Abstract
We provide a portrait of the bacterial transcription apparatus in light of the data emerging from structural studies, sequence analysis and comparative genomics to bring out important but underappreciated features. We first describe the key structural highlights and evolutionary implications emerging from comparison of the cellular RNA polymerase subunits with the RNA-dependent RNA polymerase involved in RNAi in eukaryotes and their homologs from newly identified bacterial selfish elements. We describe some previously unnoticed domains and the possible evolutionary stages leading to the RNA polymerases of extant life forms. We then present the case for the ancient orthology of the basal transcription factors, the sigma factor and TFIIB, in the bacterial and the archaeo-eukaryotic lineages. We also present a synopsis of the structural and architectural taxonomy of specific transcription factors and their genome-scale demography. In this context, we present certain notable deviations from the otherwise invariant prote-ome-wide trends in transcription factor distribution and use it to predict the presence of an unusual lineage-specifically expanded signaling system in certain firmicutes like Paenibacillus. We then discuss the intersection between functional properties of transcription factors and the organization of transcriptional networks. Finally, we present some of the interesting evolutionary conundrums posed by our newly gained understanding of the bacterial transcription apparatus and potential areas for future explorations.
Keywords: RNA polymerase, Beta barrel, Two component system, Activators, Transcription factors, Mobile elements, ATPases
1. Introduction
Of the several control steps in the flow of information from a gene to its RNA or protein product, regulation at the transcriptional level is a fundamental mechanism shared by all organisms. Transcription regulation is central to the process by which organisms convert the constant sensing of environmental changes and intracellular fluxes of metabolites to homeostatic responses (Watson, 2004). The general paradigms for the mechanism of transcription initiation and regulation first emerged from pioneering studies on gene expression in bacteria and phages (Jacob and Monod, 1961; Ptashne, 2004). Transcription in bacteria and most DNA viruses which infect them was found to be catalyzed by a single multi-subunit RNA polymerase. It is recruited to conserved DNA sequence elements upstream of genes, termed the promoter, by means of a DNA-binding protein, the σ factor, which specifically recognizes these sequences. The σ factor and the RNA polymerase, together, constitute the “basal transcription apparatus” that is required for the baseline transcription of all genes (Fig. 1). In particular, the σ factor is identified as a “general” or “basal” transcription factor (TF) (Watson, 2004). Early studies, especially in the Bacillus subtilis sporulation model, suggested that there might be several alternative sigma factors beyond the commonly used version, which might recruit the catalytic core of the RNA polymerase to specific sets of genes to result in temporally and spatially distinct alternative transcriptional programs (Ju et al., 1999; Stragier and Losick, 1996). This emerged as a general mechanism for regulating the broad changes in gene expression, which correlate with the different developmental or differentiation states of a bacterium. Starting with the classical studies of Jacob and Monod it became apparent that functionally linked groups of genes are simultaneously co-regulated by dedicated regulators. These functionally linked genes often occur as collinear groups (operons) on the chromosome, and encode components of a common pathway for the utilization of a particular metabolite (e.g. lactose), or constitute interacting components of a macromolecular complex or developmental pathway (e.g. lytic or lysogenic development of phages) (Jacob and Monod, 1961; Ptashne, 2004). Studies on the dedicated regulators of operons indicated that they are DNA-binding proteins that bind specific DNA sequences associated with the operon, which are distinct from the promoter, and act as transcription regulatory switches. These proteins, termed the “specific TFs” (as opposed to the general TFs mentioned above), belong to two distinct regulatory types: (1) repressors, which negatively regulate transcription of their target gene and (2) activators, which positively regulate transcription of their target genes (activators). Affinities of the specific TFs for their target sequences on DNA are often dependent on their binding to low-molecular weight compounds (effectors) or phosphorylation and other post-transcriptional modifications. Thus, specific TFs are integral elements of the apparatus which “converts” an intrinsic or extrinsic sensory input to a transcriptional response.
Fig. 1.
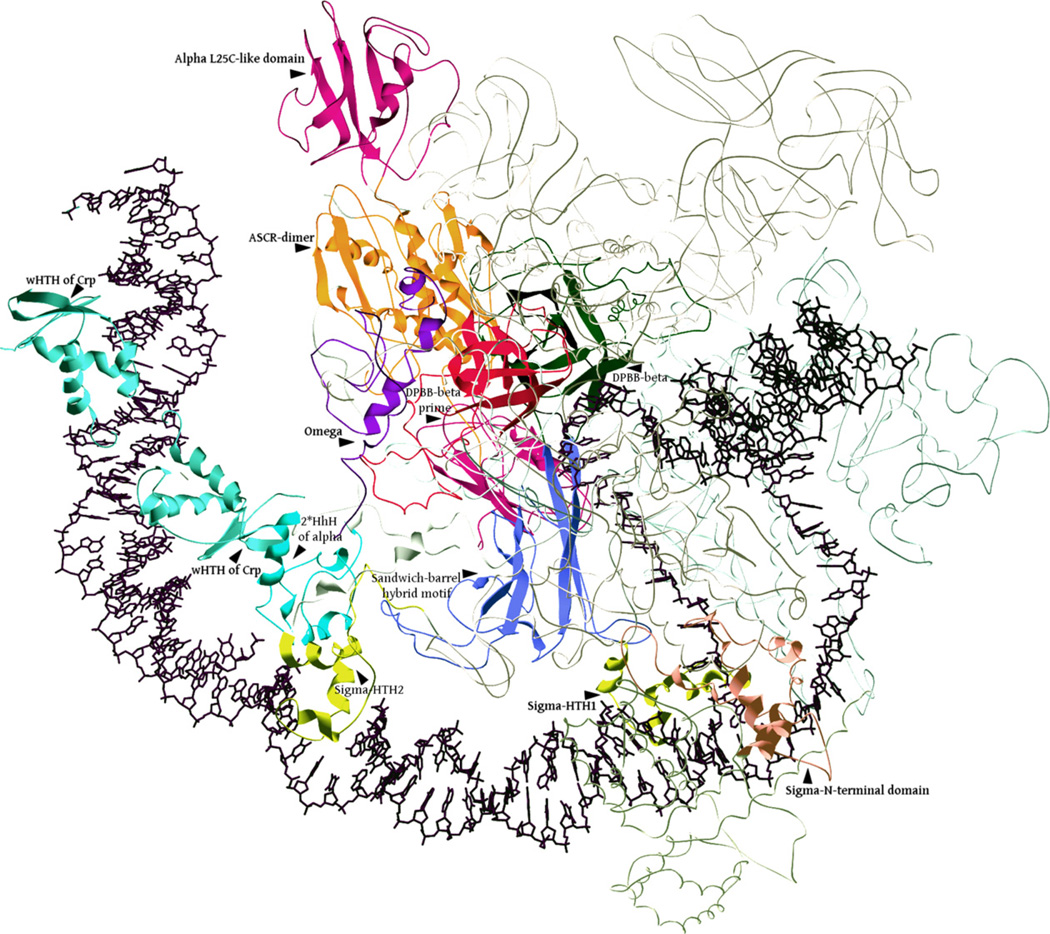
Structure of the bacterial transcription initiation complex. The cartoon representation was derived from an EM structure of the initiation complex (PDB: 3iyd) in association with DNA that contains the α, β, β′, ω, σ70 and the wHTH domains of CRP (CAP) transcription factor. For increased clarity, only the key globular domains of these proteins are shown and labeled. The remaining parts of the structure are shown as coils.
An explosion of structural studies, primarily by means of X-crystallography and site-direct mutagenesis, supplemented by NMR spectroscopy and electron microscopy, have in the past 20 years revealed the nature of these interactions at the molecular level (Harrison, 1991; Latchman, 1997). Not only have the structures of exemplars of most of the DNA-binding and effector-binding domains of TFs and RNA polymerase subunits become available, but also structures of entire complexes, such as the transcription initiation complex have been published (Feklistov and Darst, 2011; Hudson et al., 2009). These efforts allow us to subject the transcription apparatus to “microscopic scrutiny” and interpret various observations stemming from functional and evolutionary studies in atomic detail. On the other hand, there have also been major advances in terms of our “macroscopic” understanding of transcription regulation. At the “systems” level the total set of regulatory interactions mediated by the binding of general and specific TFs, either singly or in combination, to promoters and regulatory elements in operons can be conceptualized as a network, termed the transcriptional regulatory network (Madan Babu et al., 2007). The nodes of the network represent genes and TFs and edges represent regulatory interactions. Advances in genomics over the past two decades have made the reconstruction and analysis of such networks a reality. Studies on these networks have shown that at an abstract level they have architectures which can be approximated by scale-free networks which are also found in non-biological systems such as the internet (Barabasi and Bonabeau, 2003). They are characterized by the recurrence of small patterns of interconnections, called network motifs, which were first defined in Escherichia coli (Madan Babu et al., 2007; Shen-Orr et al., 2002). The study of the transcription network and its motifs are beginning to reveal the genome-scale principles of the associations between TF, their response to external or internal changes and the mode of alteration of gene expression (i.e. activation or repression) (Babu et al., 2004). In this article we mainly focus on the TF nodes of the transcription regulatory network, but interpret some of the observations on these nodes in light of our current knowledge of the architecture of the transcription network.
Our primary objective here is to provide a portrait of the transcription apparatus as from the vantage point of the wealth of data coming from structural studies, sequence analysis and comparative genomics. Due to constraints on space this portrait would necessarily be rendered in broad strokes, yet we attempt to bring out key features that are commonly overlooked by workers less familiar with evolutionary considerations. We hope that these considerations will provide a distinct perspective that could inspire a more natural vision of the transcription apparatus.
2. Basic anatomy of the RNA polymerase
In bacteria the DNA-dependent RNA polymerase is a six subunit complex, comprised of two identical α subunits and one subunit each of β, β′, σ and ω (Feklistov and Darst, 2011; Hudson et al., 2009; Iyer et al., 2004a; Watson, 2004). Most bacteria have a single gene for each of the RNA polymerase subunits. In some instances the genes for two subunits are fused; e.g. the endosymbiotic gam-maproteobacterium Wolbachia and several epsiloproteobacteria such as Helicobacter and Wolinella. Certain lineages of symbionts or parasites with degenerate genomes and the chloroflexi are an exception in that the ω subunit is currently undetectable. Highly degenerate, cooperative intracellular symbionts like Sulcia (a bacteroidetes) and Hodgkinia (an alphaproteobacterium), which live in close association with each other have individually lost several components of essential functional systems, but complement each other by exchanging components such as tRNA synthetases and ribosomal subunits (McCutcheon et al., 2009). Even these organisms encode their own α,β, β′ and σ subunits, though it appears that they share a common σ subunit (encoded by Sulcia). The active site for the nucleotidyltransferase activity of the RNA polymerase is constituted by residues from both the β and β′ subunits that together are termed the catalytic subunits (Cramer et al., 2001; Iyer et al., 2003; Opalka et al., 2010; Vassylyev et al., 2002). The a subunit does not directly contribute in any way to the catalytic activity but is still absolutely required for the effective polymerase function both in the initiation and elongation steps. The σ factors are primarily needed for the initiation step to bind to the promoter. However, they have also been found to remain associated with the elongating polymerase and cause pausing at promoter proximal sites by rebinding DNA sequences resembling the −10 sites of the promoter (Mooney et al., 2005). The ω subunit is the least understood of the subunits and is an entirely α-helical protein that is asymmetrically positioned in the complex. It primarily contacts the catalytic domain of the β′ subunit and additionally has more limited contacts with the two α subunits, the σ factor and specific activator TFs (Cramer et al., 2001; Vassylyev et al., 2002; Fig. 1). The organizational logic of the bacterial RNA polymerase became clear with the sequence-structure analysis of the crystal structures of the holoenzyme complexes and cryo-EM structure of the initiation complex (Fig. 1; Cramer et al., 2001; Hudson et al., 2009; Iyer et al., 2003; Opalka et al., 2010; Vassylyev et al., 2002). Given that it is best understood in terms of the constituent conserved domains and their functional properties, we consider below the major subunits and their key structural features.
2.1. The α subunits
The α subunit is comprised of three domains: The N-terminal unit has an α-subunit-core-related (ASCR) domain (Iyer et al., 2003) into which is inserted a distinctive domain. Structure comparison searches using the DALI program with this domain retrieved the C-terminal domain of the bacterial ribosomal subunit L25 (PDB: 1feu, Z > 3) and related proteins such as YbbR. Further, visual examination of the topologies and reciprocal structure-similarity searches with DALI confirmed that they share a common fold (Fig. 2). The C-terminal module (CTD) is comprised of two HhH motifs (Mah et al., 2000) (Fig. 2). In the transcriptional complex the two α-subunits dimerize via their ASCR domains, while the L25-like domains point in opposite directions (Fig. 1). The C-terminal HhH motifs contact the minor groove of DNA in a manner similar to HhH motifs found in several other DNA-binding proteins (Fromme et al., 2004). The HhH motifs of the C-terminal domain of a also contact the second helix-turn-helix (HTH) domain of the σ-factor, which binds the −35 promoter element in the major groove adjacent to the contact of the HhH motifs (Fig. 1). Similarly, the HhH motifs contact the specific activator TFs that bind their target elements upstream of the promoter (Fig. 1; Hudson et al., 2009). The α-dimer is asymmetrically positioned with respect to the homologous catalytic domains of the β and β′ subunits (see below). The ASCR domain from one of the α-subunits primarily contacts the catalytic domain of the β subunit, whereas that from the second α-subunit mainly contacts the catalytic domain of the β′ subunit (Fig. 1). The newly identified L25-like domain from only one of the subunits makes a second major contact with the β catalytic domain, while the equivalent domain from the other α-subunit makes a distinct contact with the β′ subunit far away from its catalytic domain. The HhH motifs of the α-subunits do not notably alter the curvature of the path of DNA at the points of their individual DNA contacts. However, the layout of the α-dimer is such that it can accommodate the specific TFs that bind target sequences to bend the DNA upstream of the promoter. Thus, the interaction of the α-dimer with both the specific and basal TFs appears to be critical for effective engagement of the transcription initiation site by the RNA polymerase (Fig. 1).
Fig. 2.
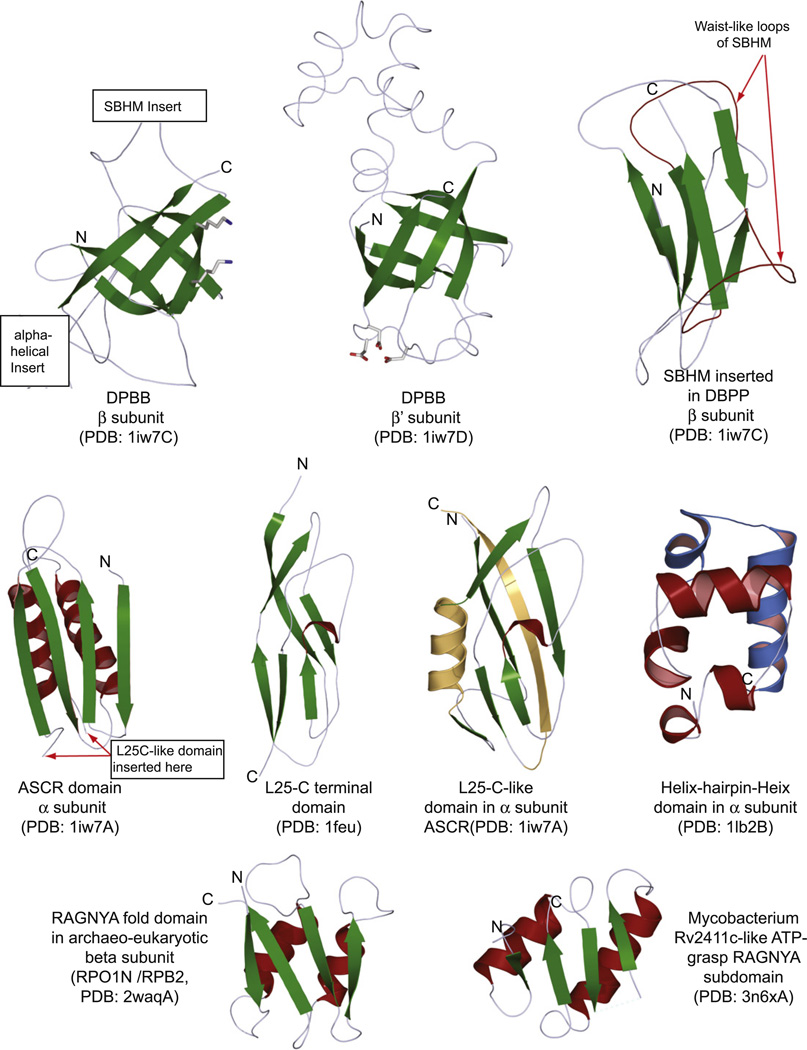
Structures of key conserved domains of the β, β′ and α subunits. Strands are colored green, whereas helices are colored red or blue. Only the core conserved regions of the domains are shown. Inserts in domains are mostly suppressed or excised as depicted. The C-terminal domain of the ribosomal L25 protein is also depicted to illustrate its structural relationship with the conserved domain inserted into the ASCR domain of the α subunit (L25C–like domain). Structural elements in the L25C–like domain of the α subunit that are not present in the ribosomal L25 protein are colored orange.
