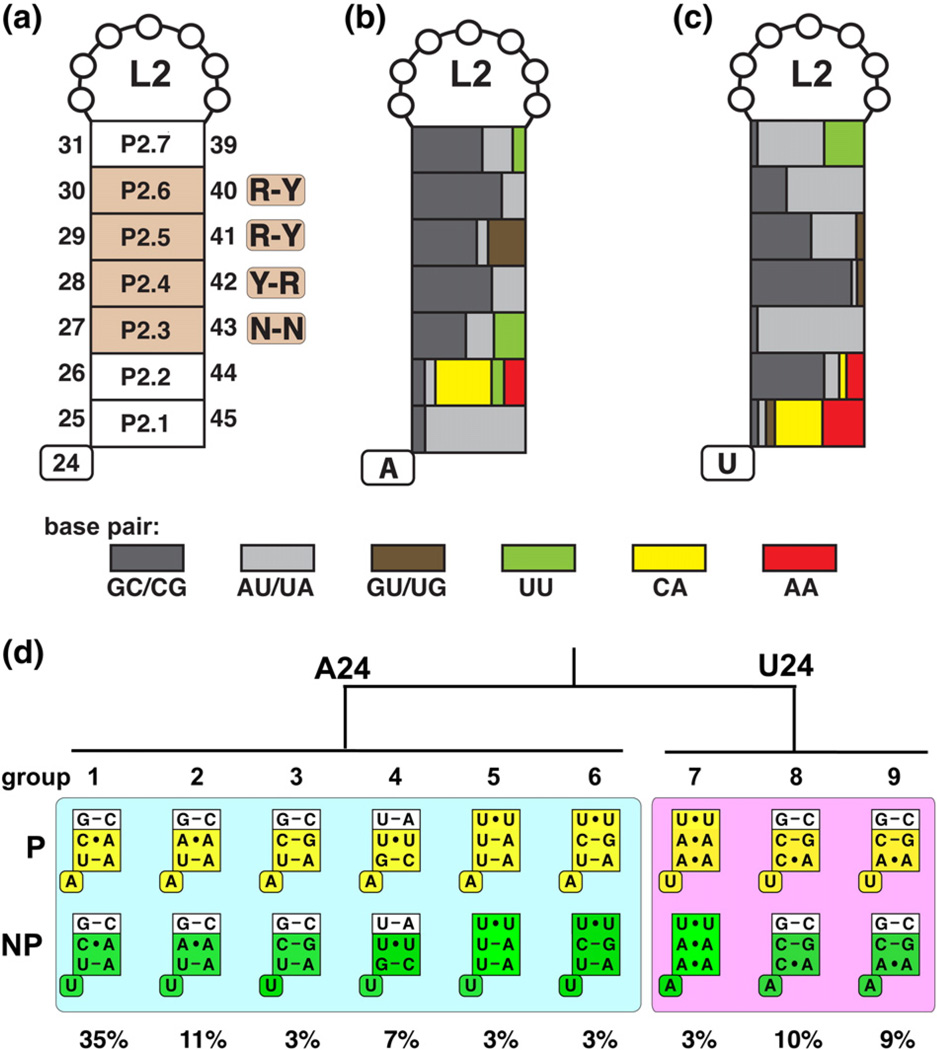
To experimentally determine whether there is functional covariation between the base of P2 (position 25) and position 24, we found it necessary to first further analyze the composition of P2. Examination of P2 revealed a marked tendency for non-canonical base pairs proximal to the junction (Fig. 3a). The base-pairing nomenclature in P2 used in this work is numbered P2.1 (positions 25–45, proximal to the three-way junction) through P2.7 (positions 31–39, proximal to L2). The central base pairs of P2 are composed of conserved pairing interactions that conform to Y–R, R–Y, and R–Y at P2.4, P2.5, and P2.6, respectively (Fig. 3a). Non-canonical base pairing is not observed at these positions, and these form the stable core of P2 as supported by temperature-dependent unfolding of the xpt-pbuX aptamer by chemical probing.36 P2.3 is also stable at temperatures in excess of 70 °C in a ligand-independent manner; however, the presence of non-canonical base pairing in natural sequences must be considered prior to classification of P2.3 as part of the rigid core. In contrast, P2.1 and P2.2 pairing interactions are significantly less stable and their formation is ligand dependent.36 This part of the P2 helix behaves as if it is part of J1/2 and J2/3 in its folding properties and thus should be considered part of the junction. Notably, these positions are highly enriched in non-canonical base pairs that may be important for imparting this behavior.
Fig. 3.
Sequence composition of the P2 stem based on a structurally backed multiple sequence alignment. A naming scheme and indication of the position as well as conservation is represented in (a) with purine (R) or pyrimidine (Y) bias indicated. (b) and (c) are color coded according to the type of base-pairing enrichment for A24 variants and U24 variants, respectively. (d) P2 sequence variants that represent a majority of all purine riboswitch aptamer domains. Aptamer variants from phylogenetically represented sequences (P) and non-phylogenetic variants (NP) with single base substitutions at position 24 are represented in yellow and green, respectively. The boxed pairs represent the P2.1, P2.2, and P2.3 pairs, consistent with Fig. 2. The abundance of each variant from the sequence alignment is indicated.
The 302 sequences in the structure-backed alignment were sorted into two groups based on the identity of position 24 (A24 or U24) and further divided into variant classes based on the composition of P2.1, P2.2, and P2.3 (Fig. 3b–c). Non-canonical pairing in the A24 group (1A–6A) occurs at P2.2 and sometimes at P2.3, while in the U24 group (7U-9U), it is enriched in non-canonical pairs P2.1 and infrequently at P2.3 (Fig. 3d). P2.7 also contains a significant fraction of non-canonical U-U base pairing as revealed in structures of the Vibrio vulnificus add and B. subtilis pbuE riboswitch aptamers.19,37 However, P2.7 was not used as a sorting criterion since there are little functional differences between aptamers with G–C or U-U pairs at this position (Table S2).
Using this classification scheme, we identified nine variant groups that account for ~84% of all sequences (Fig. 3d). Of these nine variants, 1A is the most abundant (35% of sequences) and corresponds to the sequence composition in the B. subtilis xpt-pbuX guanine riboswitch. The remaining variants are less abundant with each representing between 3% and 11 % of all sequences. Many aptamer sequences at 1 % abundance or less were grouped together as a set of lowly populated variants that account for 16% of all sequences and were not further considered. The covariation of positions 24 and 25 as well as the enrichment of non-canonical base pairing at the base of P2 raises the possibility that these nucleotides could influence the effector's interaction with the adjacent binding pocket. We refer to this region of the aptamer as the P2 tune box, which is defined as nucleotide 24 of J1/2 and the first two pairs of P2 (dashed box, Fig. 1a). To gain insight into the impact of variation in the P2 tune box on functional aspects of the aptamer domain, we measured rate constants for ligand binding as well as equilibrium dissociation constants.
