Abstract
Salvia miltiorrhiza Bunge is one of the most renowned traditional medicinal plants in China. Phenolic acids that are derived from the rosmarinic acid pathway, such as rosmarinic acid and salvianolic acid B, are important bioactive components in S. miltiorrhiza. Accumulations of these compounds have been reported to be induced by various elicitors, while little is known about transcription factors that function in their biosynthetic pathways. We cloned a subgroup 4 R2R3 MYB transcription factor gene (SmMYB39) from S. miltiorrhiza and characterized its roles through overexpression and RNAi-mediated silencing. As the results showed, the content of 4-coumaric acid, rosmarinic acid, salvianolic acid B, salvianolic acid A and total phenolics was dramatically decreased in SmMYB39-overexpressing S. miltiorrhiza lines while being enhanced by folds in SmMYB39-RNAi lines. Quantitative real-time PCR and enzyme activities analyses showed that SmMYB39 negatively regulated transcripts and enzyme activities of 4-hydroxylase (C4H) and tyrosine aminotransferase (TAT). These data suggest that SmMYB39 is involved in regulation of rosmarinic acid pathway and acts as a repressor through suppressing transcripts of key enzyme genes.
Introduction
S. miltiorrhiza Bunge, called ‘Dan-Shen’ in Chinese, is one of the most widely used traditional herbal medicines for the treatment of a variety of conditions, such as cardiovascular and cerebrovascular diseases [1]–[3], breast cancer [4] and hepatitis [5], [6]. The bioactive components of S. miltiorrhiza are divided into two groups, the water-soluble phenolic acids and the lipid-soluble tanshinones [7], [8]. The water-soluble phenolic acids mainly contain caffeic acid, danshensu ((r)-a,3,4-trihydroxybenzenepropanoic acid), 4-coumaric acid, t-cinnamic acid, rosmarinic acid (RA) and salvianolic acid B (SAB) [9], [10]. The lipid-soluble tanshinones include tanshinone I, tanshinone IIA, dihydrotanshinone I, cryptotanshinone and etc., which belong to a group of diterpenes with an abietane-type skeleton [11], [12]. In recent years, the water-soluble phenolic acids have attracted attention for their marked pharmacological activities coupled with their traditional use from herbs steeped in boiling water in China.
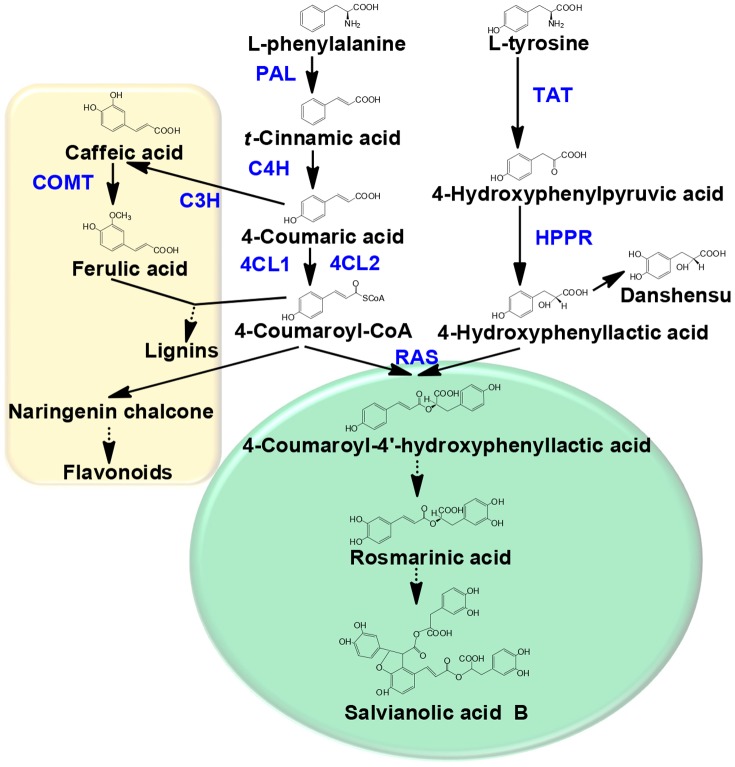
Water-soluble phenolic acids in S. miltiorrhiza are produced through the phenylpropanoid pathway and the biosynthetic pathway of RA is well characterized in plants. RA biosynthesis starts with the aromatic amino acids L-phenylalanine and L-tyrosine, which are separately converted to intermediate precursors 4-coumaroyl-CoA and 4-hydroxyphenyllactic acid through two parallel pathways. These two intermediate precursors are then covalently coupled by several biological reactions and generated RA [13], [14]. SAB is another important phenolic acid and an index chemical in the quality control of Dan-Shen [15]. It is deemed to be derived from RA, but the detailed pathway has not been characterized to date [16], [17]. Biosynthesis of most other phenolic acids are also closely related to RA pathway. For example, caffeic acid and ferulic acid are intermediate precursors of lignin biosynthetic pathway [18], which shares the upstream pathway with RA production. And t-cinnamic acid and 4-coumaric acid are the common intermediate precursors of many metabolites, such as RA, lignin and flavonoids (Fig. 1).
Figure 1. Phenolic acids biosynthetic pathway in S. miltiorrhiza.
Multiple enzymatic steps are represented by dotted lines. The ‘circle’ and ‘square’ are used to distinguish between the downstream pathway of RA and other branches of phenylpropanes metabolism. C3H, coumarate 3-hydroxylase; C4H, cinnamic acid 4-hydroxylase; 4CL, 4-coumaric acid CoA-ligase; COMT, caffeic acid O-methyltransferase; HPPR, 4-hydroxyphenylpyruvate reductase; PAL, phenylalanine ammonia-lyase; RAS, rosmarinic acid synthase; TAT, tyrosine aminotransferase.