2.2. The catalytic subunits β and β′
The β and β′ subunits share a homologous core comprised of a domain with the double-ψ-β-barrel fold (DPBB) (Castillo et al., 1999; Hulko et al., 2007; Iyer et al., 2003) (Figs. 2 and 3). The DPBB domains from the two subunits are closely appressed against each other with each of them providing key residues to the active site. The DPBB of the β′-subunit bears an absolutely conserved DxDxD signature (where x is any amino acid), which chelates a Mg2+ ion that is required for directing the phosphate of the incoming nucleotide to react with the 3′ hydroxyl of the initial nucleotide (Fig. 2). The DPBB of the β-subunit contains two absolutely conserved lysines that appear to stabilize the hypercharged reaction intermediate and interact with the negatively charged backbone of the elongating RNA-chain (Cramer et al., 2001; Iyer et al., 2003; Fig. 2). Studies have suggested that homologs of the DPBB domains of the β and β′ subunits are also found in the eukaryotic RNA-dependent-RNA polymerases (RdRPs), which are involved in amplification of the siRNA pathway and related families proteins found in several bacteria and bacteriophages (Iyer et al., 2003; Ruprich-Robert and Thuriaux, 2010; Salgado et al., 2006; Figs. 2 and 3). In these proteins the DPBBs which are equivalent to β and β′ are fused together in a single polypeptide, with the cognate of the β DPBB being the N-terminal domain and the one equivalent to the β′ DPBB being the C-terminal domain, connected by a long helical linker. In addition to the RdRP-like proteins there are other single polypeptide RNA polymerases such as those encoded by the fungal killer plasmids (e.g. the Kluyveromyces killer plasmid) and a group of bacterial proteins typified by Corynebacterium glutamicum NCgl1702, both of which are closer to the cellular DNA-dependent RNA polymerases (Iyer et al., 2003). Our analysis of the domain architectures and gene-neighborhoods suggests that most of these single polypeptide RNA polymerases are likely to be components of mobile selfish elements (Supplementary material): As noted previously several prokaryotic RdRP-like proteins are encoded by bacteriophages (Iyer et al., 2003), and might mediate transcription in these viruses. Of the remaining bacterial RdRP-like proteins, we observed that a subset typified by RUMTOR_01356 (gi: 153815131) are encoded by a predicted mobile element, which additionally code for at least three other proteins (Fig. 3, Supplementary material) – two nucleases of the restriction endonuclease fold, one of which is related to the previously characterized VRR-Nuc family (Iyer et al., 2006) and a third small α-helical protein. These RdRP-like proteins display fusions to two N-terminal transcription factor-related helix-turn-helix (HTH) domains that are predicted to bind DNA (Fig. 3, Supplementary material). The cyanobacterial RdRP-like proteins are typically fused to a SMF/DprA-like Rossmann fold domain (Fig. 3, Supplementary material; 94% probability of match to SMF using the HHpred program) that is predicted to bind DNA (Aravind et al., 2005; Smeets et al., 2006). In several bacteria this domain plays an important role in the uptake of DNA during transformation. Additionally, some of the cyanobacterial RdRP-like proteins display a fusion to one or more RNAseH domains (e = 10−18 in iteration 2 using PSI-BLAST). The genes for the RdRP-like proteins in certain Gram-positive bacteria are also present in a predicted mobile element which additionally encodes a nuclease with an UvrC-Intron homing endonuclease (URI) domain (Fig. 3, Supplementary material). The NCgl1702 like RNA polymerases are encoded by distinct mobile elements that also encode a DNA-pumping ATPase of the HerA-FtsK superfamily (Fig. 3, Supplementary material) that is similar to those encoded by certain conjugative transposons and related mobile elements (Iyer et al., 2004b). Based on the domain architectures and gene-neighborhood contexts (e.g. RNaseH fusion, presence of DNA-binding HTH and SMF domains, endonucleases), we propose that the action of these single polypeptide RNA polymerases aids in the replication of these selfish elements by synthesizing a RNA primer. This priming reaction might be initiated by the nicking action of nucleases encoded by some of these mobile elements or as these mobile elements are being taken up by a target cell.
Fig. 3.
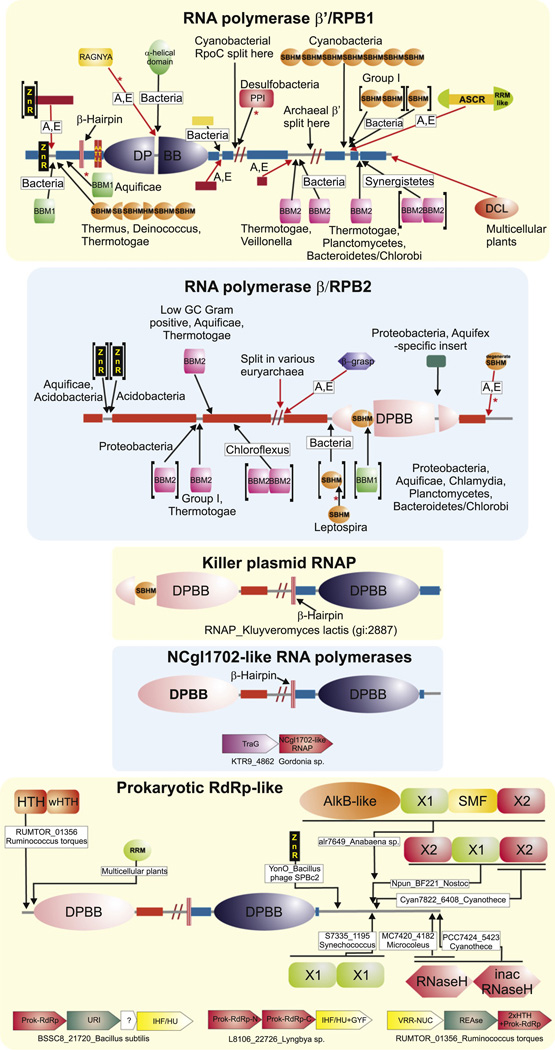
Domain architectures of the RNA polymerase β and β′ subunits, yeast killer plasmid RNA polymerase, NCgl1702-like RNA polymerases and the prokaryotic RdRP-like RNA polymerases. For the β and β′ subunits, the domain architecture reconstructed to the last universal common ancestor is shown in the center and inserts in various lineages are shown around this core. Archaeo-eukaryotic domain inserts are indicated with a red arrow and bacterial inserts are marked with a black arrow. Lineages in which the inserts are observed are indicated near the arrows or architecture. Red asterisks indicate new domains discovered in this study. Bacterial inserts, on occasions, differ within members of a closely related bacterial lineage. For a more detailed discussion of these variations, refer to Lane and Darst (2010a). A similar representation is used for the prokaryotic RdRP-like proteins, where lineage-specific inserts are marked with a representative gene and species name around a core conserved architecture. Genes in operons are shown as box-arrows with the arrow head pointing from the 5′ to the 3′ direction of the coding sequence. Operons are labeled with the gene name of the polymerase gene and species name. Refer to the supplement for more detailed domain architectures and gene neighborhoods. Standard abbreviations are used for domain and lineage names. The DCL domain is an RNA binding domain which is also found in a stand-alone form in bacteria and in several eukaryotic rRNA biogenesis proteins. Other abbreviations: A, E: archaea and eukaryotes, ASCR: alpha subunit core related, ATL: AT-Hook like motifs, PPI: peptidyl prolyl isomerase, ZnR: zinc ribbon.
We interpret the above single polypeptide RNA polymerases in selfish elements as late-surviving representatives of different stages of the ancient diversification of RNA polymerases among early replicons leading to the ancestral RNA polymerase of cellular forms. First, these enzymes suggest that the common ancestor of the DNA-dependent-RNA polymerases and the RdRP-like proteins emerged as a single protein, with adjacent copies of the DPBB domain, which corresponded to the β and β′ catalytic domains. The evolution of both the RdRP-like proteins of the mobile elements and the cellular RNA polymerases of extant cellular organisms is dominated by the accretion of several accessory domains on either side of the two DPBBs, as well as even insertion within the DPBBs themselves (Iyer et al., 2003, 2004a; Lane and Darst, 2010a; Opalka et al., 2010). For example, we observed that the cyanobacterial RdRP-like proteins show an extraordinary diversity of architectures (Fig. 3, Supplementary material), including accretion of an AlkB-like 2-oxoglutarate and iron dependent dioxygenases (e = 10−12 in iteration 3 using PSI-BLAST) that might modify methylated DNA or RNA (Iyer et al., 2010). The emergence of β and β′ subunits of cellular RNA polymerases were accompanied by an entirely different set of accretions. The RNA polymerase of the fungal killer plasmids contains several of these accretions and insertions (Fig. 3, see below), which suggest that the split of the ancestral protein into two distinct subunits happened only after these initial accretion events. Crystal structures of the bacterial RNA polymerase complexes throw considerable light on the significance of these inserts. One key insert, also called the “flap domain”, is that of the sandwich-barrel-hybrid motif (SBHM) domain in the DPBB of the β-subunit (Figs. 2 and 3). This insert is present in the fungal killer plasmids, but is absent in the RdRP-like proteins and the NCgl1702-like RNA polymerases (Fig. 3). Thus it was likely to have been acquired at some point when the enzyme was still a single subunit polymerase with fused β and β′ cognates. In bacteria it interacts specifically with the σ-factor (Fig. 1)(Kuznedelov et al., 2002; Murakami et al., 2002), while its cognates in archaea and eukaryotes interact with TFIIB (Kostrewa et al., 2009), suggesting that the emergence of this insert was the critical determinant that allowed the ancestral RNA polymerase of cellular life forms to be recruited to the basal TF that recognized the promoter. This region forms a part of the RNA-exit channel (Toulokhonov et al., 2001) and also makes notable contacts with regulatory proteins such the anti-σ factors (Pineda et al., 2004), the bacteriophage anti-termination proteins (Yuan et al., 2009) and the elongation factor NusA (Toulokhonov et al., 2001), suggesting that it is a nexus point for various transcription regulatory events.
N-terminal to the β′-DPBB domain, the ancestral version of all RNA-polymerases (including the RdRP-like enzymes, Salgado et al., 2006) had a distinctive bihelical extension preceded by two extended segments forming a standalone β-hairpin. Specifically in DNA-dependent RNA polymerases of cellular life-forms (but not RdRP-like proteins, NCgl1702-like and killer plasmid RNA polymerases) the first long helix of this extension acquired a distinctive insert in the form two flap-like structures resembling the AT-hook DNA-binding motif (Iyer et al., 2003). The above-mentioned β-hairpin and the AT-hook-like structures contact the template strand at the transcription start site and appear to be critical for melting dsDNA to allow the polymerase catalytic domains to access their template (Vassylyev et al., 2007; Westover et al., 2004). Thus the β-hairpin is likely to have been a template strand binding element that had already emerged in the common ancestor of all RNA polymerases (including RdRP-like proteins), while the AT-hook-like flaps were an innovation that augmented this interaction in the common ancestor of the DNA-dependent RNA polymerases of cellular forms. Based on comparisons of the structures of the RdRP and the cellular RNA polymerases it is also clear that the common ancestor of all RNA polymerases had a segment in the extended conformation at the C-terminus of the β DPBB that formed a brace to hold the β′ DPBB. This feature might have been a key element that held the two DPBB domains in close proximity in the ancestral polymerase. C-terminal to the β′ DPBB there is a conserved extension that folds back and interacts with the β DPBB, which is shared by all cellular RNA polymerases and the versions encoded by the killer plasmids. We posit that this region might shield part of the active site and potentially exclude solvent from the active site to favor a more processive catalytic activity.
Both the β and the β′ subunits of the bacterial RNA polymerase have several insertions of additional domains that are not found in the archaeo-eukaryotic RNA polymerases and vice versa (Lane and Darst, 2010a,b). The β DPBB shows entirely distinct inserts in the bacterial and the archaeo-eukaryotic lineages: The bacteria acquired an all α-helical insert (Figs. 1 and 3). In contrast, our structure similarity searches with the DALI program revealed that the β′ DPBB in archaeo-eukaryotic lineage acquired, in the equivalent position, an unrelated insert of a RAGNYA fold domain that is closely related in structure to the ATP-binding version found in the ATP-grasp module (DALI Z scores > 3) (Balaji and Aravind, 2007) (Fig. 2). In both cases the inserts are spatially directed in a manner similar to the SBHM of β DPBB and respectively recruit the ω-sub-unit in bacteria or its cognate RBP6 in archaea and eukaryotes by contacting them equivalently in the loop between their two conserved helices (Minakhin et al., 2001). Given the nucleic acid-binding properties of certain representatives of the RAGNYA fold (Balaji and Aravind, 2007), it would be of interest to investigate if it might have an additional role in binding the emerging transcript in the archaeo-eukaryotic polymerases. The other major divergent inserts include multiple SBHM domains and two small domains respectively known as the β-β′-motif-1 (BBM1) and the β-β′-motif-2 (BBM2) (Iyer et al., 2003, 2004a). The latter domains are comprised of long extended segments forming a highly curved hairpin, which is bounded on either side by helical segments. Several of the SBHM domains show dramatic differences between various bacterial lineages in terms of their presence or absence as well as in the number of copies in which they are present (Iyer et al., 2003, 2004a; Lane and Darst, 2010a). Archaea, eukaryotes and the killer-plasmid β subunit have a previously unreported C-terminal degenerate SBHM which appear to have been lost in the bacterial forms (Fig. 3; region 1154–1198, chain B, pdb: 1K83). The functions of the SBHM domains still remain incompletely understood. The conserved SBHMs found at the C-terminus of the bacterial β′ subunit have been shown to interact with the transcription elongation factors of the GreA/B family (Chlenov et al., 2005; Lamour et al., 2008). A set of lineage-specific SBHM inserts seen in the N-terminus of the β′ subunit of the Thermus-Deinococcus lineage and Thermotoga are known to contact the σ-factor (Chlenov et al., 2005; Vassylyev et al., 2002). Based on this, we suggest that the lineage-specific SBHM inserts might have significance in mediating interactions with transcription regulators that allow for control processes unique to specific groups of bacteria. Remarkably, we observed that the β′ subunit of the delta-proteobacterial lineage of desulfobacterales show an insertion downstream of the catalytic DPPB domain that can be unified with the parvulin-like peptidyl prolyl isomerase in sequence searches (PSI-BLAST iteration 2, E values < 10−25; see Supplementary material for sequence). It would be of interest to investigate if this domain might provide an in-built prolyl isomerization chaperone function for the RNA polymerase in these organisms.
2.3. The ω subunit
The α-helical ω subunit, which is a cognate of RPB6 in the archaeo-eukaryotic lineage, was until recently an enigma. For a long time it was even considered an impurity that associates with the purified RNA polymerase complex. However, number of studies have confirmed its role as a major player in the assembly of the β′ subunit into the RNA polymerase complex by preventing its aggregation (Mathew and Chatterji, 2006; Minakhin et al., 2001). Specifically in bacteria, the ω subunit is the focus of the stringent response, in which the metabolite (p)ppGpp produced by the SpoT/ RelA-type enzymes causes a drastic global shift in the transcription profile from growth- and cell-division- related genes to amino acid synthesis genes. It appears that the ω subunit is the binding-site for (p)ppGpp and mediates the sensitivity of the polymerase to this metabolite (Mathew and Chatterji, 2006). While there is no comparable stringent response in archaea and eukaryotes, the RBP6 subunit is likely to play a comparable role as the bacterial ω in assembly of the RNA polymerase by interacting with the insert domain in DPBB of the β′ subunit.