Phylogenetically represented P2 tune box variants generally confer kinetic control
The relationship between the ligand-binding properties and regulatory control is complicated by the fact that many riboswitches operate co-transcriptionally.38–40 The consequence of this observation is that the riboswitch has only a limited amount of time to make a regulatory decision, as directed by ligand binding, before it is no longer capable of influencing the transcription or translation machinery. Thus, many riboswitches are under “kinetic control” in which ligand binding to the aptamer domain does not reach equilibrium prior to when a regulatory decision must be made.40 For transcriptional regulation, this is the time that RNA polymerase (RNAP) requires to transcribe mRNA from the 3′-side of the aptamer domain to the intrinsic terminator (called ΔtRNAP).40 If the aptamer is not at equilibrium with respect to ligand binding when the regulatory decision is made, then the concentration of ligand required to achieve the half-maximal regulatory response (EC50) is greater than that to half-saturate the aptamer (Kd) under equilibrium conditions. This is in contrast to “thermodynamic control” in which Kd≊EC50 The equilibrium binding constant (Kd) and the rate constants for association (Kon) and dissociation (Koff) must be measured to determine the likelihood that a specific aptamer imparts kinetic or thermodynamic control. A recent study of two adenine-responsive purine riboswitches determined that they can experience either mode of control.41
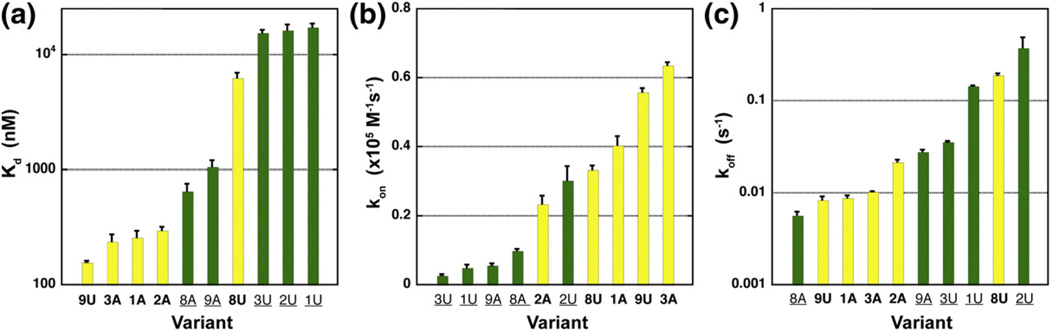
In total, five of the nine phylogenetically represented variant groups were evaluated, representing over 68% of sequences in the alignment (groups 1, 2, 3, 8 and 9; row “P”, Fig. 3d). Additionally, a non-phylogenetic (“NP”, Fig. 3d) variant was created for each of the five natural variants tested by replacing position 24 with the non-naturally occurring base, either A or U. To simplify our analysis, we placed each P2 tune box sequence into a “chassis” aptamer domain derived from the well-characterized B. subtilis xpt-pbuX guanine riboswitch.15–17,36 This ensures that the binding properties of each P2 tune box can be compared directly to other variants. The xpt aptamer domain was modified to add a base pair to the middle of P3 (pair 58b–68b, Fig. 1a) to reflect that the 302 chosen sequences all have seven base pairs in this helix. Addition of this pair minimally altered the Kd, Kon, and Koff values from those measured for the wild-type aptamer and two other tune box variants (Table S2). In addition, we introduced the C74U mutation that alters the specificity of the aptamer for adenine, including the fluorescent derivative 2AP.16
We observe that phylogenetically observed P2 tune box sequences have a strong propensity to impart higher 2AP affinity than their non-natural counterparts (Fig. 4a; Table 1). Equilibrium dissociation constants (Kd) were determined by monitoring fluorescence of a fixed concentration of 2AP with increasing concentrations of aptamer RNA. We observed over a 100-fold range in Kd (160 nM to 17 µM) with most natural variants binding in the low nanomolar range (160–300 nM) (Fig. 4a; Table 1). The only exception is variant 8U, which binds 2AP with a Kd of ~6.2 µM. This variant is found in ~10% of sequences in our alignment yet displays a Kd ~ 10-fold higher than the non-natural variant 8A.
Fig. 4.
Summary of measured binding parameters for P2 tune box variants. Equilibrium dissociation constants (a), bimolecular association rate constants (b), and dissociation rate constants (c) for five phylogenetic aptamer variants and their corresponding non-phylogenetic variant containing an A-to-U or U-to-A mutation at position 24.
Table 1.
Binding parameters for P2 tune box variants
| Variant | Kd (nM) | koff (S−1) |
kon (×105 M−1 s−1) |
ΔtB (s) |
|---|---|---|---|---|
| 1Aa | 260±40 | 0.0088±0.0005 | 0.40±0.03 | 130 |
| 1U | 17,000±1200 | 0.15±0.001 | 0.050±0.008 | 8 |
| 2Aa | 300±20 | 0.022±0.001 | 0.23±0.02 | 53 |
| 2U | 11,000±3800 | 0.38±0.11 | 0.30±0.04 | 3 |
| 3Aa | 240±40 | 0.010±0.001 | 0.64±0.01 | 111 |
| 3U | 16,000±800 | 0.036±0.001 | 0.027±0.004 | 32 |
| 8A | 700±10 | 0.0057±0.0005 | 0.099±0.005 | 201 |
| 8Ua | 6300±600 | 0.19±0.01 | 0.33±0.01 | 6 |
| 9A | 1100±100 | 0.028±0.001 | 0.057±0.005 | 41 |
| 9Ua | 160±10 | 0.0084±0.0007 | 0.56±0.01 | 137 |
These variants are phylogenetically represented.
The kinetic properties of naturally occurring P2 tune box sequences skew towards both faster association rates (Kon) and slower dissociation rates (Koff). Kinetic measurements were made by monitoring 2AP fluorescence with a stopped-flow fluorometer using established methods. 17 Natural variants display kon values greater than all non-natural variants with the exception of variant 2A (Fig. 4b). This variant displays an observed association rate only slightly less than its non-natural variant 2U. Additionally, kon measurements do not resolve why 8U is selected over 8A given that kon for 8U is only ~3.4-fold lower than that for 8A. Dissociation rate constants were measured and an ~66-fold range in activities was observed (Fig. 4c). While most natural variants have low koff values, 8U displays a koff ~ 33-fold greater than the non-phylogenetic variant 8A.