MYB proteins are one of the largest transcription factor families in plants. Among approximately 1700 transcription factor genes identified in the Arabidopsis thaliana genome, 339 belong to MYB members [19]. Based on the numbers of imperfect repeats (one, two, three or four) in the DNA-binding domain, MYB transcription factors are classified into four subfamilies and refered as 1R-MYB, 2R-MYB, 3R-MYB and 4R-MYB, respectively [20]. MYB proteins in plants are mainly 2R-MYB type, namely R2R3 MYBs, e.g. about 137 R2R3 members in Arabidopsis and 88 ones in rice [20]. The N-terminal DNA-binding domain (in the R2 and R3 repeats) of R2R3 MYB proteins is highly conserved; however the C-termininal amino acid sequences are various and have been considered to be responsible for their different regulating functions [21]. R2R3 MYBs have been categorised into 22 subgroups on the basis of conserved amino acid sequence motifs present C-terminal to the MYB domains [22], [23]. Amongst the 22 subgroups, members of subgroup 4 are suggested to act as transcriptional repressors of phenylpropanoid pathway and reduce production of phenylpropanoid metabolites by repressing transcripts of key enzymes. Two examples are AmMYB308 and AmMYB330 from Antirrhinum majus, which reduced contents of phenolic acids and lignins when over expressed in tobacco plants. Besides, overexpression of these two genes down-regulated several key enzyme genes in the phenylpropanoid pathway, such as 4-coumaric acid CoA-ligase (4CL), cinnamate-4-hydroxylase (C4H) and cinnamyl alcohol dehydrogenase (CAD) [24]. The Arabidopsis AtMYB4 knock-out mutant exhibited an increase in the accumulation of sinapate esters and up-regulation of the C4H gene [25]. Overexpression of two subgroup 4 members from maize, ZmMYB31 and ZmMYB42, in A. thaliana down-regulated both the A. thaliana and the maize caffeic acid O -methyl-transferase (COMT) genes and reduced lignin content in the transgenic plants [26], [27]. Overexpression of FaMYB1 from strawberry severely repressed accumulation of cyanidin 3-rutinoside (an anthocyanin) and of quercetin-glycosides (flavonols) in flowers of transgenic tobacco lines [28]. MdMYB6 from Malus × domestica led to a decreased anthocyanin production when over expressed in Arabidopsis [29].
Although biosynthetic pathway of RA has been characterized, little is known about transcription factors that function in this pathway. As the RA pathway shares the common phenylpropanoid pathway with other phenylpropanoid metabolites, it will be very interesting to investigate whether phenolic acids derived from the RA pathway can be regulated by subgroup 4 R2R3 MYB transcription factors. In this study, we isolated a gene of subgroup 4 R2R3 MYB member from S. miltiorrhiza and studied its roles in regulating the RA pathway through overexpression and RNAi-mediated silencing.
Materials and Methods
Plant materials
Mature seeds of S. miltiorrhiza were collected from Dan-shen cultivation base of Shaanxi Tasly plant medicine Co. Ltd. (Shangluo, P.R.China). They were used to get sterile plantlets as described by Yan and Wang [30]. Sterile plantlets were cultured on half strength Murashige and Skoog (MS) medium [31], supplemented with 30 g l−1 sucrose and 7 g l−1 agar. Leaves of these plantlets were used for the gene transformation. Two-year-old flowering S. miltiorrhiza were obtained from the medical plants garden of Northwest A&F University (May 14, 2011) and used for the analysis of tissue-specific expression pattern of SmMYB39. The field studies did not involve any endangered or protected species, and sample collection was authorized by the Agricultural Bureau of Shangluo and the management department of the medical plants garden of Northwest A&F University.
Isolation of a subgroup 4 R2R3 MYB homolog from S. miltiorrhiza
Total RNA was isolated from sterile plantlets of S. miltiorrhiza using the RNAprep pure Plant Kit (TIANGEN, Beijing, China) and then reversely transcribed according to the manufacturer's instruction of PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan) to generate cDNA. Degenerate primers were designed from conserved DNA-binding domains of subgroup 4 R2R3 MYB repressors in other plant species with sequences SmMYBP1: 5′-AGGTC(A/C/G/T)CC(G/T)TGCTG(C/T)GAGAA-3′ and SmMYBP2: 5′-G(A/G)CT(A/G)TG(C/G)AG(C/T)TTGATGATG-3′. The 50 µl PCR mixture contained 25 µl Premix Taq (Takara, Shiga, Japan), 2 µl first-strand cDNA and 0.4 µM each primer and the PCR reaction was performed as follows: pre-heating at 94°C for 5 min, 30 cycles at 94°C for 50 s, 55°C for 50 s and 72°C for 30 s, then an extension at 72°C for 10 min. The amplified fragments were subcloned into the pMD19-T vector (Takara, Shiga, Japan) and sequenced by Sangon Biotech Co., Ltd. (Shanghai, China)
Rapid amplification of cDNA ends (RACE) was performed to get full-length cDNAs of R2R3 MYB homologs under the manufacturer's instruction (Clontech, Palo Alto, CA, USA). Gene specific primers SmGSP1 (5′-TTGAGATCGGGGCGGAGGTAGTTGA-3′) and SmGSP2 (5′-ACAAACAAAGGGGCGTGGACTAAGGAAG-3′) were designed for 5′ RACE and 3′ RACE, respectively, based on the obtained conserved sequences. The PCR condition for 5′ RACE was as follows: 5 cycles at 94°C for 30 s, 72°C for 3 min; 5 cycles at 94°C for 30 s, 70°C for 30 s, 72°C for 3 min; 27 cycles at 94°C for 30 s, 65°C for 30 s, and 72°C for 3 min. The PCR condition for 3′ RACE consisted of 5 cycles at 94°C for 30 s, 72°C for 3 min; 5 cycles at 94°C for 30 s, 70°C for 30 s, 72°C for 3 min; 32 cycles at 94°C for 30 s, 68°C for 30 s, and 72°C for 3 min. The amplified fragments were subcloned into the pMD19-T vector and sequenced. The full-length cDNA sequence was assembled based on the 3′- and 5′- RACE sequences using the Lasergene 7.1 software (DNASTAR, Inc., Madison, USA). The assembled sequence was named SmMYB39 and confirmed by PCR amplification and sequencing.