2.4. σ-factors
The most prevalent r-factor that is conserved in all bacterial genomes is σ70, which initiates transcription of all or the majority of promoters in any given bacterium. Most bacteria, except symbionts and parasites with extremely reduced genomes, encode at least one alternative σ-factor (see Supplementary material). The majority of these alternative σ-factors are relatively close paralogs of σ70 and are collectively referred to as the σ70-family (Gruber and Gross, 2003; Paget and Helmann, 2003). The remaining alternative σ-factors belong to the σ54-family that bear multiple conserved HTH domains, but are only very distantly related to the σ70 family. Traditionally, the primary structure of the σ70-family has been divided into 4 regions, numbered 1–4, which were mapped on the basis of their functional properties and sequence conservation (Gruber and Gross, 2003; Paget and Helmann, 2003). While the structure-based dissection of the domains of the σ70-family partly confirms this nomenclature, it provides a more natural way of visualizing these σ factors; hence, our discussion entirely follows the structural paradigm. The conserved core of σ70-family proteins contains an N-terminal domain in the form of a 4-helical bundle, which is comprised of the only helix in region 1, which is conserved throughout the family, and the entire conserved region 2. The N-terminal domain of the primary σ-factor from several bacterial lineages usually contains a large helical insert of variable size (Iyer et al., 2004a). The N-terminal 4-helical bundle inserts deeply into the DNA at the −10 element of the promoter and fosters melting of the double helix around the transcription start site (Feklistov and Darst, 2011) (Fig. 1). The primary σ-factor contains a further α-helical domain, N-terminal to the first core domain (mapping to the reminder of region 1), which functions as a negative regulator of its DNA-binding activity (Barne et al., 1997). This additional N-terminal domain is entirely absent in the alternative σ-factors and also the primary σ-factor of the bacteroidetes-chlorobium-gemmatimonad lineage (Iyer et al., 2004a). The first domain of the conserved core of the σ factor is immediately followed by the first HTH domain (domain 2 of the conserved core) that maps to the earlier defined region 3 (Aravind et al., 2005). It binds the extended −10 element that is upstream of the −10 element (Barne et al., 1997; Campbell et al., 2002). Binding of this element by this HTH domain is particularly important in transcription initiation through promoters lacking the −35 element. This HTH domain has completely degenerated in most members of the extracellular function (ECF; see below) clade of the σ70-family (Gruber and Gross, 2003). Remarkably, we observed that in the Dictyoglomus lineage a further HTH domain is inserted between helix-2 and helix-3 of this HTH domain and is predicted to make a unique lineage-specific contact upstream of the extended −10 element (Supplementary material). The C-terminal-most domain (domain 3) of the conserved σ core is the second HTH domain that interacts with the α-subunit and binds the −35 element (Gruber and Gross, 2003; Paget and Helmann, 2003).
Bacteriologists usually classify the α70-family in groups 1–5 (Gruber and Gross, 2003; Paget and Helmann, 2003). It should be emphasized that this classification is partly inaccurate and misleading because groups 2 and 3 are not evolutionarily monophyletic assemblages within the σ70 family. Group 1 contains the classical σ70 and is typically present in a single copy in all bacterial genomes. Group 2 consists of σ factors closely related to σ70; however, these function as alternative σ factors, for example in the initiation of the transcriptional programs associated with stationary phase and stress response (e.g. σS of E. coli). Examination of the phylogenetic trees of σ-factors (Gruber and Gross, 2003; Paget and Helmann, 2003) suggests that group 2 σ-factors arose repeatedly through lineage-specific duplications of the primary σ factor. The group 3 σ factors are a heterogeneous, non-monophyletic assemblage comprised of several distinct families that are involved in initiating transcription of multi-gene batteries associated with major conditional and developmental programs such as heat shock response (e.g. E. coli RpoH gene product), flagellar gene expression and motility (e.g. E. coli FliA product), sporulation in firmicutes (B. subtilis SigE, SigF and Sig G) and stress response (e.g. B. subtilis SigB) (Gruber and Gross, 2003; Paget and Helmann, 2003). The group 4 or the ECF σ factors are a monophyletic clade of fast-evolving σ factors. They are typically associated with an anti-σ factor that might be a membrane protein with an extracellular domain (Helmann, 2002). The anti-sigma factor is dissociated from the cognate r upon receiving a sensory stimulus, typically from the extracellular environment allowing the σ factor to initiate a transcriptional program. The group 4 σ factors are major regulators of transcription in response to extrinsic sensory inputs such as iron availability, mis-folded proteins in the periplasm, redox stress and host-derived signals in the case of pathogenic bacteria. However, a subset of these σ factors might also respond to intracellular sensory stimuli as seen in the case of the redox based regulation of σR of Streptomyces coelicolor (Helmann, 2002; Paget et al., 1998) or down-stream of two-component regulatory systems (see below) as seen in the case of σE from the same organism (Helmann, 2002; Paget et al., 1999). Phylogenetic analysis shows that the recently defined group 5 sigma factors typified by TxeR of Clostridium difficile are merely a highly divergent group of ECF σ factors. Like them, they have been found to initiate the transcription of a small group of genes related to toxin and bacteriocin production (Mani and Dupuy, 2001). The ECF σ factors in particular are greatly expanded in bacteria with complex metabolic and developmental features (see below for genomic scaling). Thus, the ECF r-factors might be seen in functional terms as intermediates between specific TFs and conventional σ-factors.
The σ54-family is typically present in a single copy per genome and is sporadically distributed across the bacterial tree (Supplementary material) – it is present in proteobacteria and their closest relatives (the group-I bacteria) and firmicutes among the group-II bacteria (Iyer et al., 2004a). However, it is absent in most major group-II clades such as actinomycetes and cyanobacteria. The presence of the σ54-family is strictly correlated with the presence of a distinctive class of specific TFs, namely the NtrC family of ATPases (also called enhancer-binding proteins) (Ammelburg et al., 2006; Aravind et al., 2005; Hong et al., 2009). A structure of a complete σ54-family protein is as yet unavailable. Analysis of the structurally characterized fragments along with sequence profile analysis suggests that σ54 is comprised of four distinct conserved regions (Supplementary material). The N-terminal-most of these is a well-conserved α-helical segment, which binds the AAA+ domain of the NtrC-like protein and regulates its ATPase activity during the assembly of the σ54 initiation complex (Doucleff et al., 2005). The second domain is a conserved HTH domain (75–92% probability matches to different HTH profiles using the HHpred program), which has been shown to interact with the RNA-polymerase core, though it could potentially make additional DNA contacts. The third conserved element is also a HTH domain that is likely to contact the −12 element of the σ 54-dependent promoters (83–87% probability matches to different HTH profiles using the HHpred program; Supplementary material). The C-terminal-most domain is yet another HTH domain (84% match using HHpred to a HTH profile), which contacts the −24 element of these promoters (Doucleff et al., 2005). As in the case of the σ70 the two C-terminal HTHs respectively contact the 5′ and 3′ elements in an N- to C-terminal polarity (Hong et al., 2009). Furthermore, σ54 also interacts with the SBHM domain inserted into the β subunit just as the σ70 family (Wigneshweraraj et al., 2003). These observations suggest that there could be a potential common origin for the two families of σ-factors.
2.5. The Gram positive RNA-polymerase delta subunit and related proteins
Gram-positive bacteria display a unique RNA polymerase subunit termed delta (RpoE), which has been shown to bind the RNA polymerase catalytic complex, reduce its affinity for nucleic acids and increase transcription specificity by promoting recycling (Lopez de Saro et al., 1999; Motackova et al., 2010). Specifically, the subunit inhibits the downstream propagation of the transcription bubble at the −10 region, with its acidic C-terminal tail mimicking RNA and interacting with the RNA polymerase catalytic complex. The delta subunit contains a novel winged HTH (wHTH) domain that is fused to a highly acidic C-terminal low-complexity tail (Motackova et al., 2010). We have recently shown that this wHTH domain is widely distributed in bacteria (also fused to restriction endonuclease domains) and eukaryotes (chromatin proteins like HB1 and ASXL1/2/3) and have accordingly termed it the HB1, ASXL, Restriction Endonuclease (HARE)-HTH domain (Aravind and Iyer, 2012). Certain proteobacteria also contain a version of the HARE-HTH domain comparable to delta that instead has an acidic low-complexity tail at the N-terminus. Most remarkable are the proteins found sporadically in actinobacteria, firmicutes and proteobacteria that combine a C-terminal HARE-HTH to: (1) a N-terminal module containing two or more repeats of the specialized helix-hairpin-helix (HhH) domain found in the CTD of the bacterial RNA polymerase α-subunit; (2) Two additional HTH modules that are specifically related to those found in the region 3 and 4 of the sigma factors (Aravind and Iyer, 2012). Thus, these proteins combine parts of the architecture of the RNA polymerase α and σ subunits with the HARE-HTH in a single polypeptide (Fig. 1).The bacterial proteins that combine the RNA polymerase α-subunit CTD module, the σ-factor region 3 and 4 HTH domains with the HARE-HTH are striking because an examination of the RNA polymerase holoenzyme complex with the transcription start site (TSS) shows that these modules indeed occupy successive sites on the DNA just upstream of the TSS (Fig. 1). Thus, these proteins are predicted to function as mimics of the α and σ subunits, with the C-terminal HARE-HTH, potentially occupying yet another site upstream of the TSS. Accordingly, these proteins could possibly function as a novel inhibitor of TSS-binding by the bacterial RNA polymerase, which might either function as a negative transcriptional regulator, or a suppressor of improper transcription initiation.
3. Specific TFs and a structural portrait of their DNA-binding domains
Specific TFs are best classified on the basis of their DNA-binding domains. The two prokaryotic superkingdoms are set apart from the eukaryotes by a remarkable difference in terms of the DNA-binding domains of their specific TFs. Most specific TFs of prokaryotes contain a version of the helix-turn-helix DNA-binding domain (Fig. 3; Aravind et al., 2005). In contrast, eukaryotes show an enormous diversity of DNA-binding domains in their transcription factors (Iyer et al., 2008). In many eukaryotic lineages HTH DNA-binding domains are prevalent in specific TFs (e.g. Homeo or POU domains), but these HTH families are distinct from those found in bacteria and show only a distant sequence relationship to them. Additionally, eukaryotes possess large numbers of Zn-chelating DNA-binding domains such as the C2H2 Zn-finger, the C6 fungal-type Zn-finger and the WRKY Zn finger, which are rare or entirely absent in the prokaryotic superkingdoms (Iyer et al., 2008). The dominance of the HTH-containing specific TFs across bacteria considerably aids their computational detection as high-sensitivity sequence profiles have been developed for the HTH domain (Aravind and Koonin, 1999a; Babu et al., 2004). Thus, in conjunction with sequence similarity-based clustering, searches with such profiles allow rather accurate estimates of the specific TF complement of a given prokaryotic organism from its genome sequence. In this article we summarize the various structural variations of the HTH domain that are observed among bacterial specific TFs and briefly discuss the major families which contain each HTH type.
3.1. Tri-helical HTH domains
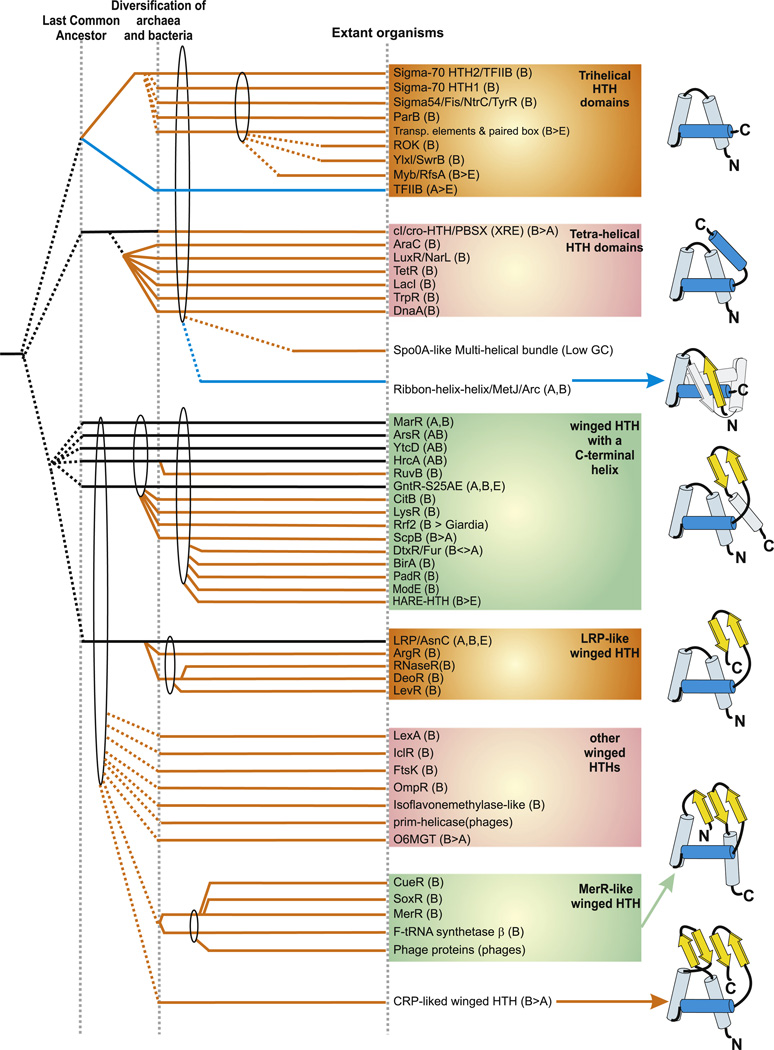
The simplest version of the HTH domain, the basic tri-helical version, is comprised entirely of the three core helices with no additional elaborations (Fig. 4). This configuration appears to be closest to the ancestral state of the HTH and is widely seen across the three super-kingdoms of life. The third helix of this unit, like in most other HTH domains plays a key role in contacting DNA via insertion into the major groove, and is called the recognition helix (Brennan and Matthews, 1989; Clark et al., 1993). This simplest version is seen in the Fis family of transcription factors (typified by the E. coli protein Fis), the 1st HTH domain of the σ70 family and the three HTH domains of the σ54 family (Fig. 5). The Fis family HTH domains are typically found fused to the C-termini of the AAA+ domains of the NtrC-like proteins which bind “enhancer elements” which are located at much greater distances from the promoter than conventional target sites bound by specific TFs (Morett and Bork, 1998; Rombel et al., 1998). Also displaying this type of HTH domains are the bacterial TFs of the Rok and YlxL/SwrB families. The Myb/SANT domain, which is very common in eukaryotic TFs and chromatin proteins is also a typical tri-helical HTH domain (Aravind et al., 2005). In bacteria the Myb/SANT domain is less prevalent than in eukaryotes and is found in TFs typified by the RsfA proteins, which are pre-spore transcription factors in firmicutes (Juan Wu and Errington, 2000) and the proteobacterial GcrA-like transcription factors (Holtzendorff et al., 2004). More recently, using sequence profile searches we uncovered several proteins in bacteria with multiple Myb/SANT repeats (e.g. ND049; gi: 34335384, recovered with e = 10−7 in an RPS-blast search with Myb/SANT profile), which are specifically related to those seen in eukaryotes (e.g. Fig. 5). We observed that these versions are encoded in operons with integrases, endonucleases and DNA methylases in bacteriophages (e.g. gp65 of Listeria phage B054) and bacterial genomes (e.g. A33_2137; gi: 254286508 in Vibrio cholerae) or are fused to endonuclease domains of the HNH and the LAGLIDADG superfamilies. These observations suggest that they are DNA-binding domains of phages or novel mobile selfish elements, wherein they help recognize integration sites. The versions derived from such selfish elements appear to have given rise to the Myb/SANT domain of the eukaryotic transcription factors. The 2nd HTH domain of σ70 family is a derived version of the trihelical HTH class, which shows an additional N-terminal helix also observed in the archaeo-eukaryotic TFIIB proteins (Fig. 4).
Fig. 4.
Higher order evolutionary relationships of bacterial specific transcription factors containing a HTH domain. The horizontal lines represent temporal epochs corresponding to major transitions in evolution of bacteria, namely the last universal common ancestor and the diversification of archaea and bacteria. Solid lines reflect the maximum depth of time to which a particular family can be traced. Broken lines indicate an uncertainty with respect to the exact point of origin of a lineage. The ellipses encompass groups of lineages from which a new lineage with relatively limited distribution could have potentially emerged. Lineages of archaeal origin are colored blue, those of bacterial origin are colored orange and those present in archaea and bacteria are colored black. The phyletic distribution of the lineages are also shown in brackets, where A: Archaea; B: bacteria and E: eukaryotes. The “>” reflects lateral transfer with the arrow head pointing to the potential direction of transfer. Also shown to the right are cartoon representations of the major structural types of HTH domains found in bacterial transcription factors. The TFIIB lineage of archaeo-eukaryotic HTHs is shown to illustrate its relationship with the sigma factor.
Fig. 5.