To further understand the relationship between binding and transcription kinetics, we applied a simple model originally developed to study the pbuE adenine-responsive riboswitch.39 Assuming that a 90% fraction bound state must be achieved to exert the full regulatory response, the time required for the aptamer to reach equilibrium with respect to ligand binding, ΔtB, must be known. This time interval can be estimated by ΔtB = 1.15/koff, allowing for identification of the mode of regulatory control when compared with ΔtRNAP. In this model, thermodynamic control is defined as when ΔtRNAP is equal to or greater than ΔtB, while kinetic control is best described when ΔtB is much smaller than ΔtRNAP. The low intracellular concentrations of ligand and mRNA results in the association rate constant for ligand binding having little influence over the regulatory response in comparison to the dissociation rate constant according to the relaxation equation
| (1) |
where the time constant t equals the time required for e−1 of a reaction to occur.39
ΔtB was calculated for all variants measured (Table 1) for comparison with a theoretical ΔtRNAP. For an average expression platform of ~ 150 nt and a transcription rate of bacterial RNAP of ~0 nt/s,42,43 ΔtRNAP is estimated to be approximately 3 s. Note that transcription factors such as NusA, programmed pauses in the riboswitch, and intracellular nucleotide pool can significantly alter ΔtRNAP such that 3 s represents a minimum value.44 In a recent study of the folding of the Escherichia coli btuB adenosylco-balamin riboswitch, pause sites in the expression platform significantly increase ΔtRNAP, assisting in the proper folding of the aptamer domain.45 Notably, aptamer variants containing phylogenetically represented P2 tune boxes display substantially slower dissociation rates resulting in high ΔtB values— greater than 50 s. These RNAs are highly likely to operate under kinetic control, given the above caveats. Conversely, non-phylogenetically represented variants tend to have smaller ΔtB values, suggesting that they might be under near thermodynamic control in the cell. The only natural variant to display a large koff and thus a short ΔtB (6 s) is variant 8U and, therefore, may operate under weak thermodynamic control.
Nucleotides in the P2 tune box display functional covariation
The above data argue that nucleotide covariation within the P2 tune box has a significant impact upon the aptamer domain's binding properties. To determine whether this sequence element also impacts the regulatory properties of the riboswitch, we investigated the ability of variants 1A, 1U, 9A, and 9U to regulate expression of a reporter gene in a ligand-dependent manner. To perform this analysis, we fused the B. subtilis xpt-pbuX aptamer domain onto the B. subtilis metE SAM-I riboswitch expression platform to decouple potential sequence-specific aptamer domain/expression platform interactions. The metE expression platform is highly functional in both in vitro and in vivo assays and can host a diverse array of biological and SELEX-derived aptamers (P. Ceres and R.T.B., unpublished results). Any differences in regulatory activity observed between aptamer variants can be directly attributed to differences in the binding properties of 2AP to the aptamer and its ability to influence the secondary structural switch of the expression platform. This method has been previously used to analyze the regulatory properties of the tetrahydrofolate riboswitch.46
Each chimeric riboswitch was tested for its ability to repress expression of GFPuv in E. coli as a function of 2AP concentration in a defined medium. In the context of the chimeric riboswitch, only the two natural aptamer variants can functionally repress green fluorescent protein (GFP) expression. The wild-type xpt-pbuX aptamer, corresponding to the variant 1A, exhibits an ~8-fold decrease in GFP expression with an EC50 (defined as the concentration of ligand in the media required to elicit a half-maximal regulatory response) of 600±80 uM 2AP (Fig. 5). The phylogenetically represented P2 tune box variant 9U sequence shows a similar degree of repression (620±20 µM 2AP). Conversely, the non-natural P2 tune box variants 1U and 9A are highly deleterious to regulatory activity; both variants exhibit very little repression activity. These data show that a deleterious mutation in nucleotide 24 can be compensated for by a change in the first two pairs of the P2 helix. Thus, in a regulatory context, the identities of the unpaired nucleotide at position 24 and the two base pairs in P2 proximal to the junction are dependent upon each other—clearly supporting the statistical coupling analysis. This result demonstrates how small changes in helix composition adjacent to the effector-binding site can dramatically impact riboswitch regulation in vivo, providing a potential mechanism for riboswitch activity tuning.
Fig. 5.
In vivo regulation of gene expression by a purine/ SAM hybrid riboswitch. Aptamer variants 1A, 1U, 9A, and 9U were fused to the Bacillus subtilis metE expression platform and were assayed for the ability to terminate transcription in response to increasing 2AP concentrations. Transcriptional termination was measured by a decrease in GFP signal corrected by optical density.
Structures of different P2 tune-box variants are equivalent
To determine whether the binding and potential regulatory differences between variants might be due to alterations in the structure of the binding pocket, we solved the crystal structure of variants 1U, 9A, and 9U. Variant 9U was selected as this aptamer is well represented in natural sequences yet contains significant differences as compared to variant 1A (“wild-type” xpt-pbuX) such as an A.A at the base of P2 and a uracil at position 24 (Fig. 3d). Additionally, variants 1U and 9A were analyzed to allow a comparison to non-natural variants. To confirm that the C74U mutation was non-perturbing to the structure, we determined the structures of the 9U and 1U variants as both C74:HX and U74:2,6-diaminopurine (DAP) complexes (note that DAP is an analog of 2AP and recognizes the aptamer in an identical fashion16,17). Each of the RNAs crystallized under similar conditions used previously and with nearly identical unit cell parameters.16,17,47 Furthermore, data were collected for all structures to between 1.32 and 1.7 Å resolution, with high-quality crystallographic and model statistics (see Table S3). The Luzzati cross-validated coordinate error of each structure is below 0.2 Å, enabling us to accurately compare small differences between each of these structures.