Genomic DNA was isolated from sterile plantlets of S. miltiorrhiza using the Genomic DNA Isolation Kit (Cowin Biotech, Beijing, China). Gene-specific primers SmMYBP3 (5′-ATGGGAAGGTCTCCTTGCTGTG-3′) and SmMYBP4 (5′-TCATTTCATCTCCAATCTTCTGTAA-3′) designed from the obtained full-length cDNA sequence were used to amplify the genomic clone of SmMYB39. Then the PCR products were cloned into pMD19-T vector and sequenced.
Bioinformatics analysis
Self Optimized Prediction Method with Alignment (SOPMA, http://npsa-pbil.ibcp.fr/) was used to predict secondary structure of the deduced amino acids sequence of SmMYB39. SWISS-MODEL program (http://swissmodel.expasy.org/) was used to create a 3-D structural model of SmMYB39 protein sequence based on the known crystal structure of a c-MYB from Mus musculus (name not assigned, PDB ID: 1H88, Chain ID: C) [32]–[34]. BLAST search is publicly available at the National Center for Biotechnology Information (NCBI) web site (www.ncbi.nlm.nih.gov/BLAST/), which was used to do homology search. Homologous R2R3 MYB protein sequences were used to perform a phylogenetic analysis using the PhyML method and tools available at Phylogeny.fr: (http://www.phylogeny.fr/) [35].
Subcellular localization analysis
A vector pTF486 containing the open reading frame of eGFP was used in this study [36]. Total RNA was isolated from sterile plantlets of S. miltiorrhiza using the RNAprep pure Plant Kit (TIANGEN, Beijing, China) and then reversely transcribed according to the manufacturer's instruction of PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan) to generate cDNA. The cDNA was used as the template for the subsequent RCR. The whole coding sequence of SmMYB39 was amplified with primers SmGFPP1-Sal I (5′-ACGCGTCGACATGGGAAGGTCTCCTTGCTGTG-3′) and SmGFPP2-BamH I (5′-CGCGGATCCACCACCACCACCACCTTTCATCTCCAATCTTCTGTAATCC-3′) using Pfu DNA Polymerase (Fermentas, Glen Burnie, USA). The amplification sequence was ligated with Sal I and BamH I -digested pTF486 vector to generate a SmMYB39-GFP fusion construct under the control of cauliflower mosaic virus 35S (CaMV 35S) promoter. The construct was confirmed by sequencing and used for transient transformation of onion epidermis via a gene gun (Bio-Rad, Hercules, CA, USA). After 24 h of incubation, GFP fluorescence in transformed onion cells was observed under a confocal microscope (Nikon A 1, Tokyo, Japan).
Construction of plant expression vectors and plant transformation
Total RNA was isolated from sterile plantlets of S. miltiorrhiza using the RNAprep pure Plant Kit (TIANGEN, Beijing, China) and then reversely transcribed according to the manufacturer's instruction of PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan) to generate cDNA. The cDNA was then used as a template for the construction of plant expression vectors.
The whole coding sequence of SmMYB39 was amplified with primers SmMYBP5-BamH I (5′-CGCGGATCCATGGGAAGGTCTCCTTGCTGTG-3′) and SmMYBP6-BamH I (5′-CGCGGATCCTCATTTCATCTCCAATCTTCTGTAA-3′) using Pfu DNA Polymerase (Fermentas, Glen Burnie, USA). The PCR products were cloned into pBluescript KS+ vector and then subcloned into the BamH I -digested binary vector pBI111L [37]. The direction of the inserted sequence in the construct was identified by digesting with restriction enzymes and sequencing (see Fig. S1A).
For construction of the RNAi plasmid, a 259 bp fragment from the 3′ end of SmMYB39 cDNA was PCR amplified using gene-specific primers (SmMYBP7-Xho I: 5′-CCGCTCGAGTGATCCCACTACGCATCGC-3′ and SmMYBP8-EcoR I: 5′-CCGGAATTCGGTATTTGTACAGCTGCAATCTTTG-3′; SmMYBP9- Hind III: 5′-CCCAAGCTTTGATCCCACTACGCATCGC-3′ and SmMYBP-Cla I: 5′-CCATCGATGGTATTTGTACAGCTGCAATCTTTG-3′) and ligated into the pKANNIBAL vector containing the pyruvate orthophosphate dikinase (PDK) intron [38] both in the sense and antisense orientations. An interfering box containing CaMV 35S promoter, the sense and antisense fragments on either side of the PDK intron and OCS terminator was excised from pKANNIBAL with Not I (Takara, Shiga, Japan) and cloned into the pART27 vector (see Fig. S1A) [39]. The recombinant plasmid was then identified by sequencing.