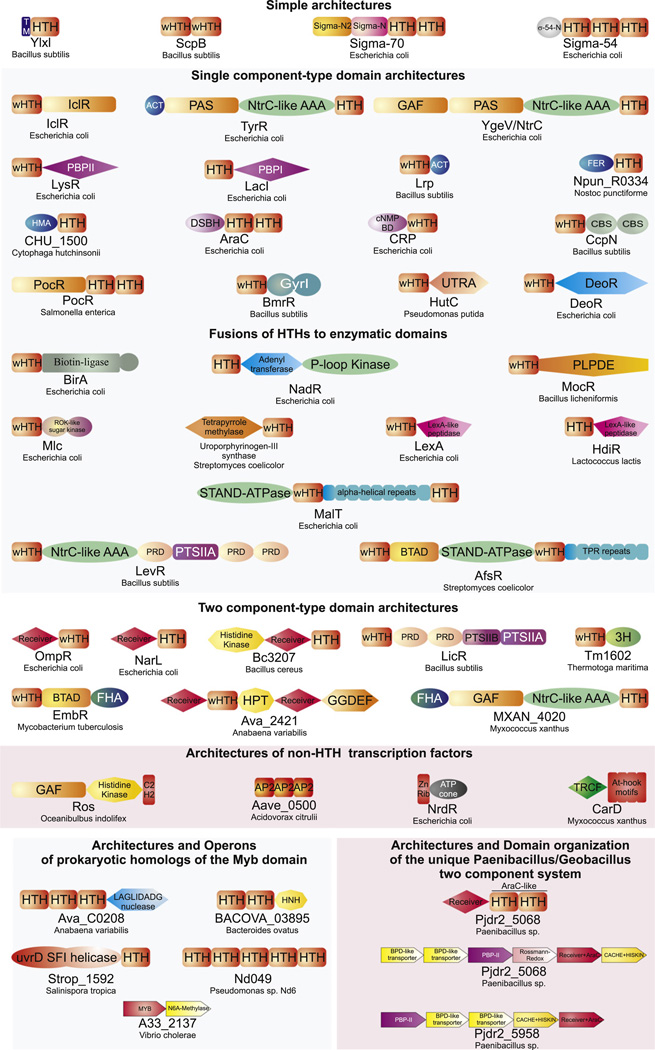
Examples of domain architectures of bacterial transcription factors described in the text. Proteins are labeled with their gene and species names. The domains are not drawn to scale. Standard nomenclatures were mostly used to depict the various domains. Some additional abbreviations include: TM: transmembrane, σ-54 N: globular domain found at the N-terminus of σ54, Sigma-N2 and SigmaN: Conserved N-terminal domains found in σ70, BTAD: conserved domain found in bacterial signaling proteins, ZnRib: Zinc ribbon, FER: classical Ferredoxin domain of the RRM fold.
3.2. Tetra-helical HTH domains
The tetra-helical version of the HTH domain is an elaboration of the basic tri-helical version and is characterized by an additional C-terminal helix which packs against the shallow cleft formed due to the open configuration of the tri-helical core (Fig. 4). Several major families of bacterial transcription factors contain this version of HTH, which can be differentiated on the basis of their sequence features. The cI-like family, typified by the phage lambda cI protein is one of the major families with this type of DNA-binding domain. Several distinct subfamilies can be recognized within this family. The largest of these is the repressor subfamily typified by the protein PbsX (Xre) from the B. subtilis prophage 168, which appears to represent the prototypical repressor-type specific TFs in bacteria (Wood et al., 1990). Another major assemblage within the tetra-helical class of HTHs contains the 6 major families of exclusively prokaryotic TFs. These are AraC, LuxR, LacI, DnaA, TrpR and TetR families. The first four of these families are nearly panbacterial in their distribution suggesting that these HTH families had probably diverged from each other even in the common ancestor of all bacteria (Fig. 4). The latter two lineages are more limited, being most prevalent in proteobacteria and firmicutes. DnaA is usually found in a single copy in all bacterial genomes, with a tetrahelical HTH occuring at the C-terminus of the AAA+ domain. The DnaA protein is primarily required in replication initiation, but it also functions as a transcription factor (Fujikawa et al., 2003; Messer and Weigel, 2003). Additionally, sporadic versions of the tetrahelical HTH are also seen in several phage transposases related to the Mu transposase, which in some cases also function as TFs (Wojciak et al., 2001).
3.3. Winged HTH domains
The winged HTH (wHTH) domains are distinguished by the presence of a C-terminal β-strand hairpin unit (the wing) that packs against the shallow cleft of the partially open tri-helical core (Brennan, 1993; Fig. 4). The simplest versions of the wHTH domains contain a tight helical core similar to basic tri-helical version followed by the two-strand hairpin. However, many wHTH domains display further serial elaborations of the β-sheet (Fig. 4) (Aravind et al., 2005). In the 3-stranded version, the loop between helix-1 and helix-2 of the HTH assumes an extended configuration and is incorporated as the 3rd strand in the sheet, via hydrogen-bonding with the basic C-terminal hairpin. In the 4-stranded version, the linker between helix-1 and helix-2 also forms a hairpin with two β-strands, and along with the C-terminal wing forms an extended β-sheet (Fig. 4). The wing often provides an additional interface for substrate contact, typically by interacting with the minor groove of DNA through charged residues in the hairpin (Brennan, 1993; Clark et al., 1993; Swindells, 1995). Majority of bacterial TFs contain the wHTH as their DNA-binding domains. Fourteen major families of prokaryotic TFs, namely the HARE-HTH (see above), BirA, ArsR, GntR, DtxR-FurR, CitB, LysR, ModE, MarR, PadR, YtcD, Rrf2, ScpB and HrcA-RuvB families, are unified by the presence of a characteristic helix after the wing, and comprise the largest monophyletic assemblage within the wHTH superclass (Fig. 4). Of these the DtxR–Fur family appears to have specialized early in bacterial evolution in regulating metal-dependent transcription of genes (Hantke, 2001); here the wing is incorporated into a large sheet formed with additional C-terminal strands. Another major monophyletic assemblage within the wHTH superclass includes the DNA-binding domains of the DeoR, ArgR, LevR and Lrp-AsnC families of TFs. These families are unified by overall sequence similarity, and a conserved pattern with a conserved glutamine or arginine residue between helix-1 and helix-2 of the HTH domain (Aravind et al., 2005). There are other distinct families of wHTH TFs in bacteria, namely the LexA, OmpR, and IclR families, with 2- or 3-stranded wHTH domains, but they do not appear to belong to any of the aforementioned assemblages (Fig. 4). Of these the classical representatives of the LexA family appear to be involved in regulating responses to DNA damage in diverse lineages of bacteria (Peat et al., 1996), whereas the OmpR-like TFs are one of the largest group of specific TFs that function downstream of histidine kinases (Itou and Tanaka, 2001).
Distinct from all the above families is the Crp family that is typified by the presence of a 4-stranded version of the wHTH domain (Fig. 4). This family has a pan-bacterial distribution and is typically fused to a C-terminal cNMP-binding domain (Korner et al., 2003). These TFs appear to have specialized early on as the primary cyclic nucleotide dependent regulators in bacteria. Beyond these classical wHTH domains there are several modified versions which display highly derived version of the wHTH (Fig. 3). These include the MerR-like family, which contains a truncated form of the 3-stranded wHTH domain with a deletion of the first helix. Instead, these proteins show an additional helical element C-terminal to the wing. The MerR family has vastly proliferated into several distinct subfamilies, like the SoxR and CueR subfamilies (Brown et al., 2003). A similar form of wHTH is also observed in the phage lambda excisionase and terminase proteins and the phage Mu-repressor family.
3.4. The Ribbon-helix-helix or MetJ/Arc domain
The MetJ-Arc family (also known as ribbon-helix-helix/RHH family) of TFs is a uniquely prokaryotic family of TFs typified by the methionine operon repressor MetJ and the bacteriophage repressor Arc (Aravind and Koonin, 1999a; Aravind et al., 2005). They function as obligate dimers, which pair through a single N-terminal strand, and possess a C-terminal helix-turn-helix unit (Fig. 4). The organization of the C-terminal helical unit is identical to corresponding unit in the HTH domain, and it shows the characteristic conserved sequence features of the HTH domain. The sheet formed by the N-terminal strands of the domain is inserted into the major groove of DNA (Gomis-Ruth et al., 1998). Mutagenesis experiments have shown that even single mutations in the N-terminal strand convert the strand of the RHH domain to a helix, and result in a structural packing that is closer to the canonical HTH domain (Cordes et al., 1999). This result, together with the notable structural and sequence similarities with the HTH domains, suggest that the RHH domain was derived from the HTH domain through conversion of the N-terminal helix to a strand (Aravind et al., 2005). Concomitant with this modification, the N-terminal strand, which came to lie atop the recognition helix, appears to have taken up the primary DNA-binding role in this domain. They are most frequently found as transcriptional regulators of the mobile toxin–antitoxin operons (Anantharaman and Aravind, 2003). Hence, it is possible that they were originally derived in such toxin–antitoxin systems, through rapid divergence from a conventional HTH. This appears to have happened early in the evolution of one of the prokaryotic lineages (Fig. 4), after which they were widely disseminated across the bacteria and archaea due to the extensive horizontal mobility of toxin–antitoxin systems.
3.5. Other DNA binding domains found in bacterial specific TFs
A small set of non-HTH DNA-binding domains are found in bacteria specific TFs. While the C2H2 Zn-finger is probably the most prevalent DNA-binding domain of eukaryotic specific TFs, it is rare in prokaryotes. The Ros/MucR family of TFs is typified by the Ros protein of Agrobacterium tumefaciens, which regulates the expression of virulence genes on the Ti plasmid (Chou et al., 1998), and MucR, which regulates the exopolysaccharide biosynthesis in various rhizobia (Keller et al., 1995). These proteins contain a single copy of the C2H2 Zn-finger and, unlike their eukaryotic counterparts, have only 9–10 residues between the two pairs of metal-chelating ligands (Esposito et al., 2006). These TFs are currently known only from proteobacteria. The Zn-ribbon is an ancient nucleic-acid-binding domain that is found in large number of nucleic acid metabolism proteins (Aravind and Koonin, 1999a; Krishna et al., 2003). While it is found in the core transcriptional machinery, for example, as a domain of the β′ subunit and occasionally inserted into the β subunit (in aquificae and acidobacteria) of the RNA polymerase (Iyer et al., 2004a; Lane and Darst, 2010a; Fig. 3), it rarely used as the primary DNA-binding domain in a specific TF. Zn-ribbon TFs in bacteria are typified by the E. coli NrdR protein which is a regulator of the ribonucleotide reducatase operons (Grinberg et al., 2006). Here it combined with a C-terminal ATP-cone domain which acts a nucleotide sensor (Fig. 5). A few other specific TFs with the Zn-ribbon fused to other sensor domains (e.g. CBS domains) are also encountered in prokaryotes (Aravind and Koonin, 1999a). The AT-hook is a very common DNA-binding motif in eukaryotes that specifically contacts the minor groove (Aravind and Landsman, 1998). In bacteria a small number of TFs with the AT-hook are currently know. The best example of this is the CarD protein from Myxococcus xanthus and other myxobacteria, which is known to function as a light-induced transcription factor (Penalver-Mellado et al., 2006). Here, the AT-hooks, which bind the target sequences, are combined with a TRCF-like domain (Fig. 4) (Subramanian et al., 2000). In the transcription repair-coupling helicase (TRCF) the same domain is fused to a superfamily-II helicase module and facilitates interaction with the RNA-polymerase holoenzyme (Westblade et al., 2010). Outside of myxobacteria the CarD orthologs merely contain a TRCF-like domain but not AT-hooks (Subramanian et al., 2000). In these organisms it is likely that these proteins associate with the RNA polymerase but do not bind DNA. Hence, these versions might not function as bona fide specific TFs. The AP2 domain is a DNA-binding domain which is found specific TFs of several eukaryotic lineages such as plants, stramenopiles and apicomplexans (Balaji et al., 2005). In bacteria they are typically found associated with integrases and transposases of selfish elements such as phages and transposons. However, in course of this study we have identified versions in bacteria that resemble eukaryotic versions from plants, stramenopiles and apicomplexans in having multiple tandem copies of the AP2 domain and are independent of integrase or transposase catalytic domains (Fig. 4, Supplementary material). We predict that these versions are likely to function as novel specific TFs and might have been the progenitors of the TFs observed in the above-stated eukaryotic lineages.
3.6. RNA regulators of transcription that interact with the RNA polymerase
The E. coli 6S RNA was discovered over 40 years ago and remained mysterious in function until recently. It was shown to be the prototype of a class of widely conserved non-coding bacterial RNAs that directly interact with the RNA polymerase to regulate transcription (Wassarman, 2007; Willkomm and Hartmann, 2005). These RNAs are about 185 nucleotides in length and fold through complementary base-pairing to give rise to a structure, which contains a large central bulge which is believed to resemble the open promoter at the transcription start site. In E. coli the 6S RNA has been shown to associate with the σ70-containing holoenzyme and repress transcription from specific promoters in the stationary phase (Wassarman, 2007). While the 6S RNA homologs from other bacteria also associated with the RNA polymerase complex, their targets and the phase of the life-cycle in which they act remain unclear. Some organisms, like B. subtilis, possess multiple 6S RNA homologs suggesting that there might be alternative regulation of transcription in different developmental phases by distinct 6S RNAs (Willkomm and Hartmann, 2005). The 6S RNA has been shown to potentially interact with the β, β′ and σ subunits suggesting that it might interact in the region of the conserved SBHM in β (the so-called flap domain) (Wassarman, 2007). Its structural similarity to the open promoter has also been interpreted as a means of mimicking the former and thereby withholding the holoenzyme from the actual promoter. While most non-coding RNAs in bacteria work at the level of translation regulation (Gottesman, 2004), it is conceivable that there are other RNAs which operate similarly to the 6S RNA to regulate transcription.
4. An overview of the domain architectures of bacterial specific TFs
The above DNA domains are combined with other domains in the same protein giving rise to a remarkable array of domain architectures (Fig. 5). Despite the diversity, all the architectures can be classified into a small number of generic architectural classes, the members of each class being unified by certain general organizational and functional principles. Hence, in the case of bacterial TFs these organizational principles serve as strong predictors of function (Aravind et al., 2005). These architectural classes illustrate how natural selection has convergently engineered similar functional solutions using a relatively small repertoire of domains, with the most populated classes representing particularly successful functional solutions.
4.1. Specific TFs with simple domain architectures
The simplest architectures are the standalone copies of the DNA-binding domain as typified by proteins related to the cI repressors and Fis. These proteins are usually almost entirely comprised of just a standalone HTH, and might, at best, have some small extensions that play a role in dimerization or interactions with other components of the basal transcriptional machinery (Aravind et al., 2005). A family of bacterial proteins typified by the B. subtilis sigma D regulator YlxL (SwrB) (Kearns and Losick, 2005) contains a HTH domain fused to a N-terminal transmembrane region (Fig. 5). These HTH proteins might regulate transcription under the influence of signaling events associated with the cell membrane. The next level of architectural diversification involves tandem duplications of HTH domains. Beyond the σ-factors, such versions are encountered in a few bacterial DNA-binding proteins like ScpB that could potentially function as TF in addition to having a role as co-factors for the chromosome-condensing SMC proteins (Mascarenhas et al., 2002; Soppa et al., 2002).
4.2. TFs displaying single component-type domain architectures
The single-component systems are defined as those signaling systems in which the transcription DNA-binding domain and the stimulus sensor module are combined into a single protein. These architectures are by far the most prevalent class in bacteria. Their simplest versions are no different from the above class in that they are simply comprised of DNA-binding domain that not only binds DNA but also directly interacts with small-molecule effectors. These minimal one-component regulators are prototyped by the MetJ-type RHH transcription factor, which, in addition to binding DNA, also senses S-adenosyl methionine directly (Augustus et al., 2010). A more typical form of the one component system combines a HTH domain with a small molecule binding domain (SMBD, Fig. 5; Aravind et al., 2010). More complex architectures may involve multiple SMBDs or even additional domains such as the NtrC-like AAA+ ATPase domain. The most common SMBDs fused to HTHs in the single component systems are drawn from a relative small set of ancient protein folds (Fig. 5): (1) The PAS-like fold, with representatives such as the PAS domain, the GAF domain, and the ligand-binding domains of the IclR-type transcription factors (Aravind et al., 2010). (2) The periplasmic-binding protein types I and II domains, which include the ligand-binding domains of the LysR family (Tam and Saier, 1993; Tyrrell et al., 1997; Vartak et al., 1991). (3) The ferredoxin-like fold, which includes the ACT domain and related ligand-sensing domains of the Lrp-like transcription factors and the classic ferredoxins, which are fused to HTH domains in cyanobacterial proteins (Aravind and Koonin, 1999b; Brinkman et al., 2003; Bull and Cox, 1994). (4) The double-stranded β-helix domain (cupin), which contains the AraC-type ligand-binding domains, as well as the cNMP-binding domains found in Crp/Cap/Fnr family TFs (Anantharaman et al., 2001; Kannan et al., 2007). (5) The CBS domain that occurs as an obligate dyad (Bateman, 1997). (6) The GyrI domain, which contains two copies of the SHS2 structural module, appears to be one of the principal ligand-binding domains of the MerR family (Heldwein and Brennan, 2001; Anantharaman et al., 2001; Kannan et al., 2007). (7) The UTRA domain, which is found in the HutC/FarR group of GntR family transcription factors and possesses the same fold as chorismate lyase (Anantharaman and Aravind, 2003). (8) The DeoR ligand-binding domain, which shares a common α/β fold (the ISOCOT fold), with enzymes of the phosphosugar isomerase family such as ribose phosphate isomerase (Anantharaman and Aravind, 2006). Several distinct clades of specific TFs, often defined by a specific architectural theme can be identified within this mélange of bacterial one-component systems. For example, the AraC family contains a duplication of the tetra-helical version of the HTH domain (Fig. 5) and typically occurs fused to the sugar-binding cupin domain suggesting that the entire clade predominantly functions as sugar-sensing transcription factors.