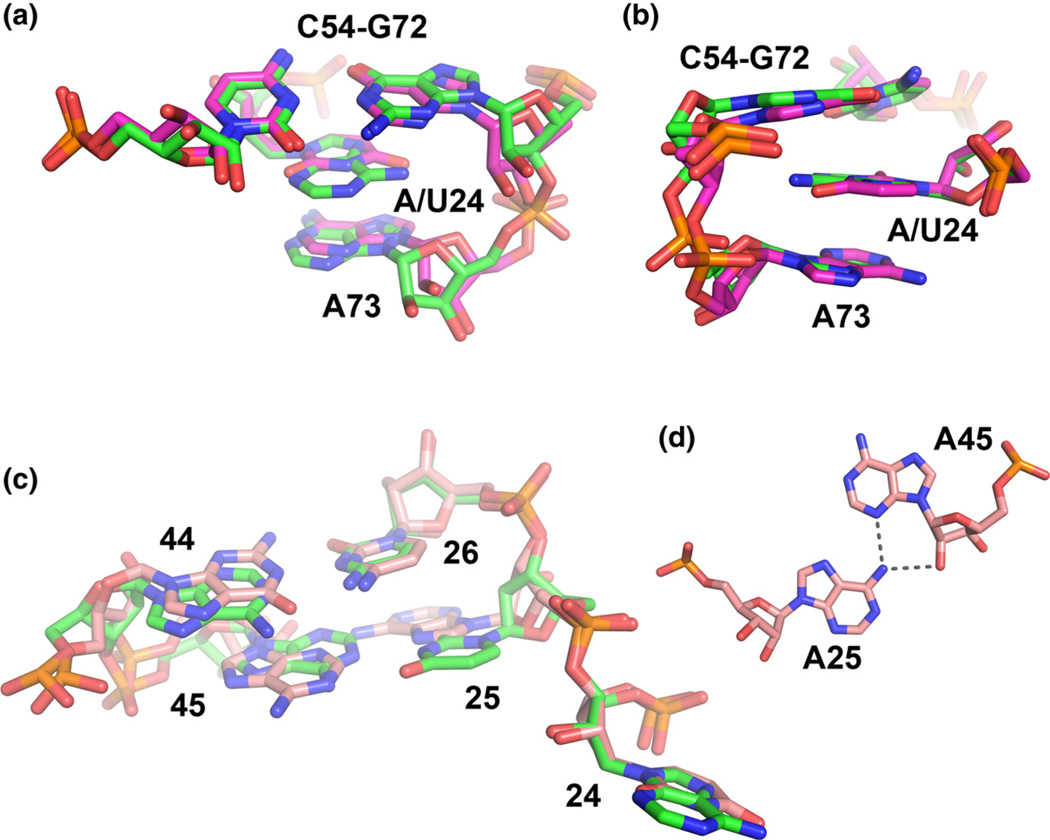
As expected, the global geometry of all RNAs and the pattern of hydrogen bonding interactions between the bases and ligand in the binding pocket is unaltered between the different variants (global pairwise r.m.s.d. between any of the structures does not exceed 0.33 Å). Changing the specificity of the binding pocket through the C74U mutation is also non-perturbing to RNA structure and interactions with the ligand, consistent with previous observations.17 Thus, the use of the C74U mutation in conjunction with 2AP binding does not appreciably alter the complex. At the local level, the sequence variations in the P2 tune box perturb the structure but in a fashion that does not alter interactions with the ligand. In the 1U and 9U structures, U24 is positioned in the same orientation as adenosine, inserting itself between G72 and A73 (Fig. 6a). However, the pyrimidine ring does not stack upon A73, as is observed for the A24 variant. Also, both the 1U, 9U(C74:HX) and 9U(C74U:DAP) structures exhibit small differences in the backbone geometry as compared to the 1A, 9A(C74:HX) and 9A(C74U: DAP) structures. These differences involve the A73 base, which is pulled into the helix by ~ 1 Å in the A24 variants, allowing it to more efficiently stack upon pyrimidine 74 as well as a 1.6-Å displacement of the phosphate of A73 relative to the U24 variant (Fig. 6b).
Fig. 6.
Structural analyses of select tune box variants, (a) Perspective showing the superimposition of the 1A and 9U structures around nucleotide 24. In both structures, the nucleotide is positioned between A73 of J3/1 and the first base pair of P3. (b) Side view of the perspective shown in (a), emphasizing the stacking relationships.(c) Superimposition of the entire P2 tune box of the 1A and 9U structures. Note the local differences in the positioning of the P2.1 pair between the two variants, but the backbone in the area is nearly identical (d) Hydrogen bonding between nucleotides 25 and 45 in the 9A (shown) and 9U variants.
The alternative pairing in P2 of the 9A/9U variants also introduces small changes in the geometry of the RNA. The first pair, which is changed from a standard Watson–Crick A–U pair (1A, 1U) to an A.A mismatch, forms a sheared A.A mismatch in which A25(N6) forms two hydrogen bonds with A45(N3) and A45(02′) (Fig. 6c and d). This type of pair is isosteric with the commonplace G.A sheared pair, as exemplified by that within GNRA-type tetraloops.48 The second pair is changed from the single-bond A26–C44 base pair found in the wild-type xpt aptamer to a standard Watson-Crick C–G base pair. The introduction of the sheared A.A mismatch in 9A/9U alters the position and conformation the backbone at nucleotide 25, particularly at C5′ (Fig. 6c). Thus, one potential explanation for the compensatory nature of positions 24 and 25 is that the change in backbone geometry at nucleotide 25 in J1/2 facilitates the positioning of U24 and stabilizes changes in the backbone around nucleotide 73 in J3/ 1. While all of these differences are small, it must be emphasized that they are significantly larger than the coordinate error of any of the individual structures (Table S3) and thus likely reflect real differences in the local architecture of the aptamer.
Probing the conformational flexibility of xpt variants using SHAPE
The above crystal structures inform only the ligand-bound state of the aptamer. However, the structure and conformational flexibility of the unbound state clearly have an impact upon the binding properties of the aptamer. To address this aspect of the RNA, we employed a chemical probing technique called selective 2′-hydroxyl acylation analyzed by primer extension or SHAPE.49,50 This technique, which has emerged as a major method for probing RNA structure and folding, uses an acid anhydride reagent (in this study, N-methylisatoic anhydride, or NMIA) that reacts with 2′-hydroxyl groups in the backbone. The product of the reaction is an adduct that is subsequently read out by sequencing the reverse-transcribed cDNA. Importantly, it has been demonstrated that the reactivity of an individual 2′-hydroxyl group in the RNA is largely dependent upon its conformational flexibility.49,51Studies that have compared the reactivity patterns of small model RNAs have shown that it correlates to the NMR order parameter S2, another measure of local conformational dynamics in a molecule52 as well as 2AP fluorescence.53 We have previously employed this method to investigate the structure and folding of several riboswitch aptamers, including the B. subtilis xpt-pbuX guanine riboswitch.36,54,55
The variants utilized in this study were probed with NMIA at 25 °C under the buffer conditions used in the binding experiments in the presence and absence of 2AP (Fig. 7). In each RNA, we observe regions that display strong differences in their reactivity pattern between the unbound and bound states: nucleotides within J1/2, J2/3, and the base of P2. Furthermore, all RNAs have nearly identical patterns of reactivity in the presence of a saturating amount of 2AP (1 mM) such as a strong site of reactivity at U48 that is flipped out towards solvent in crystal structures. This pattern is consistent with productive binding of the ligand. This demonstrates that all of the RNAs interact with the ligand in the same fashion and adopt the same final structure.
Fig. 7.
SHAPE chemical probing identifies regions of the aptamer domain that are conformationally flexible. (a) Aptamer variants 1 A, 1U, 9A, and 9U were probed in the absence of NMIA and ligand (control) or in the presence of NMIA with and without ligand (lanes −2AP and +2AP, respectively). Conformationally flexible regions are indicated by primer extension stops with the intensity reflecting the proportion of the population that is conformationally dynamic (b) SHAPE probing of aptamer variants 2A, 2U, 3A, 3U, 8A, and 8U. Control and sequencing lanes, although omitted from these panels, were run alongside these lanes.
In the free state, all of the RNAs display differences in their reactivity patterns in two key regions. Within J2/3, most RNAs show reactivity of nucleotides 43–53 to varying extents, with nucleotides U48 and U51 showing the greatest amount of reactivity in all variants. The high degree of similarity of the reactivity pattern of these nucleotides, as determined through quantification of band intensities, reveals that the degree of conformational flexibility of J2/3 is not significantly affected by mutations at position 24 or the 25–45/26–44 base pairs in P2. The only variant with any significant differences in reactivity in this region is 9A (Fig. 7a), which has increased reactivity throughout this region as well as J3/1 (data not shown).