Constructs were introduced into Agrobacterium tumefaciens EHA105 by electroporation (Eppendorf Multiporator, Eppendorf, AG, Germany). Kanamycin-resistant colonies were verified by PCR-amplification, and PCR-positive colonies were used in the subsequent plant transformation. Transgenic plantlets were obtained as described by Song et al. [40]. A single clone of A. tumefaciens EHA105 harboring the SmMYB39-overexpression or SmMYB39-RNAi vector was inoculated into 10 ml liquid LB medium that contained 20 mg l−1 rifampicin and 50 mg l−1 kanamycin, and then grown on a shaker (180 rev. min−1) at 28°C for 16–18 h in the dark. Cells were collected by centrifugation (at 2292 g for 10 min) when the OD600 reached 0.6, and were re-suspended in 20–30 ml liquid MS medium. Sterile leaves were cut into 0.5×0.5 cm discs and pre-cultured for 1 day on the MS basal medium supplemented with 1.0 mg l−1 6-Benzylaminopurine (6-BA) and 0.1 mg l−1 1-naphthlcetic acid (NAA). Then the discs were submerged with shaking in the bacterial suspension for 25–30 min. Excess bacteria were later blotted, and the discs were transferred to the same media type and cultured for 3 days. They were then moved to a selection medium (MS basal medium supplemented with 1.0 mg l 6-BA, 0.1 mg l−1 NAA, 200 mg l−1 cefotaxime sodium and 50 mg l−1 kanamycin). After four cycles of selection (10 days each), the regenerated buds were transferred to the 1/2-strength MS basal medium supplemented with 30 mg l−1 kanamycin for root formation and elongation. Buds regenerated from leaf discs that had not been submerged in bacteria but were only cultured on the MS basal medium supplemented with 1.0 mg l−1 6-BA and 0.1 mg l−1 NAA were used as the untransformed control. Rooted plantlets were cut from their internodes into segments and were cultured on the 1/2-strength MS basal medium for propagation. Two- month-old plantlets were used for analysis.
Identification of transgenic plantlets by PCR
DNA was isolated according to the manufacturer's instruction of Genomic DNA Isolation Kit (Cowin Biotech, Beijing, China) from two-month-old kanamycin-resistant plantlets and used as template in PCR analysis for detecting transgenic lines (see Fig. S1B). Two pair primers, Sm35S1F (5′-GATTGATGTGATATCTCCACTGACG-3′) and SmMYB39-11R (5′-GAGACTACAAGCGAAGCACAAGG-3′), NPTII-1F (5′-GCTATGACTGGGCACAACAGACAAT-3′) and NPTII-1R (5′-GCGATACCGTAAAGCACGAGGA-3′), were designed for SmMYB39-overexpressing lines identification. Sm35S1F located at the 35S promoter of pBI111L vector and SmMYB39-11R was designed from the SmMYB39 gene. Both NPTII-1F and NPTII-1R located at the neomycin phosphotransferase II (NPTII) gene of pBI111L vector (see Fig. S1A). Primers used for the selection of SmMYB39-RNAi lines were Sm35S2F (5′-AAGGACAGTAGAAAAGGAAGGTGGC-3′) and SmMYB39-22R (5′-ATAGGGAGGGCTGATTCTGAGGTC-3′), NPTII-2F (5′-CCGCAACTTCTTTACCTATTTCCG-3′) and NPTII-2R (5′-CGATACCGTAAAGCACGAGGAA-3′). Sm35S2F located at the 35S promoter of the interfering box and SmMYB39-22R was designed from the 259 bp fragment of SmMYB39. NPTII-2F and NPTII-2R were designed from the NPTII gene of pART27 vector (see Fig. S1A). All PCR products were verified by sequencing.
Analysis of phenolic acids contents by HPLC
Compound extraction and analysis followed the methods described by Liang et al. [41] with minor modifications. The dried plantlet was ground to powder with mortar and pestle and sieved through a 0.45-mm screen. The sample powder (50 mg) was extracted with 10 ml 70% methanol under sonication for 45 min, and then centrifuged at 13201 g for 10 min. The supernatant was diluted with 70% methanol to 10 ml total volume and filtered through a 0.22 µm microporous membrane (Jinteng, Tianjin, China) before analysis. Contents of phenolic acids were determined by a Waters HPLC system (Waters, Milford, MA, USA) equipped with a 1525 binary pump, a manual sample injector, and a Waters 2996 photodiode array detector. Chromatography separation was performed with a C18 column (Waters, SunFire C18, 4.6 mm×250 mm, 5 µm particle size) at 30°C with a sample injection volume of 20 µl. Empower 2 software was used for data acquisition and processing. Detection at 280 nm using a flow rate of 1.0 ml min−1 over a gradient of acetonitrile (Fisher Scientific, Springfield, NJ, USA) (buffer A) against 0.02% phosphoric acid (Kermel, Tianjin, China) solution (buffer B) was used as follows: 0–10 min, 5–20% A (v/v); 10–15 min, 20–25% A (v/v); 15–20 min, 25% A (v/v); 20–25 min, 25–20% A (v/v); 25–28 min, 20–30% A (v/v); 28–36 min, 30% A (v/v); 37–44 min, 100% A (v/v). Standards of phenolic acids were purchased from the National Institute for the Control of Pharmaceutical and Biological Products (Beijing, China).