A variation on the single-component theme is the fusion of the DNA-binding domain to an enzymatic domain, which catalyzes a reaction pertaining to the biochemical pathway regulated by the specific TF (Fig. 4). By this action these TFs are major players in the phenomenon of feedback regulation of metabolic pathways, in which the concentrations of the metabolites produced by the pathway regulate the activity of the TF. The archetypal representative of this architectural theme is the biotin operon repressor, BirA, which contains an N-terminal HTH domain fused to a C-terminal biotin ligase domain (Wilson et al., 1992). In the presence of biotin the enzymatic domain synthesizes the co-repressor, and the HTH domain represses the transcription of the biotin biosynthesis genes (Wilson et al., 1992). Comparative genomics suggests that architectures involving fusions to a range of enzymes from cofactor, nucleotide, amino acid and carbohydrate metabolism are fairly common in bacteria (Fig. 5; Aravind and Koonin, 1999a; Aravind et al., 2005). Some notable fusions include combination of the HTH with nicotinamide mononucleotide adenylyl transferase and a P-loop kinase in NadR, with the pyridoxal-phosphate dependent amino-transferase domain (TFs of the GntR family) and sugar kinases (Rok family) (Fig. 4; Singh et al., 2002). Some of these architectures, like BirA are widely distributed in the prokaryotic genomes and appear to be ancient, while others like the fusion of an OmpR family wHTH with the uroporphyrinogen-III synthase are found only in actinobacteria. These observations suggest that the combinations of HTHs with enzymatic domains have been repeatedly selected for throughout bacterial evolution. Yet another variation on the theme of enzyme-linked HTH domains is provided by the LexA protein, the repressor of several bacterial DNA repair genes (Fig. 4). It contains a protease domain of the signal peptidase fold fused to a wHTH domain. The protease domain catalyzes an auto-catalytic cleavage in response to a DNA-damage signal and triggers dissociation of its wHTH domain from target sequences, thereby allowing transcription of DNA repair genes (Peat et al., 1996). Architectures analogous to LexA are also seen in the repressors typified by the heat-response transcription factor HdiR from the Lactococcus lactis, where a LexA-like protease domain is fused to a cI-like HTH instead of the wHTH seen in LexA (Savijoki et al., 2003). This implies that the mechanism of transcription regulation with a proteolytic processing step was innovated at least twice independently.
4.3. TFs with specialized architectures involving ATPase domains
Two other specialized classes of domain architectures arise through fusions of the HTH domains with either of two types of P-loop NTPase domains, namely the NtrC-like AAA+ domains (Zhang et al., 2002) and the related STAND (signal transduction ATPases with numerous domains) NTPase domain (Ammelburg et al., 2006; Leipe et al., 2004). These NtrC-like TFs typically sense various sensory inputs via their effector-binding domains and associate as a ring-shaped multimer with σ54 via their AAA+ ATP-ase domains (Wigneshweraraj et al., 2008). The AAA+ ATPase domains of these proteins perform an ATP-dependent chaperone-like activity that converts the “closed” σ54-containing transcription complexes to an “open” configuration, which is favorable for transcription initiation (Wigneshweraraj et al., 2008). The NtrC-like AAA+ domains are fused to at least two different types of HTH domains. The classical versions like NtrC and TyrR are fused to a C-terminal basic tri-helical HTH domain of the Fis family (Wang et al., 2001). The second version typified by the Bacillus levanase operon regulator, LevR, instead contains an N-terminal wHTH domain (Aravind et al., 2005). Structural comparisons suggest that core NTPase module of the STAND superfamily has been derived from the Orc/Cdc6 family of AAA+ domains. These two share a unique configuration of the dyad of helices occurring after the core NTPase strand-2 and a distinctive winged HTH (wHTH) occurring C-terminal to AAA+ module (part of the HETHS module (Leipe et al., 2004)). Given that the Orc/CDC6 family of AAA+ NTPases is ancestrally present in the archaeo-eukaryotic lineage, it is likely that the STANDs emerged from them early in archaeal evolution. Indeed, most archaea show lineage-specific expansions of the basal versions of the STAND NTPases encoded by mobile elements (the MJ-, PH- and SSO-type ATPases) that still retain several features of the ancestral AAA+ ATPases (Leipe et al., 2004). These archaeal versions are often linked in the same polypeptide with restriction endonuclease fold domains and are likely to catalyze the ATP-dependent assembly of complexes on DNA that allow the replication of the mobile elements that encode them. Hence, they are likely to retain the ancestral function of the Orc/Cdc6 family in assembling complexes on DNA.
However, from such precursors a distinct lineage of STAND NTPases with signaling functions arose in bacteria (Leipe et al., 2004). As a rule they are large multi-domain proteins that catalyze the ATP-dependent assembly of complexes in variety of signaling contexts. They typically contain superstructure-forming repeat domains, such as the WD and TPR domains, which may serve as surfaces for the assembly of multi-protein complexes (Leipe et al., 2004). The archetypal members of the architectural class combining a DNA-binding HTH and STAND NTPases are the E. coli MalT (Larquet et al., 2004; Marquenet and Richet, 2010), B. subtilis GutR (Poon et al., 2001) and Streptomyces AfsR proteins (Lee et al., 2002). The DNA-binding HTH domains in these proteins are of several distinct types. The fusions involving the OmpR family of wHTH domains (e.g. in AfsR) usually link the HTH to the N-ter-minus of the STAND NTPase domain. In contrast, fusions involving the LuxR family of HTH link it to the C-terminus of the STAND module, with a set of super-structure forming α-helical repeats occurring between these two modules (e.g. GutR and MalT; Fig. 4). The STAND-domain-containing transcription regulators integrate signaling inputs sensed via their super-structure forming domains with an NTP-dependent switch provided by the STAND. The energetically demanding use of NTPs in STAND signaling suggests these switches are likely to control expression of metabolic states that might impose a high cost on the cell (Marquenet and Richet, 2010). The STAND regulators are particularly prevalent in developmentally or organizationally complex bacteria like cyano-bacteria and actinobacteria.
4.4. Specific TFs with architectures pertaining to two-component, phosphotransfer and serine/threonine kinase signaling systems
The core of the two component phospho-relay system comprises of a histidine kinase and the receiver domain, which is phosphorylated on a conserved aspartate. These represent one of the most prevalent signaling systems of the bacterial world (Pao and Saier, 1995; Ulrich and Zhulin, 2007; West and Stock, 2001). A large subset of the receiver components are specific TFs that convert the sensory input received from the histidine kinase into a transcriptional response (Ulrich and Zhulin, 2007). These TFs are typified by fusions of the receiver domain to a HTH domain. Two of the most common architectures, seen in the majority of bacteria, involve combinations of a single N-terminal receiver domain to either a LuxR-like tetrahelical HTH domain (e.g. UhpA and NarL) or wHTH domain (e.g. OmpR and PhoB) (Fig. 5). Less frequent fusions involving HTH domains of the AraC and the CitB families are seen in certain bacteria. Other than these simple architectures, several more complicated architectures involving multiple receiver domains or even fusions to additional histidine kinase (e.g. B .cereus protein BC3207) and NtrC-like AAA+ ATPase (e.g. E. coli NtrC) domains are also observed (Fig. 5). The PTS sugar-transport systems use a phosphorelay cascade to transfer a phosphate from phosphoenol pyruvate to a histidine on the PTS regulatory domain (PRD), which often co-occurs in the same polypeptide with HTH domains (Barabote and Saier, 2005; Stulke et al., 1998). The PRDs receive the phosphates from the HPr and EIIB proteins of the PTS system, and depending on their phosphorylation state regulate transcription. Architectures involving the PRD domain are analogous to those involving the receiver domain of the two-component system (Barabote and Saier, 2005). The simplest versions contain an N-terminal wHTH domain fused to a C-terminal PRD domain (Aravind et al., 2005). The more complex forms contain more than one PRD domains, or fusions to NtrC-like AAA+ domains and PTS system EIIB domains, which determine sugar specificity (Fig. 5). The B. subtilis LicR protein contains an N-terminal HTH fused to two PRDs and both EIIB and EIIA components of the PTS system, indicating that it is a multi-functional protein that directly regulates both sugar uptake and transcription of sugar-utilization genes (Tobisch et al., 1999). The 3H domain, which is related to the HPr domain of the PTS system, is also found fused to a BirA-related wHTH domain in several bacterial proteins typified by Tm1602 from Thermotoga maritima (Fig. 5) (Anantharaman et al., 2001; Weekes et al., 2007). The 3H domain might represent another novel domain that may be regulated by phosphorylation on its conserved histidines, perhaps via a PTS-like system. The serine– threonine kinases are over-represented in certain organizationally complex bacteria, like the cyanobacteria, myxobacteria and the actinobacteria (Aravind et al., 2010). In the latter group there is class of proteins, typified by the protein EmbR, containing a fusion of the HTH domain with the FHA domain (Hofmann and Bucher, 1995). The FHA domain in this protein binds phosphoserine peptides, and mediates its interaction with the upstream protein kinase in regulating the biogenesis of the mycobacterial cell wall (Molle et al., 2003). The same SMBDs found in the single component systems may also occasionally be found fused to two-component and other phosphorylation-dependent regulators, where they might supply secondary allosteric inputs (Fig. 5).
5. The proteome-wide demographics and phyletic patterns of specific TFs
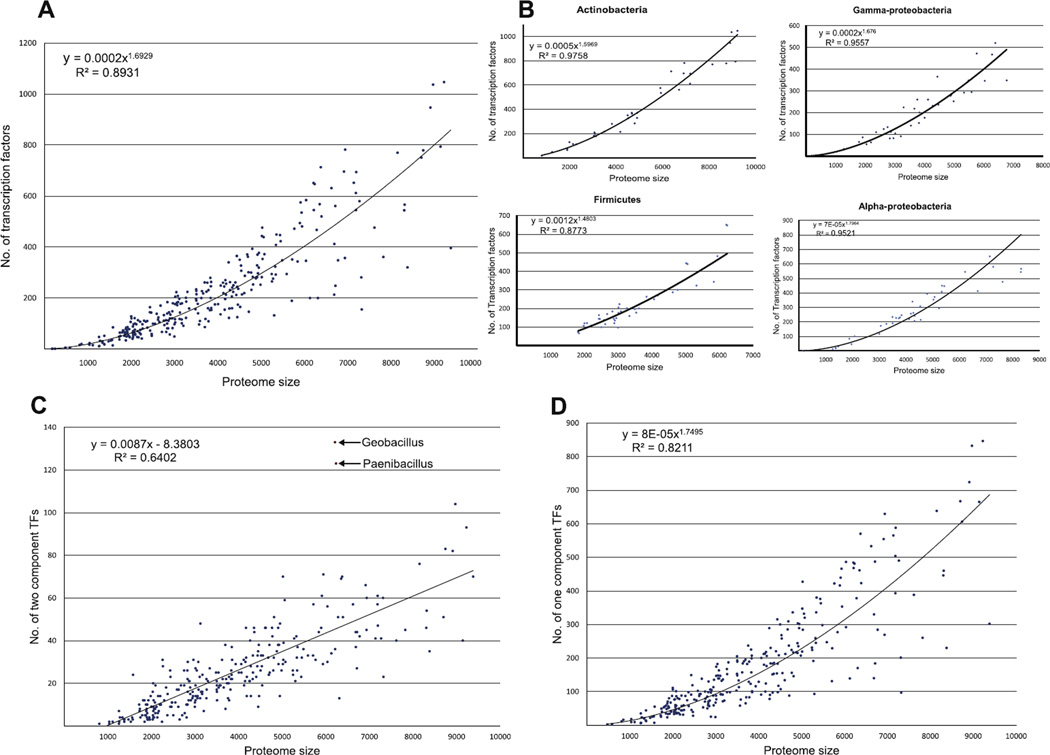
The availability of a large number and phyletic diversity of complete bacterial genome sequences allows robust estimation of the general trends in the proteome-wide distribution of TFs. Position-specific score matrices or sequence profiles for the various distinct families of DNA-binding domains found in TFs have proven to be a very effective method to detect TFs in proteomes. These sequence profiles can be used to iteratively search the target proteomes with the PSI-BLAST program (Altschul et al., 1997). Alternatively, the seed alignments for the different families can be used to generate hidden Markov models, which can be similarly used to search the proteomes with the HMMER program (Eddy, 2009). Over the years several independent studies on scaling of the number of transcription factors with proteome size in bacteria point to a very specific version of the power-law: y = a•xφ (where ‘y’ is number of TFs per proteome, ‘x’ is the proteome size, ‘a’ is a constant and φ is the power which around 1.62) (Aravind et al., 2005, 2010; van Nimwegen, 2003; Fig. 6). Interestingly, examination of individual bacterial clades shows that this form of the power-law scaling of TFs is rather invariant across lineages (Fig. 6). Thus, irrespective of whether we are looking at proteobacteria, firmicutes, actinobacteria or cyanobacteria the exponent of this power-law remains more or less the same, suggesting that this scaling stems from a rather fundamental feature of the bacterial cell. This distribution function suggests that as gene number increases, a greater than linear number of TFs are required per oper-on/gene.
Fig. 6.
Scaling of bacterial transcription factors with proteome size. All graphs show a scatter plot of number of transcription factors in a given proteome (Y-axis) versus the number of protein-coding genes in that organism (X-axis). In (A) and (B), the Y-axis is the overall number of transcription factors across bacteria and in individual lineages respectively. In (C) The Y-axis is the number of predicted two-component system proteins. Note that anomalous numbers in Geobacillus and Paenibacillus that are shown as red points. (D) The Y-axis is number of one-component system and other phospho-relay system proteins.
However, very distinct trends are observed when individual architectural classes of TFs are examined. In bacteria, two-component systems show a strong tendency for linear scaling with respect to proteome size (Fig. 6; Aravind et al., 2005, 2010). Thus there is a strong tendency across bacterial lineages to show about one copy of a two component TF for every 175 genes. This scaling trend should be considered in light of the observation that the scaling of receiver domain proteins with respect to histidine kinases is generally linear in most bacteria (Aravind et al., 2010). This suggests that each two-component system TF is strongly coupled with respect to its upstream signaling histidine kinase. This observation, together with the linear scaling of two-component system TFs with proteome size (Fig. 6), suggests that a similar constraint also operates with respect to the number of target genes downstream of the two-component TF. It implies that that two-component TFs tend to regulate their own target operons to the exclusion of other two-component TFs. This exclusivity is likely to result in a linear increase in the number of such TFs with increasing proteome size. Remarkably, the only notable exceptions to this situation is seen in certain sporulating firmicutes of the Bacillus-like clade Paenibacillus and Geobacillus, which have an anomalously large number of two-component TFs for their proteome size (one per every 47 and 50 genes respectively; Fig. 6). The excess in these organisms appear to stem from the lineage-specific expansion of a version of two-component TF that is relatively uncommon in other bacteria, namely the version combining the receiver domain to C-terminal AraC family HTH domains. Given this unusual violation of a strong trend, we propose that in these organisms these excess two component TFs do not function as distinct TFs in separate signaling processes, but more likely as alternative forms of the same TF in a single signaling process. This idea is supported by our observation that these TFs occur in a very stereotypic operon that also encodes a histidine kinase with an extracellular sensory CACHE domain, a multi-TM transporter and a PBP-II-type solute-binding protein (Fig. 5; Supplementary material). These operonic connections suggest that each isoform of this two-component system is a sensory system that recognizes alternative versions of a variable soluble secreted signal. It is conceivable that the associated transporter and PBP-II domains are involved in the transport of the cognate version of the secreted signal. We propose that the diversification of this two component system might be related to the phenomenon of “identity switching” (Ben-Jacob, 2003) and “sibling rivalry” observed in Paenibacillus, in which under nutrient-poor conditions encroaching sibling colonies are killed by a secreted toxin (Be’er et al., 2011). Such behavior would particularly benefit bacteria if they have a means of distinguishing self from non-self colonies. In light of this, it is conceivable that the expression of different alternative versions of the above two component system operon from colony to colony might provide the necessary diversity for such discrimination. This remarkable system would benefit from further experimental exploration.