In contrast, J1/2 has different patterns of reactivity between the variants. The wild-type RNA shows reactivity towards NMIA between nucleotides 21 and 26 (Fig. 7a), with nucleotides A23 and A24 consistently displaying higher reactivity than the surrounding J1/2-P2 region. Comparison of this pattern of reactivity to the other variants used in this study (Fig. 7) reveals that each RNA has a different pattern of reactivity in this region. Unfortunately, no clear trend that correlates the degree of reactivity (reflecting conformational flexibility or dynamics) to 2AP binding properties is observed in these data. Upon binding 2AP, the J1/2-P2 region of all variants did establish nearly the same reactivity pattern (except for small differences in nucleotide 23). Therefore, while variation in the P2 tune box clearly alters the local pattern of nucleotide conformational flexibility, these changes in flexibility cannot be directly interpreted in the context of the variable binding parameters they yield.
Discussion
Using a structure-based sequence alignment and an NMI scoring method, we identified functionally coupled nucleotides adjacent to the ligand-binding pocket at the base of P2 that covary in a manner that alters ligand-binding activity. These nucleotides comprise the P2 tune box and reside in the junction between the P2 stem and the ligand-binding site in the three-way junction. Measurement of the binding properties of 2AP for select variants of this region reveals that this site has a large effect on the rate of ligand dissociation (koff) and, thereby, the equilibrium dissociation constant (Kd). The association rate constant is also affected; however, due to the rapid nature of ligand binding compared to ΔtRNAP, the impact on activity resulting from alterations to kon is minimal compared to koff. This site appears to be a major source of affinity tuning immediately surrounding the binding pocket since other base-pairing regions adjacent to the binding pocket, such as the top of P1 (A21–U75, U20–A76) and the base of P3 (C54–G72), are highly conserved. These in vitro observations were supported by the in vivo activity of several variants of the P2 tune box that clearly demonstrate covariation of nucleotide 24 and the first two pairs in the P2 helix.
A study of the 11 SAM-I riboswitches in B. subtilis revealed a correlation between the regulatory activity and the gene class that they control and concluded that individual riboswitches are tuned to meet the regulatory requirements of their transcriptional unit.9 In this study, we observed no clear correlation between the classes of P2 tune boxes and nature of the transcriptional unit they regulate (data not shown). The primary difference between these two studies is that in the current work, the riboswitch sequences are obtained from a wide spectrum of species that live in different environmental conditions. It is reasonable to speculate that variability in extracellular and intracellular conditions would have a significant influence on the aptamer. Other unknown variables such as additional levels of regulation at the DNA level (by the purR repressor protein, for example56) further complicate such an analysis. Thus, while regulatory tuning is evident between riboswitches within a single organism, this observation likely cannot be extended over all sequences of a particular class of riboswitches.
The P2 tune box has a role not only in modulating the activity of the purine riboswitch but also in specificity. The purine riboswitch family is divided into three distinct binding classes: guanine,5 adenine,14 and 2′-deoxyguanosine.57 In a study that dissected the differences between the xpt-pbuX guanine and Mesoplasma florum type IA 2′-deox-yguanosine aptamers, a single nucleotide difference at the ligand-binding site determines selectivity between the two effector molecules.24 However, it was also found that nucleotides at the base of P2 are responsible for further increasing the selectivity of the RNA for 2′-deoxyguanosine. In the crystallo-graphic structure of the wild-type M. florum IA aptamer complexed to 2′-deoxyguanosine, the equivalent of nucleotide 24 (G33) directly interacts with the 25–45 pair to form a base triple25 in a very different conformation than that observed in the xpt-pbuX aptamer. These observations argue that the P2 tune box is an important feature for influencing the specificity switch from guanine/adenine to 2′-deoxyguanosine in the purine riboswitch family.
The role of the P2 tune box appears to be to modulate the stability of the ligand-aptamer complex. A detailed analysis of the crystal structures of the 1A, 1U, 9A, and 9U variants reveals that each adopts a nearly identical conformation, indicating that the P2 tune box has little impact on the final architecture of the complex. Chemical probing of the dynamics of each variant reveals that in each RNA, the most dynamic elements in the absence of ligand are J1/2 and J2/3, consistent with previous work. In each of the variants (Fig. 7), the pattern of reactivity in J2/3 is virtually identical, indicating that the P2 tune box likely does not affect the dynamics of J2/3. Instead, we observe idiosyncratic patterns of reactivity for J1/2 (nucleotides 21–23) and nucleotides 24 and 25. Further, we do not observe consistently greater reactivity in non-natural variants or other systematic differences that would distinguish the two sets of RNAs. Nonetheless, the binding data strongly suggest that the non-natural variants are less stable by virtue of faster dissociation rates that confer a kinetically controlled mode of regulation. This kinetic control can lead to novel properties of the riboswitch that cannot be imparted by thermodynamically controlled regulatory mechanisms. In a recent study of the lysine riboswitch, we demonstrate that this phenomenon resulted in a riboswitch that is sensitive to both effector and nucleotide triphosphate (NTP) concentrations under biological conditions, the latter being a proxy for the overall metabolic state of the cell.44
Tuning the dynamics of RNAs appears to be an important aspect of the biological function of a wide assortment of biological RNAs, including riboswitches.58,59 Analysis of the unbound SAM-I riboswitch aptamer using a variety of structural and computational approaches revealed that it transiently samples “bound-like conformations”.54 Similar behavior has been observed in the SAM-II,60–61 FMN, 55 and preQ1 riboswitch aptamers, 62 suggesting that the conformational selection is a broadly utilized means of efficient binding by the effector molecule. Beyond riboswitches, it has been shown that helical sequences adjacent to an internal bulge within the HIV transactivation region significantly influence the binding properties of small-molecule compounds.63 In this work, we also observe that naturally occurring P2 tune boxes tend to have faster association rates, which in the conformational selection model is the result of increased sampling of the bound-like conformations recognized by the effector. Our data provide the first mechanistic insights into how a specific sequence module can be employed to tune both binding and regulatory activities of riboswitches and establish a framework for understanding how activity tuning is achieved in other RNAs.
Materials and Methods
Bioinformatics
The starting alignment was obtained from the RNASTAR structural RNA alignment repository23 (RST00106). This alignment was chosen as a starting point because we wanted a high-quality alignment, which was backed by a crystal structure. From this starting alignment, we included all sequences that contain seven base pairs in P2 and P3 helix and a fixed number of bases in J1/2 (3 nt), L2 (7 nt), J2/3 (8 nt), L3 (7 nt), and J3/1 (2 nt).
The covariation analysis was performed using the implementation of NMI presented in Caporaso et al.35 The result of the NMI analysis is a matrix of values, where the rows and columns represent indices in the sequence alignment. Each value in the matrix is the NMI score between the first position (row) and the second position (column). The significance of each pairwise nucleotide interaction was assessed by comparing the NMI value of reference nucleotide (row) and second nucleotide (column) with the NMI value of all other nucleotides compared to the reference position (all values in a row), using a one-tailed, two-sample t test. The matrix containing the NMI values was plotted as a heat map (Fig. 3a), where cooler colors represent little or no covariation and warmer values represent higher levels of covariation. The positional entropy for the alignment for Fig. 1b was calculated using the PyCogent package.64 Figure 1b is a sequence logo representing the nucleotide conservation as well as information content at each position in the alignment.65 For each position, the height of each nucleotide symbol represents its frequency at that position, while the overall height represents the information content at that position.