Analysis of total phenolics
The extraction and analysis of total phenolics followed methods reported by Yan et al. [42]. The dried plantlet was ground to powder with mortar and pestle and 100 mg sample powder was extracted with 15 ml phosphate buffer (75 mM, adjusted to pH 7.0 with NaOH) by vortexing for 1 min. The mixture was centrifuged at 13201 g for 20 min. The supernatant was collected as total phenolic extract, which was subsequently diluted with the extraction buffer into a suitable concentration for analysis. Total phenolic content was determined with Folin-Ciocalteu reagent (Sigma-Aldrich, St. Louis, MO, USA) using gallic acid as standard. The extract solution (20 µl) was mixed in a test tube with 1.58 ml distilled water and 100 µl Folin-Ciocalteu reagent for 8 min, and then incubated with 300 µl sodium carbonate solution at 40°C for 30 min. The absorbance was measured at 765 nm against a reagent blank without the extract.
Expression analysis by quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from transgenic and control plantlets using the RNAprep pure Plant Kit (TIANGEN, Beijing, China) and then reversely transcribed according to the manufacturer's instruction of PrimeScriptTM RT reagent Kit (Takara, Shiga, Japan) to generate cDNA. The obtained cDNA was used as template for the qRT-PCR analysis. Primers were designed as described in Table 1 to detection expression levels of SmMYB39, phenylalanine ammonia lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumaric acid CoA-ligase (4CL1 and 4CL2), tyrosine aminotransferase (TAT) and hydroxyphenylpyruvate reductase (HPPR). The constitutively expressed actin gene was used as an internal control. QRT-PCR was performed according to the manufacturer's instruction (Takara, Shiga, Japan) under the following condition: 30-s pre-denaturation at 95°C, 1 cycle; 5-s denaturation at 95°C, 30-s annealing using calculated Tm, 5-s collection fluorescence from 65°C to 95°C, 40 cycles. Quantification of the gene expression was done with comparative Ct method. Experiments were performed in triplicate, and the results were represented by their means ± SD.
Table 1. Primers used for the quantitative real-time PCR analysis.
| Gene | Accession | Forward (5′–3′) | Revere (5′–3′) |
| SmMYB39 | KC213793 | GCCCTCCCTATCAACAAGAAC | ATCCCAGAAAATCGAATCCAG |
| PAL | DQ408636.1 | ACCTACCTCGTCGCCCTATGC | CCACGCGGATCAAGTCCTTCT |
| C4H | DQ355979.1 | CCAGGAGTCCAAATAACAGAGCC | GAGCCACCAAGCGTTCACCAA |
| 4CL1 | AY237163.1 | ATTCGCATTCGCATTTCTCGG | GCGGCGTAGTGCTTCACCTTT |
| 4CL2 | AY237164.1 | CGCCAAATACGACCTTTCCTC | TGCTTCAGTCATCCCATACCC |
| TAT | DQ334606.1 | TTCAACGGCTACGCTCCAACT | AAACGGACAATGCTATCTCAAT |
| HPPR | DQ099741.1 | GACTCCAGAAACAACCCACATT | CCCAGACGACCCTCCACAAGA |
| Actin | HM231319.1 | GGTGCCCTGAGGTCCTGTT | AGGAACCACCGATCCAGACA |
Analysis of C4H and TAT activities
C4H activity was measured as described by Lamb and Rubery with some modifications [43]. The enzyme extract was added to 4.8 ml reaction buffer (50 mM phosphate buffer containing 2 mM 2-mercaptoethanol, 2 mM t-cinnamic acid and 0.5 mM NADPH) and then incubated for 30 min at 30°C. The reaction was stopped with 6 M HCl and readjusted to pH 11 with 6 M NaOH. Absorbance value of the sample was measured at 310 nm.
TAT activity was determined using the method reported by Yan et al. [42]. The reaction mixture consisted of 200 µl enzyme extract, 200 µl L-tyrosine (88 mM), 200 µl α-ketoglutarate (10 mM), 100 µl pyridoxal phosphate (0.2 mM) and 3 ml kalium phosphate buffer at pH 7.5. The mixture was incubated at 37°C for 30 min and reaction was stopped with 1 ml 10 M NaOH. Initial (0 min) and final absorbance (30 min) of the reaction solution against a reagent blank at 331 nm were recorded.
Statistical analysis
Statistical analyses were carried out with SPSS software (version 18.0, SPSS, Inc, Chicago, IL, USA). Spearman's correlation was used to assess the relation between the expression levels of SmMYB39 and total phenolics content in different tissues of two-year-old flowering S. miltiorrhiza. ANOVA was used to identify metabolites accumulation, gene transcripts and enzyme activities that showed significant (P<0.05) changes in relative abundance in the transgenic S. miltiorrhiza lines.