In contrast to the above picture, the one component TFs and σ factors, scaled nonlinearly with proteome size and their distributions are best approximated by a power-law distribution comparable to that observed for the overall TF counts (Fig. 6; Aravind et al., 2005). This observation implies that as genome size increases a greater than proportional increase in the numbers of one-component transcription factors is required for controlling the newly added genes. For example, the GntR family has vastly proliferated in several bacteria giving rise to many of the major bacterial one-component transcription factors. This tendency might be related with the need to regulate specialized genes batteries by combining the distinct inputs sensed by the effector-binding domains of different sets of one-component TFs, especially in the metabolically or organizationally complex bacteria with large genomes. This proposal is also consistent with other transcription network-based observations, which suggest that one-component TFs are likely to be important for the fine tuning of gene expression in conjunction with more global changes mediated by two-component TFs (Balaji et al., 2007). Non-linear scaling of the σ factors suggests that in the more complex genomes the additional genes are distributed amongst several functionally specialized gene batteries, which are under the regulation of devoted sigma factors responding to specific conditions. Interestingly,a few genomes show a significantly greater than expected number of σ factors. The most striking example is seen in the case of Phytoplasma asteris, which, like other mycoplasmas, has a highly reduced genome with just over 700 genes (Aravind et al., 2005). Whereas, the other mycoplasmas have only a basal σ-factor, P. asteris has a recent lineage-specific expansion of 11 sigma factors that are related to the Bacillus σF. Likewise, Bacteroides thetaiotaomicron and Nitrosomonas show recent lineage-specific expansions of ECF-type sigma factors that have given rise to at least 10 closely related paralogous members in their proteomes (Aravind et al., 2005). In the case of P. asteris there is evidence that the sigma factors may constitute a novel transposon (Lee et al., 2005). While this possibility also exists in the case of the other bacteria that show a greater than expected number of sigma factors, it is likely that in the latter examples they might indeed be conventional transcriptional regulators recruited for a distinctive sensory signaling pathway.
6. The logic of the overall organization of the transcriptional regulatory interactions in bacteria
Until recently it was thought that the transcription regulatory network (TRN) of both eukaryotes and bacteria are essentially similarly organized with a comparable structure that resembles scale-free networks (Balazsi et al., 2005; Guelzim et al., 2002; Thieffry et al., 1998). However, further studies exploring their fine structure revealed that despite their superficial similarities, the organizational principles of the TRN of the model bacterium E. coli is notably different from that of the model eukaryote Saccharomyces cerevisiae (Balaji et al., 2007). Synthesizing the conclusions from this and related studies several principles pertaining to TRN organization might be discerned. In eukaryotes, highly connected TFs or hubs of the TRN, i.e. those that regulate a large number of genes are not typically those that integrate disparate transcriptional responses (Balaji et al., 2006a,b). However, in the bacterial TRN the hubs indeed function as both global regulators and integrators of diverse transcriptional responses (Balaji et al., 2007). By linking multiple TFs that regulate the same genes in the TRN one can reconstruct the underlying “co-regulatory network” (CRN), which defines how TFs intersect in their regulatory actions. In the E. coli network, the degree distribution of TFs in this CRN (i.e. the number of regulatory intersections they make with other TFs) approximately follows a power law (Balaji et al., 2007). These results are in contrast to yeast CRN, which displays a discernable central tendency in the degree distribution (Balaji et al., 2006a). These organizational differences appear to be related to the fact the bacterial genes are primarily organized as operons or regulons with their own dedicated specific TFs (Collado-Vides et al., 2009). Though S. cerevisiae and E. coli have a comparable number of predicted TFs, the organization of the bacterial genome into operons, with several genes sharing a common set of regulatory elements, effectively reduces the set of targets available for TFs. Hence, in the bacterial TRN the global TFs would also have a propensity for being required for across-operon integration of gene regulation. In the case of eukaryotes the absence of such an organization, with co-expressed genes scattered around the chromosome, might have selected for a preferred number of co-regulatory associations between different TFs to allow co-regulation of a group of genes in different sets of conditions (Balaji et al., 2007).
Further, the hubs in the TRN are enriched in specific TFs that have a dual function as both activators and repressors and are significantly underrepresented in TFs that are either dedicated activators or repressors. Similarly, even the CRN hubs are significantly enriched in TFs that can function as both repressors and activators (Balaji et al., 2007). The enrichment of the dual mode regulators in TRN hubs suggests that TFs mediating large-scale physiological state changes primarily do so by causing large-scale bi-directional changes in gene expression. Further, their prevalence in the CRN implies that these changes are likely to involve cooperative action with other TFs, wherein the dedicated activator and repressor TFs might provide further fine-tuning and amplification of the original effects. Interestingly, two-component systems tend not be pure repressors and are evenly distributed amongst activators or dual regulators. In contrast, one-component TFs depending on import of external effector metabolites by transporters are rarely dual mode regulators and are evenly distributed amongst dedicated repressors and activators (Balaji et al., 2007). Thus, the two distinct modes of signal sensing, namely via two-component systems or via one-component systems are strongly distinguished by their mode of action. The two-component TFs are also enriched in hubs when compared to one-component TFs that depend on the import of external metabolites into the cell by transporters (Balaji et al., 2006a). Hence, the former TFs appear to have been optimized for signaling larger scale changes. The latter category, in contrast usually regulate a small group of genes specifically required for processing a given metabolite, and appear to do so by merely turning them “on” or “off”. Hubs in the TRN appear to be preferentially retained across genomes at small phylogenetic distances (e.g. within a well-defined lineage such as gammaproteobacteria)(Balaji et al., 2006b). Thus, at smaller phylogenetic distances there is a stronger tendency for retention of the large-scale and bi-directional transcriptional responses. However, there is a contrasting trend across larger phylogenetic distance; there is no evidence for preferential retention of hubs amongst bacteria. At large phylogenetic distances the hubs are only about as well conserved as any other TF suggesting that there are major differences in the global regulators between major clades of bacteria (Madan Babu et al., 2006). Given the strong correlation between TFs and proteome size across all bacterial lineages (i.e. the linear scaling for two-component TFs and a gentle power-law increase for one-component TFs), it is quite likely that these features of the transcriptional network inferred from E. coli are generally relevant for bacteria. However, it must be noted that bacteria can greatly differ in terms of their signaling mechanism. Particularly, certain lineages like cyanobacteria, myxobacteria and filamentous actinomycetes display complex signaling cascades involving STAND superfamily NTPase, eukaryotetype serine/threonine kinases, and caspase-like proteases, which are rare or entirely absent in E. coli (Aravind et al., 2010). Hence, it is conceivable that certain optimizations of the TRNs in these bacteria are notably different.
7. Comparative and evolutionary perspectives
Early studies on the bacterial transcription apparatus saw it as model for all of life, indeed keeping with the adage of Monod: “anything that is true of E. coli must be true of elephants, except more so”. As subsequent studies indicated, the archaeal and eukaryotic systems are noticeably more complex than bacterial systems, they came to be seen as simplified models from which several basic mechanistic conclusions could be extrapolated to the other systems (Ptashne, 2004; Watson, 2004). This belief turned out to be partly true at least in the case of the core RNA polymerase complex (Cramer, 2002; Cramer et al., 2001; Vassylyev et al., 2002). With respect to the RNA polymerase complex, the archaea and eukaryotes share orthologs of the α, β, β′ and ω subunits with the bacteria; thus, in the last universal common ancestor (LUCA) the RNA polymerase can be reconstructed as having four distinct subunits. Comparisons with the RdRPs and the RNA polymerases of selfish elements help reconstruct the possible pre-LUCA stages in the evolution of these enzymes. The earliest precursor was probably a DPBB domain that bound nucleic acids as a dimer and probably facilitated replication or transcription as a protein cofactor (Iyer et al., 2003). Subsequently, this DPBB domain duplicated, and the two copies diverged, with each acquiring a distinct set of residues to respectively constitute the Mg2+-chelating and negative-charge-stabilizing parts of the polymerase active site. These forms probably functioned in priming replication of DNA replicons that, unlike RNAs, cannot initiate unprimed daughter strand synthesis (Iyer et al., 2005). This activity is predicted to be still retained by the versions found in the bacterial selfish elements describe above. Finally, the polymerase increased in complexity via domain accretion and became the primary catalyst of transcription. By the time of the LUCA it split up into two separate catalytic subunits and two additional subunits in the form of α and ω were added to the catalytic core. While the bacterial enzyme more or less retained this ancestral state, the archaea and eukaryotes added several additional subunits to this core, which are highly conserved in those two superkingdoms (Cramer, 2002). The transcripts produced by the RNA polymerases of the selfish elements were probably also used by RNA-dependent restriction systems (e.g. such as the CRISPR system (Makarova et al., 2011)) to control their activity. This type of an activity appears to have found a niche in the RNAi system of eukaryotes, where the polymerases were recruited as RNA-replicating enzymes that catalyze the primed or unprimed synthesis of dsRNA from diverse templates ranging from small siR-NAs to mRNA.
With regards to TFs, both general and specific, and the organization of the transcriptional network profound differences emerged in each of the three superkingdoms, whose full magnitude has only recently become clear with availability of genomic data across a wide phylogenetic spectrum. In terms of the actual protein components there are four major areas of difference between the bacterial and archaeo-eukaryotic systems: (1) the subunit complexity of the RNA polymerase, (2) the nature of basal TFs, (3) the specific TFs and (4) the role of chromatin-associated proteins (Iyer et al., 2008). In regard to basal TFs, the archaeo-eukaryotic system possesses two distinct TFs, namely TFIIB and the TATA-binding protein (TBP) that apparently have no orthologs in the bacteria (Aravind and Koonin, 1999a; Burley, 1996). However, reanalysis of the structures of the respective RNA polymerases complexes with the basal TFs suggests that the picture might be more complex. Firstly, the TFIIB protein contains two HTH domains, by means of which it makes a direct contact with the promoter elements on either side of the TBP-binding site (Nikolov et al., 1995). This contact of the promoter region by means of the two HTH domains of the archaeo-eukaryotic TFIIB is reminiscent of similar situation in bacteria, wherein the two HTH domains of the σ-factor mediate two major DNA contacts associate with the two separated promoter elements (Hudson et al., 2009). Furthermore, both TFIIB and σ-factor also make comparable contacts with the conserved SBHM domain (the so called “flap” region) of the β (or its ortholog’s) catalytic region. This observation suggests that the SBHM insert of the RNA polymerase in the LUCA was already recruiting the primary basal TF that was bound to the promoter. Further, in light of the above, it is likely that the basal TF in the LUCA was potentially comprised of two HTH domains contacting DNA; hence, TFIIB and the r-factor are likely to be ancient orthologs. Thus, the RNA polymerase complex in the LUCA can be reconstructed as having not just the four universally conserved subunits but also a two-HTH-domain basal TF that enabled it to become the primary catalyst of transcription. In bacteria the basal TF evolved into the σ factor by accretion of an additional N-terminal helical domain, which performed the function of −10 element recognition and initiation of promoter melting. On the other hand in the archaeo-eukaryotic lineage the RNA polymerase complex appears to have recruited a new promoter-binding protein in the form of TBP (Cramer, 2002; Cramer et al., 2001; Vassylyev et al., 2002). Given the relationship of TBP to the RNA-binding domains of certain RNAse III family nucleases it is conceivable that it was recruited independently from an ancestral RNA-binding domain (Aravind and Koonin, 2001).
The specific TFs appear to have followed a different evolutionary course. In this case it is the eukaryotes that possess very different specific TFs, but bacteria and archaea share several families of specific TFs, especially those with HTH domains (Aravind and Koonin, 1999a; Aravind et al., 2005; Iyer et al., 2008). Though several of the specific TF families shared by bacteria and archaea can be easily explained as arising from relatively recent lateral transfer between the prokaryotic super-kingdoms, some others like the MarR, ArsR, YctD, Lrp, HrcA and GntR families appear to show distinct pan-archaeal and pan-bacterial groups. This suggests that they were present from very early in the evolution of each of the prokaryotic super-kingdoms (Aravind et al., 2005). As a corollary we are presented with an apparent evolutionary conundrum because the evolutionary picture of these specific TFs is not congruent with that of the basal TFs and the RNA polymerase complex, which point to a greater and hence possibly much earlier divergence. This paradox is further accentuated by the fact that the specific TFs of bacteria and the archaea interact with the RNA polymerase core in very distinct ways – for example, the archaeo-eukaryotic orthologs of the α-subunit lack the HhH motifs (CTD) found in the C-terminus of the bacterial α subunit that interact with the specific TFs. While number of scenarios can be conceived to account for this situation, the one that resorts to least number of unusual events depends on two considerations: (1) The core RNA polymerase and basal TF represent a tightly interacting system (in terms of interactions between both the polymerase subunits and between the polymerase and the basal TF) that does not tolerate much xenologous displacement following lateral transfer. (2) The specific TFs interact less tightly and do not require conserved interfaces for these interactions. Thus, they are liable to lateral transfer. Hence, the families of specific TFs, which are shared widely by the two prokaryotic superkingdoms, might be interpreted as very early lateral transfers that happened between the two prokaryotic superkingdoms. The spread of these TFs through lateral transfer could be related to their adaptive value given that they (usually one-component TFs) often confer ability to alter gene expression in response to specific environmental compounds (Madan Babu et al., 2006). The origin of eukaryotes through the symbiosis of an archaeal and bacterial progenitor resulted in a compartmentalized cell. This appears to have rendered most prokaryote-type one-component systems ineffective (Aravind et al., 2005). Furthermore, emergence of histone- modification-mediated chromatin-based gene repression (see below) in the eukaryotes appears to have made the prokaryote-type repressors superfluous. As consequence, early in eukaryotic evolution there appears to have been massive loss of the specific TFs inherited from the two prokaryotic progenitors, clearing the way for the recruitment and innovation of new types of eukaryote-specific TFs (Iyer et al., 2008). Our studies suggest that some of these eukaryotic TFs might have been recruited from DNA-binding domains that were already present in bacterial TFs (e.g. AP2 and Myb) but with a marginal phyletic spread. However, in certain eukaryotic lineages they expanded to give rise to some of the largest families of paralogous specific TFs encoded by those genomes.
Finally, while bacteria possess chromatin proteins that package genomic DNA in a functionally analogous manner to the eukaryotes, with some exceptions, they do not possess the remarkable array of chromatin-remodeling and modifying enzyme complexes that are conserved throughout eukaryotes (Iyer et al., 2008). These eukaryotic complexes include Swi2/Snf2 ATPases (a specific version of the superfamily-II helicases), acetylases, methylases, ubiquitin-conjugating enzymes, deacetylases, demethylases and deubiquitinating enzymes, which remodel chromatin proteins in an ATP-dependent manner or modify histone-side chains covalently or remove such covalent modifications. Bacteria possess two kinds of Swi2/Snf2 ATPases, RapA/HepA and restriction-modification system associated Swi2/Snf2 ATPases. The RapA/HepA protein is highly conserved in bacteria and associates with dsDNA and the RNA polymerase. In bacteria the RNA polymerase after performing a single or a limited set of transcription cycles become incapable of further activity unless it is taken off the template and allowed to re-associate with σ and this recycling is catalyzed by the RapA/HepA Swi2/Snf ATPase (Nechaev and Severinov, 2008; Shaw et al., 2008). Thus, this bacterial Swi2/Snf2 ATPase is mechanistically similar to the eukaryotic Swi2/Snf2 ATPases in reorganizing protein-DNA contacts in an ATP-dependent manner which might involve their helicase activity. However, the bacterial version appears to be functionally distinct in that it appears to play no such role with respect to the bacterial chromatin. The Swi2/Snf2 ATPases associated with the restriction-modification systems appear to be required for remodeling the protein-DNA complexes in facilitating restriction enzymes that cut sites distant to their recognition site (Iyer et al., 2006). Thus, while these systems again mechanistically resemble their eukaryotic counterparts they do not appear to have any dedicated transcription related function. Likewise, while some bacteria possess chromatin-modifying SET domain methylases (e.g. in Chlamydia) (Koonin et al., 2001), which might function in conjunction with a SWIB domain protein (also found in eukaryotic chromatin remodeling complexes) and a topoisomerase (Aravind et al., 2011). However, this does not appear to be a widely used regulatory mechanism. Similarly, covalent modification of chromatin proteins, like that seen in eukaryotes, is not prevalent in bacteria.