Binding constant measurements
Aptamer domain RNAs were prepared in 10 mM K+ Hepes (pH 7.5), 100 mM KCl, and 10 mM MgCl2 and were thermally denatured at 90 °C for 1 min followed by a 10-min incubation on ice to promote proper refolding. Increasing concentrations of RNA were rapidly mixed at 30 °C with a limiting concentration of the fluorescent nucleobase 2AP (50 nM) on an SX.17MV stopped-flow spectrofluorometer (Applied Photophysics). 2AP binding was detected using a 335-nm long-pass filter and an excitation wavelength of 318 nm. A total of 4000 data points were collected for each injection. The instrument has a dead time of 2 ms. Observed rate data were fit to the model
| (2) |
where Ft is the observed fluorescence at time point t, F∞ is the fluorescence at equilibrium, and kobs is the measured rate constant. To determine the association rate constant, we determined kobs at four different 2AP concentrations and we fit the plot of kobs versus [2AP] to
| (3) |
The raw fluorescence data are shown in Fig. S1 and the kobs versus [2AP] data and fits are shown in Fig. S2.
Dissociation rate constants were measured using stopped-flow fluorescence and the non-fluorescent competitor ligand DAP. A limiting amount of 2AP was pre-incubated with an excess of aptamer domain RNA such that 80–100% of the ligand was bound by RNA; however, the absolute concentration of RNA was variable due to changes in the equilibriumdissociation constant between aptamer variants. The aptamer-2AP complex was rapidly mixed with 10 mM DAP (5 mM final concentration) and the observed fluorescence as a function of time was fit to the equation
| (4) |
where Ft is the observed fluorescence at time point t, F∞ is the fluorescence at equilibrium, F0is the initial fluorescence, and kobs is the observed dissociation rate constant. The raw fluorescence data are shown in Fig. S3.
Equilibrium dissociation constants were measured using a Safire-ll fluorescence plate reader (Tecan) under identical buffer conditions at room temperature. Increasing concentrations of aptamer RNA were titrated against 50 nM 2AP and fluorescence quenching was measured after a 10-min incubation. Data were fit by a two-state binding model
| (5) |
where F is the observed fluorescence, Fmin is the minimal observed fluorescence, ΔF is the fluorescence change, and Kd is the apparent dissociation constant. The raw data and fits to Eq. (5) are shown in Fig. S4.
In vivo activity of aptamer variants
The regulatory activity of the 1A, 1U, 9A, and 9U aptamer variants was fused to the B. subtilis metE riboswitch expression platform using a strategy previously employed to measure the activity of a cobalamin riboswitch66 and tetrahydrofolate aptamer domain.46 After the resultant chimeras were cloned into the leader sequence of an mRNA encoding gfpuv using standard molecular biological techniques, they were transformed into E. coli strain BW25113(Δnep)67. nep encodes the purine base efflux pump.68,69 Single colonies were grown overnight in 3 ml of a rich defined medium supplemented with 100 µg/ml ampicillin. This starter culture was used to inoculate 100 ml of the same defined media and was allowed to grow to early exponential phase (OD600=0.1–0.5) at 37 °C. 2AP was added to 3-ml aliquots and the cells were allowed to grow for an additional 6 h at 37 °C. We used 300 µl of each culture to measure the OD600 and fluorescence intensity of the cells in an Infinity M200 Pro fluorescence plate reader (Tecan). Fluorescent measurements were taken at an excitation wavelength of 395 nm and average fluorescence was taken from 513 to 515 nm, the maximum emission for GFP. Optical-density-normalized fluorescence values were plotted against the ligand concentration and were fit to a standard two-state binding equation to determine the EC50 (concentration of 2AP in the medium required to elicit half-maximal repression of GFP expression). Background cellular fluorescence was determined by performing an identical 2AP titration into cells carrying pBR322 (the parental vector of the GFP reporter). An average of the background was subtracted from the values of the cultures containing the plasmids with the riboswitch constructs to calculate the cell-normalized fluorescence.
Crystallographic data collection and analysis
RNA variants used in this study were transcribed in vitro and purified using precisely described methods.15,17 Briefly, RNA was transcribed off a double-stranded DNA template using T7 RNAP and purified using 12% denaturing polyacrylamide gel electrophoresis. RNA was gel extracted into 0.5× TE buffer. Crystals were obtained by the hanging drop vapor diffusion method. One microliter of RNA/ligand stock (500 µM RNA, 1 mM HX or 2AP) was mixed with 1 µl of mother liquor (10 mM Hepes, pH 7.5, 15–20% polyethylene glycol 3000, 8–12 mM cobalt hexamine, and 450 to 740 mM ammonium acetate). The well contained 500 µl of mother liquor. Crystals grew to a maximum size in about 5 days' time with a needle-like morphology. Crystals were cryoprotected in 20–30% ethylene glycol or polyethylene glycol 3000 in mother liquor and flash frozen. Data were collected either on a home X-ray source (Rigaku RU200 copper rotating anode source equipped with a Rigaku IV++ area detector) or at the National Synchrotron Light Source at Brookhaven on beamline X29. All data were indexed, integrated, and scaled using D*TREK70 in the crystal clear package (Rigaku MSC).
Molecular replacement was used to create initial electron density maps in CNS71 with the structure of the B. subtilis xpt-pbuX guanine riboswitch aptamer domain in complex with HX used as the starting model [with all ligand and solvent molecules stripped; Protein Data Bank (PDB) ID 1U8D].15 The RNA was refined with CNS using iterative rounds of rebuilding nucleotides that were different using PyMOL (Schrodinger), simulated annealing, and B-factor refinement. All models were refined using isotropic B-factors, regardless of resolution. Ligand, solute molecules, and water were subsequently built and refined to yield the final model. Coordinates of final models and structure factors have been deposited in the RCSB PDB.
SHAPE chemical probing of RNA
RNA was transcribed in vitro as described above with a structure cassette appended at the 3′ end.50 Two picomoles of RNA in 12 µl of 0.5× TE was denatured at 90 °C for 2 min and put on ice for 10 min to refold. Six microliters of folding buffer (333 mM Hepes, pH 8.0, 333 mM NaCl, and 20 mM MgCl2) and 1 mM 2AP were added to RNA mix. Nine microliters of mix was incubated with 1 µl of dimethyl sulfoxide (control) or 130 mM NMIA in dimethyl sulfoxide and incubated at 37 °C for 3 h. Three microliters of 0.3 uM 32P reverse transcription primer was added and incubated at 65 °C for 5 min followed by 35 °C for 15 min. Six microliters of enzyme mix (167 mM Tris, pH 8.3, 250 mM KCl, 1.67 mM dNTPs, 17 mM DTT, and 0.33 units of Superscript III Reverse Transcriptase) were incubated at 52 °C to extend the primer. The reaction was terminated after a 10-min extension by the addition of 1 µl of 4 M NaOH and incubation at 90 °C for 5 min. The incubation continued for an additional 5 min after the addition of 29 µl of acid stop mix [4:25 (v/v) mixture of unbuffered Tris and RNA load buffer]. Sequencing reactions were preformed similarly as above but with supplementation of the enzyme mix with 3.0 mM ddNTP. Extension products were resolved on a 12% 29:1 acrylamide:bisacrylamide gel and were subsequently imaged with a Typhoon Phosphorimager (Molecular Dynamics). Gel images were quantified with the program SAFA72 using methods consistent with prior footprinting analysis of the B. subtilis xpt guanine riboswitch aptamer domain.36
Accession numbers
Coordinates and structure factors have been deposited in the PDB under the following accession numbers: 4FE5 (variant 1A:HX), 4FEJ (variant 1U:HX), 4FEL (variant 9A: HX), 4FEO (variant 9A:DAP), 4FEN (variant 9U:HX), and 4FEP (variant 9U C74U:HX).