Results
Isolation of SmMYB39 and sequence analysis
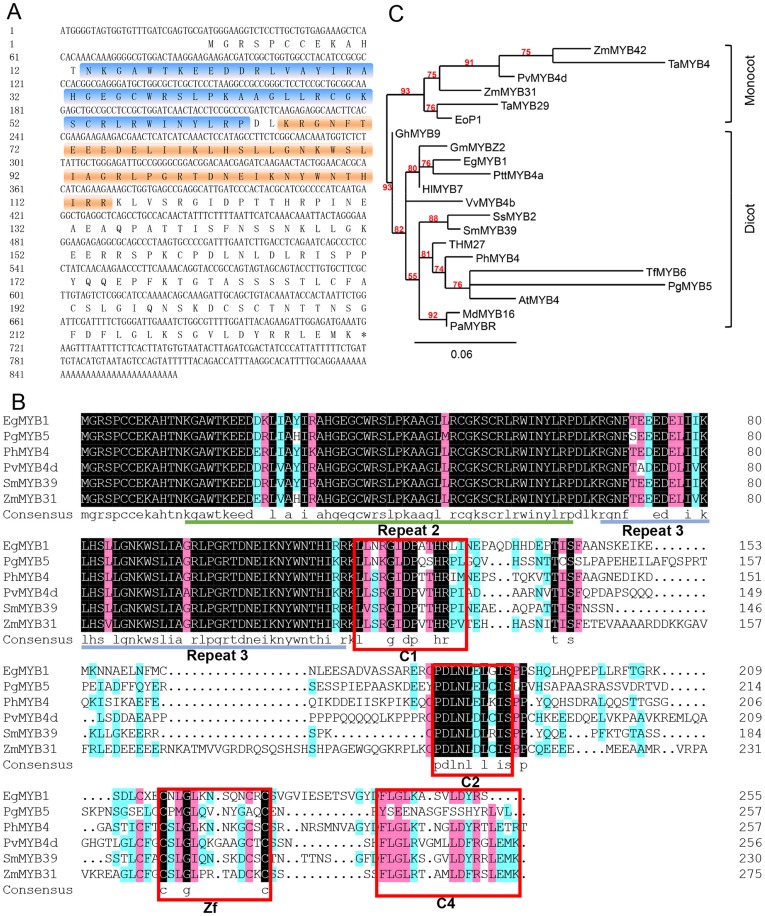
A 243-bp band was amplified with the degenerate primers. After being subcloned into pMD19-T vector, a total of six independent clones of the PCR products were sequenced; all sequences were identical and yielded a single full-length cDNA sequence (termed as SmMYB39) through RACE. SmMYB39 (Genbank accession number: KC213793) contains an open reading frame (ORF) of 693 bp in length and encodes a 25.97-kDa protein (Fig. 2A). The protein sequences alignment between SmMYB39 and five known subgroup 4 R2R3 MYB transcription factors revealed the presence of four protein motifs such as C1 (LlsrGIDPxT/SHRxI/L), C2 (pdLNLD/ELxiG/S), Zf (CX1-2CX7-12CX2C) and C4 (FLGLX4-7V/LLD/GF/YR/SX1LEMK) (Fig. 2B). They were considered as conserved protein motifs of subgroup 4 R2R3 MYBs and suggested to be essential for these proteins to act as transcriptional regulators [22], [26], [44], [45].
Figure 2. Sequence analysis of SmMYB39.
(A) Nucleotide sequence of SmMYB39 with amino acid translation. R2 and R3 repeats are highlighted in blue and orange, respectively. (B) Amino acid alignment of SmMYB39 with known R2R3-MYB regulators of phenylpropanes metabolism from other species. R2 and R3 repeats are underlined. The boxed sequences are the potential functional motifs. (C) Phylogenetic tree of SmMYB39 and known R2R3 MYB transcription factors from other plant species. Accession numbers of the proteins in the GenBank database are as follows: ZmMYB42 (NP_001106009.1, Zea mays), TaMYB4 (AEG64799.1, Triticum aestivum), PvMYB4d (AEM17351.1, Panicum virgatum), ZmMYB31 (NP_001105949.1, Zea mays), TaMYB29 (AEV91152.1, Triticum aestivum), EoP1 (ADL18407.1, Eremochloa ophiuroides), GhMYB9 (AAK19619.1, Gossypium hirsutum), GmMYBZ2 (NP_001235092.1, Glycine max), EgMYB1 (CAE09058.1, Eucalyptus gunnii), PttMYB4a (CAD98762.1, Populus tremula × Populus tremuloides), HlMYB7 (CCC14990.1, Humulus lupulus), VvMYB4b (ACN94269.1, Vitis vinifera), SsMYB2 (ABP57083.1, Solenostemon scutellarioides), THM27 (NP_001233975.1, Solanum lycopersicum), PhMYB4 (ADX33331.1, Petunia × hybrida), TfMYB6 (AAS19480.1, Tradescantia fluminensis), PgMYB5 (ABQ51221.1, Picea glauca), AtMYB4 (NP_195574.1, Arabidopsis thaliana), MdMYB16 (ADL36756.1, Malus × domestica), PaMYBR (ADY15315.1, Prunus avium).
A phylogenetic tree was constructed by the PhyML method based on an alignment of 21 R2R3 MYB protein sequences (Fig. 2C). According to the phylogenetic tree, all proteins were classified into two groups belonging to dicot and monocot plants. SmMYB39 is most closely related to SsMYB2 from Solenostemon scutellarioides, and the identity between the two amino acid sequences is 77%. It is rational as S. miltiorrhiza and S. scutellarioides belong to the same plant family (Lamiaceae). The known negative regulators of phenylpropanes metabolism, such as ZmMYB42, EgMYB1, PhMYB4 and AtMYB4, dispersedly distribute in the phylogenetic tree, suggesting that the functions of subgroup 4 R2R3 MYBs are conserved in different plant species.
The genomic sequence of SmMYB39 (Genbank accession number: KC771280) was PCR amplified from genomic DNA using gene-specific primers. Comparison of the genomic DNA and cDNA sequences revealed that SmMYB39 harbored a 69 bp intron in the R3 domain (see Fig. S2A). The secondary structure and 3-D structural model of SmMYB39 were predicted (see Text S1 and Fig. S2B, C).