8. Future directions
With the recent advances in genomics and structural studies we have come a long way in our understanding of the bacterial transcription apparatus since the proposal of the operon theory of bacterial gene regulation and the discovery of the RNA polymerase. Yet, the increasing focus on eukaryotic transcription systems has resulted in the more interesting problems in bacterial transcription regulation being neglected to a certain degree. In particular, the discoveries of a rather invariant scaling of TFs in bacterial genomes and differences in the underlying architecture of bacterial and eukaryotic TRNs emphasize the need for more studies on bacterial TRNs. These need to be directed at questions such as: (1) Why exactly do these scaling laws hold across widely different bacteria? (2) Do bacteria with more complex signaling systems (e.g. actinobacteria, cyanobacteria and myxobacteria) and architecturally complex specific TFs (i.e. the STAND domain TFs) possess differences in the organization of the TRNs? (3) Are there any discernable patterns in terms of the TRN hubs which emerge in different bacterial lineages? (4) Can the binding sites of TFs be identified on a genome scale? (6) Can a comprehensive catalogue of the effectors bound by bacterial single-component systems be developed? (7) What do the archaeal TRNs look like and do they differ in any way from the bacterial versions? These and other questions firstly require a dedicated experimental program that is ready to explore systems beyond model bacteria such as E. coli and B. subtilis. The existence of genome sequences and reverse-genetics approaches for a wide range of bacteria make these studies at least technically feasible. The computational analysis of the data emerging from such studies is likely to open unexpected vistas and offer some of the most fundamental insights into the functions and evolution of prokaryotes.
9. Material and methods
Iterative sequence profile searches were performed using the PSI-BLAST (Altschul et al., 1997) and JACKHMMER programs (Eddy, 2009) run against the non-redundant (NR) protein database of National Center for Biotechnology Information (NCBI). Similarity-based clustering for both classification and culling of nearly identical sequences was performed using the BLASTCLUST program (ftp://ftp.ncbi.nih.gov/blast/documents/blastclust.html). The HHpred program was used for profile-profile comparisons (Soding et al., 2005). Structure similarity searches were performed using the DaliLite program (Holm et al., 2008). Multiple sequence alignments were built by the Kalign (Lassmann et al., 2009) and PCMA programs (Pei et al., 2003), followed by manual adjustments on the basis of profile-profile and structural alignments. Secondary structures were predicted using the JPred program (Cole et al., 2008). For previously known domains, the Pfam database (Finn et al., 2010) was used as a guide, though the profiles were occasionally augmented by addition of newly detected divergent members that were not detected by the original Pfam models. Clustering with BLASTCLUST followed by multiple sequence alignment and further sequence profile searches were used to identify other domains that were not present in the Pfam database. Contextual information from prokaryotic gene neighborhoods was retrieved by a Perl custom script that extracts the upstream and downstream genes of the query gene and uses BLASTCLUST to cluster the proteins to identify conserved gene-neighborhoods. Phylogenetic analysis was conducted using an approximately-maximum-likelihood method implemented in the FastTree 2.1 program under default parameters (Price et al., 2010). Structural visualization and manipulations were performed using the PyMol (http://www.pymol.org) program. The in-house TASS package, which comprises a collection of Perl scripts, was used to automate aspects of large-scale analysis of sequences, structures and genome context (Anantharaman, V., Balaji, S., and Aravind, L., unpublished).
Supplementary Material
Acknowledgements
Work by the authors is supported by the intra-mural funds of the National Library of Medicine, National Institutes of Health, USA.
Footnotes
Supplementary material is also available at: ftp://ftp.ncbi.-nih.gov/pub/aravind/PROKHTH/prok_trans.html.
Appendix A. Supplementary material
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jsb.2011.12.013.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammelburg M, Frickey T, Lupas AN. Classification of AAA+ proteins. J. Struct. Biol. 2006;156:2–11. doi: 10.1016/j.jsb.2006.05.002. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. New connections in the prokaryotic toxin–antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. Diversification of catalytic activities and ligand interactions in the protein fold shared by the sugar isomerases, eIF2B, DeoR transcription factors, acyl-CoA transferases and methenyltetrahydrofolate synthetase. J. Mol. Biol. 2006;356:823–842. doi: 10.1016/j.jmb.2005.11.031. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J. Mol. Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM. The HARE-HTH and associated domains: novel modules in the coordination of epigenetic DNA and protein modifications. Cell Cycle. 2012;11:1–13. doi: 10.4161/cc.11.1.18475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. DNA-binding proteins and evolution of transcription regulation in the archaea. Nucleic Acids Res. 1999a;27:4658–4670. doi: 10.1093/nar/27.23.4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999b;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. A natural classification of ribonucleases. Methods Enzymol. 2001;341:3–28. doi: 10.1016/s0076-6879(01)41142-6. [DOI] [PubMed] [Google Scholar]
- Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Aravind L, Iyer LM, Anantharaman V. Natural history of sensor domains in bacterial signaling systems. In: Spiro S, Dixon R, editors. Sensory Mechanisms in Bacteria: Molecular Aspects of Signal Recognition. London: Caister Academic Press; 2010. [Google Scholar]
- Aravind L, Abhiman S, Iyer LM. Natural history of the eukaryotic chromatin protein methylation system. Prog. Mol. Biol. Transl. Sci. 2011;101:105–176. doi: 10.1016/B978-0-12-387685-0.00004-4. [DOI] [PubMed] [Google Scholar]
- Augustus AM, Sage H, Spicer LD. Binding of MetJ repressor to specific and nonspecific DNA and effect of S-adenosylmethionine on these interactions. Biochemistry. 2010;49:3289–3295. doi: 10.1021/bi902011f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu MM, Luscombe NM, Aravind L, Gerstein M, Teichmann SA. Structure and evolution of transcriptional regulatory networks. Curr. Opin. Struct. Biol. 2004;14:283–291. doi: 10.1016/j.sbi.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Balaji S, Aravind L. The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 2007;35:5658–5671. doi: 10.1093/nar/gkm558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Iyer LM, Aravind L, Babu MM. Uncovering a hidden distributed architecture behind scale-free transcriptional regulatory networks. J. Mol. Biol. 2006a;360:204–212. doi: 10.1016/j.jmb.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Luscombe NM, Aravind L. Comprehensive analysis of combinatorial regulation using the transcriptional regulatory network of yeast. J. Mol. Biol. 2006b;360:213–227. doi: 10.1016/j.jmb.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Aravind L. Interplay between network structures, regulatory modes and sensing mechanisms of transcription factors in the transcriptional regulatory network of E. coli . J. Mol. Biol. 2007;372:1108–1122. doi: 10.1016/j.jmb.2007.06.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balazsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli . Proc. Natl. Acad. Sci. USA. 2005;102:7841–7846. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Bonabeau E. Scale-free networks. Sci. Am. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- Barabote RD, Saier MH., Jr Comparative genomic analyses of the bacterial phosphotransferase system. Microbiol. Mol. Biol. Rev. 2005;69:608–634. doi: 10.1128/MMBR.69.4.608-634.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barne KA, Bown JA, Busby SJ, Minchin SD. Region 2.5 of the Escherichia coli RNA polymerase sigma70 subunit is responsible for the recognition of the ‘extended-10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 1997;22:12–13. doi: 10.1016/s0968-0004(96)30046-7. [DOI] [PubMed] [Google Scholar]
- Be’er A, Florin EL, Fisher CR, Swinney HL, Payne SM. Surviving bacterial sibling rivalry: inducible and reversible phenotypic switching in Paenibacillus dendritiformis. MBio. 2011;2:e00069–11. doi: 10.1128/mBio.00069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Jacob E. Bacterial self-organization: co-enhancement of complexification and adaptability in a dynamic environment. Philos. Trans. Math. Phys. Eng. Sci. 2003;361:1283–1312. doi: 10.1098/rsta.2003.1199. [DOI] [PubMed] [Google Scholar]
- Brennan RG. The winged-helix DNA-binding motif: another helix-turn-helix takeoff. Cell. 1993;74:773–776. doi: 10.1016/0092-8674(93)90456-z. [DOI] [PubMed] [Google Scholar]
- Brennan RG, Matthews BW. The helix-turn-helix DNA binding motif. J. Biol. Chem. 1989;264:1903–1906. [PubMed] [Google Scholar]
- Brinkman AB, Ettema TJ, de Vos WM, van der Oost J. The Lrp family of transcriptional regulators. Mol. Microbiol. 2003;48:287–294. doi: 10.1046/j.1365-2958.2003.03442.x. [DOI] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol. Rev. 2003;27:145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Bull PC, Cox DW. Wilson disease and Menkes disease: new handles on heavy-metal transport. Trends Genet. 1994;10:246–252. doi: 10.1016/0168-9525(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Burley SK. The TATA box binding protein. Curr. Opin. Struct. Biol. 1996;6:69–75. doi: 10.1016/s0959-440x(96)80097-2. [DOI] [PubMed] [Google Scholar]
- Campbell EA, Muzzin O, Chlenov M, Sun JL, Olson CA, Weinman O, Trester-Zedlitz ML, Darst SA. Structure of the bacterial RNA polymerase promoter specificity sigma subunit. Mol. Cell. 2002;9:527–539. doi: 10.1016/s1097-2765(02)00470-7. [DOI] [PubMed] [Google Scholar]
- Castillo RM, Mizuguchi K, Dhanaraj V, Albert A, Blundell TL, Murzin AG. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure. 1999;7:227–236. doi: 10.1016/s0969-2126(99)80028-8. [DOI] [PubMed] [Google Scholar]
- Chlenov M, Masuda S, Murakami KS, Nikiforov V, Darst SA, Mustaev A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase beta’ subunit. J. Mol. Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- Chou AY, Archdeacon J, Kado CI. Agrobacterium transcriptional regulator Ros is a prokaryotic zinc finger protein that regulates the plant oncogene ipt. Proc. Natl. Acad. Sci. USA. 1998;95:5293–5298. doi: 10.1073/pnas.95.9.5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/ fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado-Vides J, Salgado H, Morett E, Gama-Castro S, Jimenez-Jacinto V, Martinez-Flores I, Medina-Rivera A, Muniz-Rascado L, Peralta-Gil M, Santos-Zavaleta A. Bioinformatics resources for the study of gene regulation in bacteria. J. Bacteriol. 2009;191:23–31. doi: 10.1128/JB.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes MH, Walsh NP, McKnight CJ, Sauer RT. Evolution of a protein fold in vitro. Science. 1999;284:325–328. doi: 10.1126/science.284.5412.325. [DOI] [PubMed] [Google Scholar]
- Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- Doucleff M, Malak LT, Pelton JG, Wemmer DE. The C-terminal RpoN domain of sigma54 forms an unpredicted helix-turn-helix motif similar to domains of sigma70. J. Biol. Chem. 2005;280:41530–41536. doi: 10.1074/jbc.M509010200. [DOI] [PubMed] [Google Scholar]
- Eddy SR. A new generation of homology search tools based on probabilistic inference. Genome Inform. 2009;23:205–211. [PubMed] [Google Scholar]
- Esposito S, Baglivo I, Malgieri G, Russo L, Zaccaro L, D’Andrea LD, Mammucari M, Di Blasio B, Isernia C, Fattorusso R, Pedone PV. A novel type of zinc finger DNA binding domain in the Agrobacterium tumefaciens transcriptional regulator Ros. Biochemistry. 2006;45:10394–10405. doi: 10.1021/bi060697m. [DOI] [PubMed] [Google Scholar]
- Feklistov A, Darst SA. Structural basis for promoter-10 element recognition by the bacterial RNA polymerase sigma subunit. Cell. 2011 doi: 10.1016/j.cell.2011.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, Holm L, Sonnhammer EL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromme JC, Banerjee A, Huang SJ, Verdine GL. Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase. Nature. 2004;427:652–656. doi: 10.1038/nature02306. [DOI] [PubMed] [Google Scholar]
- Fujikawa N, Kurumizaka H, Nureki O, Terada T, Shirouzu M, Katayama T, Yokoyama S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acids Res. 2003;31:2077–2086. doi: 10.1093/nar/gkg309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S. The small RNA regulators of Escherichia coli: roles and mechanisms*. Annu. Rev. Microbiol. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- Grinberg I, Shteinberg T, Gorovitz B, Aharonowitz Y, Cohen G, Borovok I. The Streptomyces NrdR transcriptional regulator is a Zn ribbon/ATP cone protein that binds to the promoter regions of class Ia and class II ribonucleotide reductase operons. J. Bacteriol. 2006;188:7635–7644. doi: 10.1128/JB.00903-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber TM, Gross CA. Multiple sigma subunits and the partitioning of bacterial transcription space. Annu. Rev. Microbiol. 2003;57:441–466. doi: 10.1146/annurev.micro.57.030502.090913. [DOI] [PubMed] [Google Scholar]
- Guelzim N, Bottani S, Bourgine P, Kepes F. Topological and causal structure of the yeast transcriptional regulatory network. Nat. Genet. 2002;31:60–63. doi: 10.1038/ng873. [DOI] [PubMed] [Google Scholar]
- Hantke K. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- Harrison SC. A structural taxonomy of DNA-binding domains. Nature. 1991;353:715–719. doi: 10.1038/353715a0. [DOI] [PubMed] [Google Scholar]
- Heldwein EE, Brennan RG. Crystal structure of the transcription activator BmrR bound to DNA and a drug. Nature. 2001;409:378–382. doi: 10.1038/35053138. [DOI] [PubMed] [Google Scholar]
- Helmann JD. The extracytoplasmic function (ECF) sigma factors. Adv. Microb. Physiol. 2002;46:47–110. doi: 10.1016/s0065-2911(02)46002-x. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem. Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzendorff J, Hung D, Brende P, Reisenauer A, Viollier PH, McAdams HH, Shapiro L. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- Hong E, Doucleff M, Wemmer DE. Structure of the RNA polymerase core-binding domain of sigma(54) reveals a likely conformational fracture point. J. Mol. Biol. 2009;390:70–82. doi: 10.1016/j.jmb.2009.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson BP, Quispe J, Lara-Gonzalez S, Kim Y, Berman HM, Arnold E, Ebright RH, Lawson CL. Three-dimensional EM structure of an intact activator-dependent transcription initiation complex. Proc. Natl. Acad. Sci. USA. 2009;106:19830–19835. doi: 10.1073/pnas.0908782106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulko M, Lupas AN, Martin J. Inherent chaperone-like activity of aspartic proteases reveals a distant evolutionary relation to double-psi barrel domains of AAA-ATPases. Protein Sci. 2007;16:644–653. doi: 10.1110/ps.062478607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itou H, Tanaka I. The OmpR-family of proteins: insight into the tertiary structure and functions of two-component regulator proteins. J. Biochem. 2001;129:343–350. doi: 10.1093/oxfordjournals.jbchem.a002863. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004a;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Makarova KS, Koonin EV, Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004b;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Babu MM, Aravind L. The HIRAN domain and recruitment of chromatin remodeling and repair activities to damaged DNA. Cell Cycle. 2006;5:775–782. doi: 10.4161/cc.5.7.2629. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Anantharaman V, Wolf MY, Aravind L. Comparative genomics of transcription factors and chromatin proteins in parasitic protists and other eukaryotes. Int. J. Parasitol. 2008;38:1–31. doi: 10.1016/j.ijpara.2007.07.018. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Abhiman S, de Souza RF, Aravind L. Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 2010;38:5261–5279. doi: 10.1093/nar/gkq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Ju J, Mitchell T, Peters H, 3rd, Haldenwang WG. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 1999;181:4969–4977. doi: 10.1128/jb.181.16.4969-4977.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juan Wu L, Errington J. Identification and characterization of a new prespore-specific regulatory gene, rsfA, of Bacillus subtilis . J. Bacteriol. 2000;182:418–424. doi: 10.1128/jb.182.2.418-424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannan N, Wu J, Anand GS, Yooseph S, Neuwald AF, Venter JC, Taylor SS. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007;8:R264. doi: 10.1186/gb-2007-8-12-r264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Cell population heterogeneity during growth of Bacillus subtilis . Genes Dev. 2005;19:3083–3094. doi: 10.1101/gad.1373905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller M, Roxlau A, Weng WM, Schmidt M, Quandt J, Niehaus K, Jording D, Arnold W, Puhler A. Molecular analysis of the Rhizobium meliloti mucR gene regulating the biosynthesis of the exopolysaccharides succinoglycan and galactoglucan. Mol. Plant Microbe Interact. 1995;8:267–277. doi: 10.1094/mpmi-8-0267. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Makarova KS, Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korner H, Sofia HJ, Zumft WG. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 2003;27:559–592. doi: 10.1016/S0168-6445(03)00066-4. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Zeller ME, Armache KJ, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- Lamour V, Rutherford ST, Kuznedelov K, Ramagopal UA, Gourse RL, Severinov K, Darst SA. Crystal structure of Escherichia coli Rnk, a new RNA polymerase-interacting protein. J. Mol. Biol. 2008;383:367–379. doi: 10.1016/j.jmb.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J. Mol. Biol. 2010a;395:671–685. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 2010b;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larquet E, Schreiber V, Boisset N, Richet E. Oligomeric assemblies of the Escherichia coli MalT transcriptional activator revealed by cryo-electron microscopy and image processing. J. Mol. Biol. 2004;343:1159–1169. doi: 10.1016/j.jmb.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lassmann T, Frings O, Sonnhammer EL. Kalign2: high-performance multiple alignment of protein and nucleotide sequences allowing external features. Nucleic Acids Res. 2009;37:858–865. doi: 10.1093/nar/gkn1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchman DS. Transcription factors: an overview. Int. J. Biochem. Cell Biol. 1997;29:1305–1312. doi: 10.1016/s1357-2725(97)00085-x. [DOI] [PubMed] [Google Scholar]