Supplementary Material
Acknowledgements
R.T.B. would like to thank Drs. Jane Richardson and Laura Murray for their assistance in correcting several backbone errors in the original model of the xpt purine riboswitch (PDB ID 1U8D). This work was supported by grants from the National Institutes of Health (GM073850 and 1S10-RR026516) to R.T.B. and from the National Aeronautics and Space Administration (NNX08AP60G) and Howard Hughes Medical Institute to R.K. J.J.T. was supported by a Creative Training in Molecular Biology predoctoral training grant (T32-GM07135) and J.G.M.-V. was supported by the National Institutes of Health/CU Molecular Biophysics Training Program (T32 GM065103).
Abbreviations used
- 2AP
2-aminopurine
- DAP
2,6-diaminopurine
- GFP
green fluorescent protein
- HX
hypoxanthine
- MI
mutual information
- NMI
normalized mutual information
- NMIA
N-methylisatoic anhydride
- NTP
nucleotide triphosphate
- PDB
Protein Data Bank
- RNAP
RNA polymerase
- SAM
S-adenosylmethionine
- SHAPE
selective 2′-hydroxyl acylation analyzed by primer extension
Footnotes
Author Contributions. C.D.S. and J.W. performed the structure-based alignment of purine riboswitches and analysis of covariation patterns, C.D.S. made all binding affinity and kinetics measurements and analyzed the resultant data, J.J.T. performed the chemical probing analysis of all aptamers, J.J.T. crystallized select RNAs and collected data, J.J.T. and R.T.B. built the crystallographic models, J.G.M.-V. performed the in vivo analysis of select mutants, and all authors contributed to the writing of the manuscript.
Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jmb.2013.02.023
References
- 1.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 2.Roth A, Breaker RR. The structural and functional diversity of metabolite-binding riboswitches. Annu. Rev. Biochem. 2009;78:305–334. doi: 10.1146/annurev.biochem.78.070507.135656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barrick JE, Breaker RR. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007;8:R239. doi: 10.1186/gb-2007-8-11-r239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- 6.Mulhbacher J, Lafontaine DA. Ligand recognition determinants of guanine riboswitches. Nucleic Acids Res. 2007;35:5568–5580. doi: 10.1093/nar/gkm572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundy FJ, Henkin TM. The S box regulon: a new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol. Microbiol. 1998;30:737–749. doi: 10.1046/j.1365-2958.1998.01105.x. [DOI] [PubMed] [Google Scholar]
- 8.McDaniel BA, Grundy FJ, Artsimovitch I, Henkin TM. Transcription termination control of the S box system: direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl Acad. Sci. USA. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomsic J, McDaniel BA, Grundy FJ, Henkin TM. Natural variability in S-adenosylmethionine (SAM)-dependent riboswitches: S-box elements in Bacillus subtilis exhibit differential sensitivity to SAM In vivo and in vitro. J. Bacteriol. 2008;190:823–833. doi: 10.1128/JB.01034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winkler WC, Nahvi A, Sudarsan N, Barrick JE, Breaker RR. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- 11.Lu C, Ding F, Chowdhury A, Pradhan V, Tomsic J, Holmes WM, et al. SAM recognition and conformational switching mechanism in the Bacillus subtilis yitJ S box/SAM-l riboswitch. J. Mol. Biol. 2010;404:803–818. doi: 10.1016/j.jmb.2010.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montange RK, Batey RT. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature. 2006;441:1172–1175. doi: 10.1038/nature04819. [DOI] [PubMed] [Google Scholar]
- 13.Kim JN, Breaker RR. Purine sensing by riboswitches. Biol. Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 14.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 15.Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- 16.Gilbert SD, Reyes FE, Edwards AL, Batey RT. Adaptive ligand binding by the purine riboswitch in the recognition of guanine and adenine analogs. Structure. 2009;17:857–868. doi: 10.1016/j.str.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert SD, Stoddard CD, Wise SJ, Batey RT. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J. Mol. Biol. 2006;359:754–768. doi: 10.1016/j.jmb.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Noeske J, Richter C, Grundl MA, Nasiri HR, Schwalbe H, Wohnert J. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc. Natl Acad. Sci. USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serganov A, Yuan YR, Pikovskaya O, Polons-kaia A, Malinina L, Phan AT, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem. Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Batey RT. Structure and mechanism of purine-binding riboswitches. Q. Rev. Biophys. 2012;45:345–381. doi: 10.1017/S0033583512000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Widmann J, Stombaugh J, McDonald D, Chocho-lousova J, Gardner P, Iyer MK, et al. RNASTAR: an RNA STructural Alignment Repository that provides insight into the evolution of natural and artificial RNAs. RNA. 2012;18:1319–1327. doi: 10.1261/rna.032052.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards AL, Batey RT. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J. Mol. Biol. 2009;385:938–948. doi: 10.1016/j.jmb.2008.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pikovskaya O, Polonskaia A, Patel DJ, Serganov A. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat. Chem. Biol. 2011;7:748–755. doi: 10.1038/nchembio.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson JD, Holbrook SR, Katoh K, Koehl P, Moras D, Westhof E, Poch O. MAO: a Multiple Alignment Ontology for nucleic acid and protein sequences. Nucleic Acids Res. 2005;33:4164–4171. doi: 10.1093/nar/gki735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, et al. The RNA structure alignment ontology. RNA. 2009;15:1623–1631. doi: 10.1261/rna.1601409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cruz JA, Westhof E. Sequence-based identification of 3D structural modules in RNA with RMDetect. Nat. Methods. 2011;8:513–521. doi: 10.1038/nmeth.1603. [DOI] [PubMed] [Google Scholar]
- 29.Fodor AA, Aldrich RW. Influence of conservation on calculations of amino acid covariance in multiple sequence alignments. Proteins. 2004;56:211–221. doi: 10.1002/prot.20098. [DOI] [PubMed] [Google Scholar]
- 30.Griffiths-Jones S, Bateman A, Marshall M, Khanna A, Eddy SR. Rfam: an RNA family database. Nucleic Acids Res. 2003;31:439–441. doi: 10.1093/nar/gkg006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lemay JF, Lafontaine DA. Core requirements of the adenine riboswitch aptamer for ligand binding. RNA. 2007;13:335–350. doi: 10.1261/rna.142007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Freyhult E, Gardner PP, Moulton V. A comparison of RNA folding measures. BMC Bioinformatics. 2005;6:241. doi: 10.1186/1471-2105-6-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindgreen S, Gardner PP, Krogh A. Measuring covariation in RNA alignments: physical realism improves information measures. Bioinformatics. 2006;22:2988–2995. doi: 10.1093/bioinformatics/btl514. [DOI] [PubMed] [Google Scholar]