SmMYB39 is located in nucleus
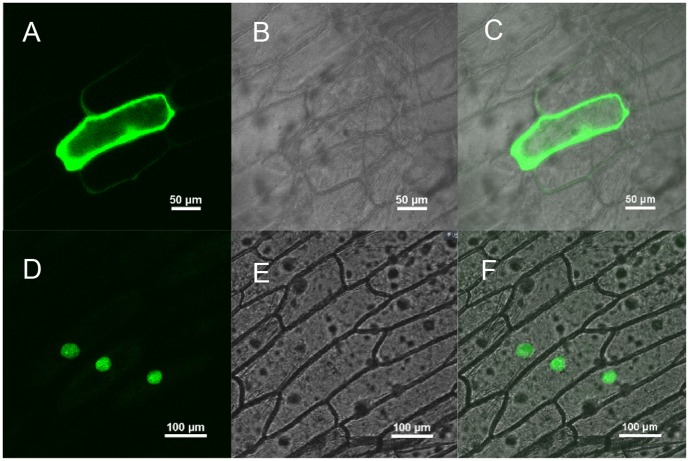
To examine the subcellular localization of SmMYB39, the open reading frame of SmMYB39 was fused to the 5′ -terminus of the GFP reporter gene under control of the CaMV 35S promoter. The recombinant constructs of the SmMYB39-GFP fusion gene and GFP alone were introduced into onion epidermal cells by particle bombardment, respectively. As showed in Fig. 3, the SmMYB39-GFP fusion protein was specifically localized in the nucleus, whereas GFP alone showed ubiquitous distribution in the whole cell. This result indicated that the SmMYB39 protein was localized in the nucleus and may act as a transcription factor in gene transcriptional regulating.
Figure 3. Nuclear localization of SmMYB39.
Confocal images of onion epidermis cells under the GFP channel show the constitutive localization of GFP (A) and nuclear localization of SmMYB39 -GFP (D). The confocal images (B) and (E) are of the same cells in (A) and (D) with transmitted light, respectively. The image (C) is the merged image of image (A) and (B), and the image (F) is the merged image of image (D) and (E).
Tissue-specific expression pattern of SmMYB39 in S. miltiorrhiza
QRT-PCR was performed to determine expression levels of SmMYB39 in different tissues (root, stem, leaf and flower) of two-year-old flowering S. miltiorrhiza. Results showed that SmMYB39 expressed in all the tissues tested, with the highest expression in stem and lowest expression in root. Total phenolics accumulation in corresponding tissues was also measured. In contrast to SmMYB39 expression, the accumulation in root was the most while in stem was the least (Fig. 4). As the result of Spearman correlation analysis, SmMYB39 expression was negative-correlated with accumulation of total phenolics (r = −0.916, P<0.01), implying that SmMYB39 may be a repressor of phenolic acids biosynthesis in S. miltiorrhiza.
Figure 4. Relative expression levels of SmMYB39 (A) and total phenolics content (B) in different tissues of S. miltiorrhiza.
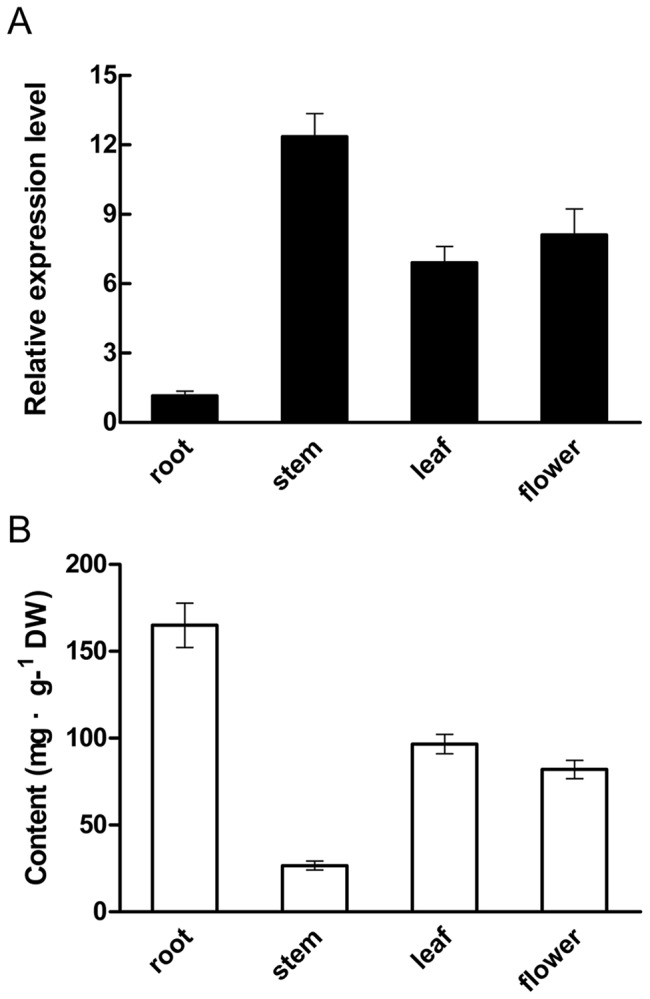
The results were analyzed using the comparative Ct method and presented as fold-changes compared with the root. The S. miltiorrhiza actin gene was used as an internal control to normalize expression levels. Data presented here are the mean of three replicates with error bars indicating ± SD.