- Lee PC, Umeyama T, Horinouchi S. AfsS is a target of AfsR, a transcriptional factor with ATPase activity that globally controls secondary metabolism in Streptomyces coelicolor A3(2) Mol. Microbiol. 2002;43:1413–1430. doi: 10.1046/j.1365-2958.2002.02840.x. [DOI] [PubMed] [Google Scholar]
- Lee IM, Zhao Y, Bottner KD. Novel insertion sequence-like elements in phytoplasma strains of the aster yellows group are putative new members of the IS3 family. FEMS Microbiol. Lett. 2005;242:353–360. doi: 10.1016/j.femsle.2004.11.036. [DOI] [PubMed] [Google Scholar]
- Leipe DD, Koonin EV, Aravind L. STAND, a class of P-loop NTPases including animal and plant regulators of programmed cell death: multiple, complex domain architectures, unusual phyletic patterns, and evolution by horizontal gene transfer. J. Mol. Biol. 2004;343:1–28. doi: 10.1016/j.jmb.2004.08.023. [DOI] [PubMed] [Google Scholar]
- Lopez de Saro FJ, Yoshikawa N, Helmann JD. Expression, abundance, and RNA polymerase binding properties of the delta factor of Bacillus subtilis . J. Biol. Chem. 1999;274:15953–15958. doi: 10.1074/jbc.274.22.15953. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Teichmann SA, Aravind L. Evolutionary dynamics of prokaryotic transcriptional regulatory networks. J. Mol. Biol. 2006;358:614–633. doi: 10.1016/j.jmb.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Madan Babu M, Balaji S, Aravind L. General trends in the evolution of prokaryotic transcriptional regulatory networks. Genome Dyn. 2007;3:66–80. doi: 10.1159/000107604. [DOI] [PubMed] [Google Scholar]
- Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli. RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–2675. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Aravind L, Wolf YI, Koonin EV. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani N, Dupuy B. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc. Natl. Acad. Sci. USA. 2001;98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquenet E, Richet E. Conserved motifs involved in ATP hydrolysis by MalT, a signal transduction ATPase with numerous domains from Escherichia coli . J. Bacteriol. 2010;192:5181–5191. doi: 10.1128/JB.00522-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas J, Soppa J, Strunnikov AV, Graumann PL. Cell cycle-dependent localization of two novel prokaryotic chromosome segregation and condensation proteins in Bacillus subtilis that interact with SMC protein. EMBO J. 2002;21:3108–3118. doi: 10.1093/emboj/cdf314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathew R, Chatterji D. The evolving story of the omega subunit of bacterial RNA polymerase. Trends Microbiol. 2006;14:450–455. doi: 10.1016/j.tim.2006.08.002. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc. Natl. Acad. Sci. USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messer W, Weigel C. DnaA as a transcription regulator. Methods Enzymol. 2003;370:338–349. doi: 10.1016/S0076-6879(03)70030-5. [DOI] [PubMed] [Google Scholar]
- Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit omega and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc. Natl. Acad. Sci. USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Kremer L, Girard-Blanc C, Besra GS, Cozzone AJ, Prost JF. An FHA phosphoprotein recognition domain mediates protein EmbR phosphorylation by PknH, a Ser/Thr protein kinase from Mycobacterium tuberculosis. Biochemistry. 2003;42:15300–15309. doi: 10.1021/bi035150b. [DOI] [PubMed] [Google Scholar]
- Mooney RA, Darst SA, Landick R. Sigma and RNA polymerase: an on-again, off-again relationship? Mol. Cell. 2005;20:335–345. doi: 10.1016/j.molcel.2005.10.015. [DOI] [PubMed] [Google Scholar]
- Morett E, Bork P. Evolution of new protein function: recombinational enhancer Fis originated by horizontal gene transfer from the transcriptional regulator NtrC. FEBS Lett. 1998;433:108–112. doi: 10.1016/s0014-5793(98)00888-6. [DOI] [PubMed] [Google Scholar]
- Motackova V, Sanderova H, Zidek L, Novacek J, Padrta P, Svenkova A, Korelusova J, Jonak J, Krasny L, Sklenar V. Solution structure of the N-terminal domain of Bacillus subtilis delta subunit of RNA polymerase and its classification based on structural homologs. Proteins. 2010;78:1807–1810. doi: 10.1002/prot.22708. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: an RNA polymerase holoenzyme-DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Severinov K. RapA: completing the transcription cycle? Structure. 2008;16:1294–1295. doi: 10.1016/j.str.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Nikolov DB, Chen H, Halay ED, Usheva AA, Hisatake K, Lee DK, Roeder RG, Burley SK. Crystal structure of a TFIIB-TBP-TATA-element ternary complex. Nature. 1995;377:119–128. doi: 10.1038/377119a0. [DOI] [PubMed] [Google Scholar]
- Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Helmann JD. The sigma70 family of sigma factors. Genome Biol. 2003;4:203. doi: 10.1186/gb-2003-4-1-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Kang JG, Roe JH, Buttner MJ. SigmaR, an RNA polymerase sigma factor that modulates expression of the thioredoxin system in response to oxidative stress in Streptomyces coelicolor A3(2) EMBO J. 1998;17:5776–5782. doi: 10.1093/emboj/17.19.5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paget MS, Leibovitz E, Buttner MJ. A putative two-component signal transduction system regulates sigmaE, a sigma factor required for normal cell wall integrity in Streptomyces coelicolor A3(2) Mol. Microbiol. 1999;33:97–107. doi: 10.1046/j.1365-2958.1999.01452.x. [DOI] [PubMed] [Google Scholar]
- Pao GM, Saier MH., Jr Response regulators of bacterial signal transduction systems: selective domain shuffling during evolution. J. Mol. Evol. 1995;40:136–154. doi: 10.1007/BF00167109. [DOI] [PubMed] [Google Scholar]
- Peat TS, Frank EG, McDonald JP, Levine AS, Woodgate R, Hendrickson WA. Structure of the UmuD’ protein and its regulation in response to DNA damage. Nature. 1996;380:727–730. doi: 10.1038/380727a0. [DOI] [PubMed] [Google Scholar]
- Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate multiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- Penalver-Mellado M, Garcia-Heras F, Padmanabhan S, Garcia-Moreno D, Murillo FJ, Elias-Arnanz M. Recruitment of a novel zinc-bound transcriptional factor by a bacterial HMGA-type protein is required for regulating multiple processes in Myxococcus xanthus . Mol. Microbiol. 2006;61:910–926. doi: 10.1111/j.1365-2958.2006.05289.x. [DOI] [PubMed] [Google Scholar]
- Pineda M, Gregory BD, Szczypinski B, Baxter KR, Hochschild A, Miller ES, Hinton DM. A family of anti-sigma70 proteins in T4-type phages and bacteria that are similar to AsiA, a Transcription inhibitor and co-activator of bacteriophage T4. J. Mol. Biol. 2004;344:1183–1197. doi: 10.1016/j.jmb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Poon KK, Chu JC, Wong SL. Roles of glucitol in the GutR-mediated transcription activation process in Bacillus subtilis: glucitol induces GutR to change its conformation and to bind ATP. J. Biol. Chem. 2001;276:29819–29825. doi: 10.1074/jbc.M100905200. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. 3rd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 2004. A Genetic Switch: Phage Lambda Revisited. [Google Scholar]
- Rombel I, North A, Hwang I, Wyman C, Kustu S. The bacterial enhancer-binding protein NtrC as a molecular machine. Cold Spring Harb. Symp. Quant. Biol. 1998;63:157–166. doi: 10.1101/sqb.1998.63.157. [DOI] [PubMed] [Google Scholar]
- Ruprich-Robert G, Thuriaux P. Non-canonical DNA transcription enzymes and the conservation of two-barrel RNA polymerases. Nucleic Acids Res. 2010;38:4559–4569. doi: 10.1093/nar/gkq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgado PS, Koivunen MR, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:e434. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savijoki K, Ingmer H, Frees D, Vogensen FK, Palva A, Varmanen P. Heat and DNA damage induction of the LexA-like regulator HdiR from Lactococcus lactis is mediated by RecA and ClpP. Mol. Microbiol. 2003;50:609–621. doi: 10.1046/j.1365-2958.2003.03713.x. [DOI] [PubMed] [Google Scholar]
- Shaw G, Gan J, Zhou YN, Zhi H, Subburaman P, Zhang R, Joachimiak A, Jin DJ, Ji X. Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli . Nat. Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Singh SK, Kurnasov OV, Chen B, Robinson H, Grishin NV, Osterman AL, Zhang H. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J. Biol. Chem. 2002;277:33291–33299. doi: 10.1074/jbc.M204368200. [DOI] [PubMed] [Google Scholar]
- Smeets LC, Becker SC, Barcak GJ, Vandenbroucke-Grauls CM, Bitter W, Goosen N. Functional characterization of the competence protein DprA/ Smf in Escherichia coli . FEMS Microbiol. Lett. 2006;263:223–228. doi: 10.1111/j.1574-6968.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Ogasawara N, Moriya S. Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol. Microbiol. 2002;45:59–71. doi: 10.1046/j.1365-2958.2002.03012.x. [DOI] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis . Annu. Rev. Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Stulke J, Arnaud M, Rapoport G, Martin-Verstraete I. PRD – a protein domain involved in PTS-dependent induction and carbon catabolite repression of catabolic operons in bacteria. Mol. Microbiol. 1998;28:865–874. doi: 10.1046/j.1365-2958.1998.00839.x. [DOI] [PubMed] [Google Scholar]
- Subramanian G, Koonin EV, Aravind L. Comparative genome analysis of the pathogenic spirochetes Borrelia burgdorferi and Treponema pallidum . Infect. Immun. 2000;68:1633–1648. doi: 10.1128/iai.68.3.1633-1648.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindells MB. Identification of a common fold in the replication terminator protein suggests a possible mode for DNA binding. Trends Biochem. Sci. 1995;20:300–302. doi: 10.1016/s0968-0004(00)89055-6. [DOI] [PubMed] [Google Scholar]
- Tam R, Saier MH., Jr Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol. Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thieffry D, Huerta AM, Perez-Rueda E, Collado-Vides J. From specific gene regulation to genomic networks: a global analysis of transcriptional regulation in Escherichia coli . BioEssays. 1998;20:433–440. doi: 10.1002/(SICI)1521-1878(199805)20:5<433::AID-BIES10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Tobisch S, Stulke J, Hecker M. Regulation of the lic operon of Bacillus subtilis and characterization of potential phosphorylation sites of the LicR regulator protein by site-directed mutagenesis. J. Bacteriol. 1999;181:4995–5003. doi: 10.1128/jb.181.16.4995-5003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- Tyrrell R, Verschueren KH, Dodson EJ, Murshudov GN, Addy C, Wilkinson AJ. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure. 1997;5:1017–1032. doi: 10.1016/s0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]
- Ulrich LE, Zhulin IB. MiST: a microbial signal transduction database. Nucleic Acids Res. 2007;35:D386–D390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nimwegen E. Scaling laws in the functional content of genomes. Trends Genet. 2003;19:479–484. doi: 10.1016/S0168-9525(03)00203-8. [DOI] [PubMed] [Google Scholar]
- Vartak NB, Reizer J, Reizer A, Gripp JT, Groisman EA, Wu LF, Tomich JM, Saier MH., Jr Sequence and evolution of the FruR protein of Salmonella typhimurium: a pleiotropic transcriptional regulatory protein possessing both activator and repressor functions which is homologous to the periplasmic ribose-binding protein. Res. Microbiol. 1991;142:951–963. doi: 10.1016/0923-2508(91)90005-u. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhao S, Somerville RL, Jardetzky O. Solution structure of the DNA-binding domain of the TyrR protein of Haemophilus influenzae. Protein Sci. 2001;10:592–598. doi: 10.1110/ps.45301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassarman KM. 6S RNA: a regulator of transcription. Mol. Microbiol. 2007;65:1425–1431. doi: 10.1111/j.1365-2958.2007.05894.x. [DOI] [PubMed] [Google Scholar]
- Watson JD. Molecular biology of the gene. fifth ed. San Francisco (Woodbury, NY): Pearson/Benjamin Cummings, CSHL Press; 2004. [Google Scholar]
- Weekes D, Miller MD, Krishna SS, McMullan D, McPhillips TM, Acosta C, Canaves JM, Elsliger MA, Floyd R, Grzechnik SK, Jaroszewski L, Klock HE, Koesema E, Kovarik JS, Kreusch A, Morse AT, Quijano K, Spraggon G, van den Bedem H, Wolf G, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Crystal structure of a transcription regulator (TM1602) from Thermotoga maritima at 2.3 A resolution. Proteins. 2007;67:247–252. doi: 10.1002/prot.21221. [DOI] [PubMed] [Google Scholar]
- West AH, Stock AM. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 2001;26:369–376. doi: 10.1016/s0968-0004(01)01852-7. [DOI] [PubMed] [Google Scholar]
- Westblade LF, Campbell EA, Pukhrambam C, Padovan JC, Nickels BE, Lamour V, Darst SA. Structural basis for the bacterial transcription-repair coupling factor/RNA polymerase interaction. Nucleic Acids Res. 2010;38:8357–8369. doi: 10.1093/nar/gkq692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- Wigneshweraraj SR, Kuznedelov K, Severinov K, Buck M. Multiple roles of the RNA polymerase beta subunit flap domain in sigma 54-dependent transcription. J. Biol. Chem. 2003;278:3455–3465. doi: 10.1074/jbc.M209442200. [DOI] [PubMed] [Google Scholar]
- Wigneshweraraj S, Bose D, Burrows PC, Joly N, Schumacher J, Rappas M, Pape T, Zhang X, Stockley P, Severinov K, Buck M. Modus operandi of the bacterial RNA polymerase containing the sigma54 promoter-specificity factor. Mol. Microbiol. 2008;68:538–546. doi: 10.1111/j.1365-2958.2008.06181.x. [DOI] [PubMed] [Google Scholar]
- Willkomm DK, Hartmann RK. 6S RNA - an ancient regulator of bacterial RNA polymerase rediscovered. Biol. Chem. 2005;386:1273–1277. doi: 10.1515/BC.2005.144. [DOI] [PubMed] [Google Scholar]
- Wilson KP, Shewchuk LM, Brennan RG, Otsuka AJ, Matthews BW. Escherichia coli biotin holoenzyme synthetase/bio repressor crystal structure delineates the biotin- and DNA-binding domains. Proc. Natl. Acad. Sci. USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciak JM, Iwahara J, Clubb RT. The Mu repressor-DNA complex contains an immobilized ‘wing’ within the minor groove. Nat. Struct. Biol. 2001;8:84–90. doi: 10.1038/83103. [DOI] [PubMed] [Google Scholar]
- Wood HE, Devine KM, McConnell DJ. Characterisation of a repressor gene (xre) and a temperature-sensitive allele from the Bacillus subtilis prophage, PBSX. Gene. 1990;96:83–88. doi: 10.1016/0378-1119(90)90344-q. [DOI] [PubMed] [Google Scholar]
- Yuan AH, Nickels BE, Hochschild A. The bacteriophage T4 AsiA protein contacts the beta-flap domain of RNA polymerase. Proc. Natl. Acad. Sci. USA. 2009;106:6597–6602. doi: 10.1073/pnas.0812832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chaney M, Wigneshweraraj SR, Schumacher J, Bordes P, Cannon W, Buck M. Mechanochemical ATPases and transcriptional activation. Mol. Microbiol. 2002;45:895–903. doi: 10.1046/j.1365-2958.2002.03065.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.