- 34.Martin LC, Gloor GB, Dunn SD, Wahl LM. Using information theory to search for coevolving residues in proteins. Bioinformatics. 2005;21:4116–4124. doi: 10.1093/bioinformatics/bti671. [DOI] [PubMed] [Google Scholar]
- 35.Caporaso JG, Smit S, Easton BC, Hunter L, Huttley GA, Knight R. Detecting coevo-lution without phylogenetic trees? Tree-ignorant metrics of coevolution perform as well as tree-aware metrics. BMC Evol. Biol. 2008;8:327. doi: 10.1186/1471-2148-8-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delfosse V, Bouchard P, Bonneau E, Dagenais P, Lemay JF, Lafontaine DA, Legault P. Riboswitch structure: an internal residue mimicking the purine ligand. Nucleic Acids Res. 2010;38:2057–2068. doi: 10.1093/nar/gkp1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garst AD, Batey RT. A switch in time: detailing the life of a riboswitch. Biochim. Biophys. Acta. 2009;1789:584–591. doi: 10.1016/j.bbagrm.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wickiser JK, Cheah MT, Breaker RR, Crothers DM. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- 40.Wickiser JK, Winkler WC, Breaker RR, Crothers DM. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- 41.Lemay JF, Desnoyers G, Blouin S, Heppell B, Bastet L, St-Pierre P, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328:504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garst AD, Porter EB, Batey RT. Insights into the regulatory landscape of the lysine riboswitch. J. Mol. Biol. 2012;423:17–33. doi: 10.1016/j.jmb.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perdrizet GA, 2nd, Artsimovitch I, Furman R, Sosnick TR, Pan T. Transcriptional pausing coordinates folding of the aptamer domain and the expression platform of a riboswitch. Proc. Natl Acad. Sci. USA. 2012;109:3323–3328. doi: 10.1073/pnas.1113086109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trausch JJ, Ceres P, Reyes FE, Batey RT. The structure of a tetrahydrofolate-sensing riboswitch reveals two ligand binding sites in a single aptamer. Structure. 2011;19:1413–1423. doi: 10.1016/j.str.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert SD, Mediatore SJ, Batey RT. Modified pyrimidines specifically bind the purine riboswitch. J. Am. Chem. Soc. 2006;128:14214–14215. doi: 10.1021/ja063645t. [DOI] [PubMed] [Google Scholar]
- 48.Leontis NB, Westhof E. Conserved geometrical base-pairing patterns in RNA. Q. Rev. Biophys. 1998;31:399–55. doi: 10.1017/s0033583599003479. [DOI] [PubMed] [Google Scholar]
- 49.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acyla-tion and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 50.Wilkinson KA, Merino EJ, Weeks KM. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 51.Wilkinson KA, Vasa SM, Deigan KE, Mortimer SA, Giddings MC, Weeks KM. Influence of nucleotide identity on ribose 2′-hydroxyl reactivity in RNA. RNA. 2009;15:1314–1321. doi: 10.1261/rna.1536209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Souliere MF, Haller A, Rieder R, Micura R. A powerful approach for the selection of 2-aminopurine substitution sites to investigate RNA folding. J. Am. Chem. Soc. 2011;133:16161–16167. doi: 10.1021/ja2063583. [DOI] [PubMed] [Google Scholar]
- 54.Stoddard CD, Montange RK, Hennelly SP, Rambo RP, Sanbonmatsu KY, Batey RT. Free state conformational sampling of the SAM-I riboswitch aptamer domain. Structure. 2010;18:787–797. doi: 10.1016/j.str.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vicens Q, Mondragon E, Batey RT. Molecular sensing by the aptamer domain of the FMN riboswitch: a general model for ligand binding by conformational selection. Nucleic Acids Res. 2011;39:8586–8598. doi: 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansen LE, Nygaard P, Lassen C, Agerso Y, Saxild HH. Definition of a second Bacillus subtilis pur regulon comprising the pur and xpt-pbuX operons plus pbuG, nupG (yxjA), and pbuE (ydhL) J. Bacteriol. 2003;185:5200–5209. doi: 10.1128/JB.185.17.5200-5209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim JN, Roth A, Breaker RR. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl Acad. Sci. USA. 2007;104:16092–16097. doi: 10.1073/pnas.0705884104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liberman JA, Wedekind JE. Riboswitch structure in the ligand-free state. Wiley Interdiscip. Rev.: RNA. 2012;3:369–384. doi: 10.1002/wrna.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haller A, Rieder U, Aigner M, Blanchard SC, Micura R. Conformational capture of the SAM-II riboswitch. Nat. Chem. Biol. 2011;7:393–400. doi: 10.1038/nchembio.562. [DOI] [PubMed] [Google Scholar]
- 61.Chen B, Zuo X, Wang YX, Dayie TK. Multiple conformations of SAM-II riboswitch detected with SAXS and NMR spectroscopy. Nucleic Acids Res. 2012;40:3117–3130. doi: 10.1093/nar/gkr1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J, Walter NG, Brooks CL., 3rd Cooperative and directional folding of the preQ1 riboswitch aptamer domain. J. Am. Chem. Soc. 2011;133:4196–4199. doi: 10.1021/ja110411m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stelzer AC, Kratz JD, Zhang Q, Al-Hashimi HM. RNA dynamics by design: biasing ensembles towards the ligand-bound state. Angew. Chem. 2010;49:5731–5733. doi: 10.1002/anie.201000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Knight R, Maxwell P, Birmingham A, Carnes J, Caporaso JG, Easton BC, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fowler CC, Brown ED, Li Y. Using a riboswitch sensor to examine coenzyme B(12) metabolism and transport in E. coli . Chem. Biol. 2010;17:756–765. doi: 10.1016/j.chembiol.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 67.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nygaard P, Saxild HH. The purine efflux pump PbuE in Bacillus subtilis modulates expression of the PurR and G-box (XptR) regulons by adjusting the purine base pool size. J. Bacteriol. 2005;187:791–794. doi: 10.1128/JB.187.2.791-794.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zakataeva NP, Gronskiy SV, Sheremet AS, Kutukova EA, Novikova AE, Livshits VA. A new function for the Bacillus PbuE purine base efflux pump: efflux of purine nucleosides. Res. Microbiol. 2007;158:659–665. doi: 10.1016/j.resmic.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 70.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 71.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., Sect. D: Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 72.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.