SmMYB39 suppresses phenolic acids biosynthesis in S. miltiorrhiza
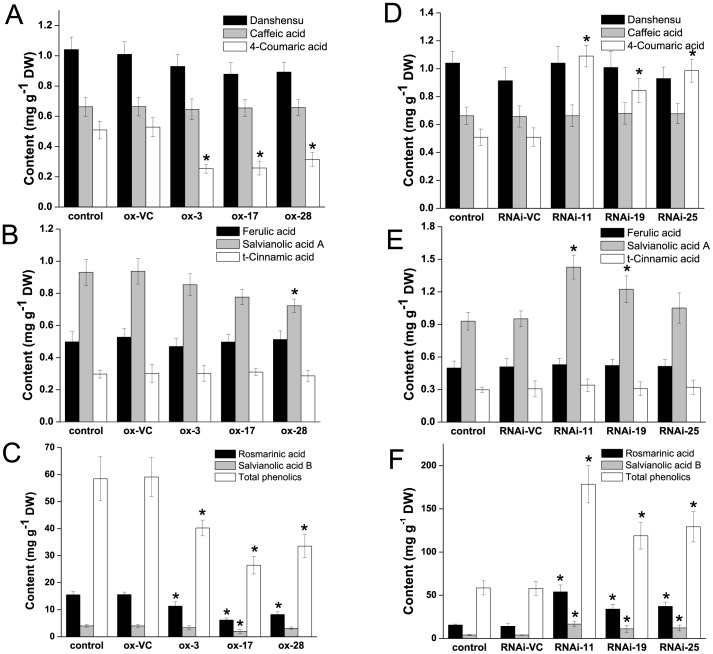
In this study, 31 transgenic S. miltiorrhiza plantlets overexpressing SmMYB39 and 25 independent SmMYB39-RNAi lines were obtained. Three overexpressing lines (ox-3, ox-17 and ox-28) and three RNAi lines (RNAi-11, RNAi-19 and RNAi-25) that presented obvious changes in phenolic acids content were selected for analysis. Expression levels of SmMYB39 in these six transgenic lines and controls were measured. The qRT-PCR results showed that the expression of the target gene was successfully regulated through genetic manipulation. Transcripts of SmMYB39 in the three SmMYB39-overexpressing lines were approximately 5.53-, 8.04- and 6.80-fold with respect to the empty-vector control (the line that only transformed with empty expression vector), respectively. While in the three SmMYB39-RNAi lines, expression levels of SmMYB39 only reached about 8.55%, 14.71% and 10.81% relative to the empty-vector control, respectively (Fig. 5). The contents of metabolites involved in the RA biosynthetic pathway were determined and the results revealed a broad reconfiguration of metabolic flux in transgenic plantlets. Compared with the untransformed plantlets and vector-control lines, accumulations of phenolic acids were dramatically (P<0.05) reduced in SmMYB39-overexpressing lines. For example, the content of 4-coumaric acid, rosmarinic acid, salvianolic acid B, salvianolic acid A and total phenolics were maximally decreased by 52.00% (in ox-3), 60.35% (in ox-17), 49.83% (in ox-17), 22.87% (in ox-28) and 55.21% (in ox-17), respectively (Fig. 6A, B, C). While in the transgenic SmMYB39-RNAi lines, production of the target metabolites was markedly (P<0.05) up-regulated. For instance, the content of 4-coumaric acid, rosmarinic acid, salvianolic acid B, salvianolic acid A and total phenolics in RNAi-11 line approximately reached 2.14-, 3.81-, 4.23-, 1.50-, and 3.08-fold of vector-control lines, respectively (Fig. 6D, E, F). Accumulations of other four tested phenolic acids, Danshensu, caffeic acid, ferulic acid and t-cinnamic acid were not affected by the genetic manipulation. Changes of metabolites content in transgenic lines implied that SmMYB39 was involved in the RA biosynthetic pathway and acted as a repressor of phenolic acids production.
Figure 5. Relative quantitative analysis of SmMYB39 expression in transgenic lines and controls of S. miltiorrhiza.
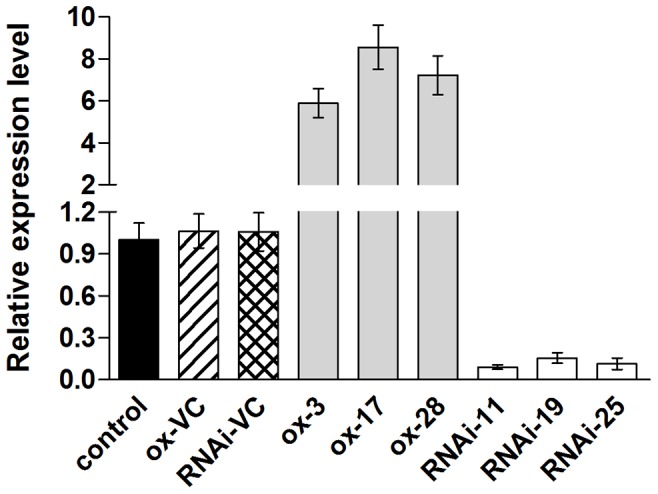
The results were analyzed using the comparative Ct method and presented as fold-changes compared with the control sample (untransformed control). The S. miltiorrhiza actin gene was used as an internal control to normalize expression levels. control: untransformed plant; ox-VC, RNAi-VC: empty vector controls of SmMYB39-overexpressing lines and SmMYB39-RNAi lines; ox-3, ox-17, ox-28: SmMYB39-overexpressing lines; RNAi-11, RNAi-19, RNAi-25: SmMYB39-RNAi lines.
Figure 6. Analysis of related phenolic compounds contents in transgenic lines and controls of S. miltiorrhiza.
Data presented here are the mean of three replicates with error bars indicating ± SD. The asterisks indicate statistically significant differences (P<0.05) compared to the empty vector control. control: untransformed plant; ox-VC, RNAi-VC: empty vector controls of SmMYB39-overexpressing lines and SmMYB39-RNAi lines; ox-3, ox-17, ox-28: SmMYB39-overexpressing lines; RNAi-11, RNAi-19, RNAi-25: SmMYB39-RNAi lines.
SmMYB39 down-regulates expression of C4H and TAT in RA pathway
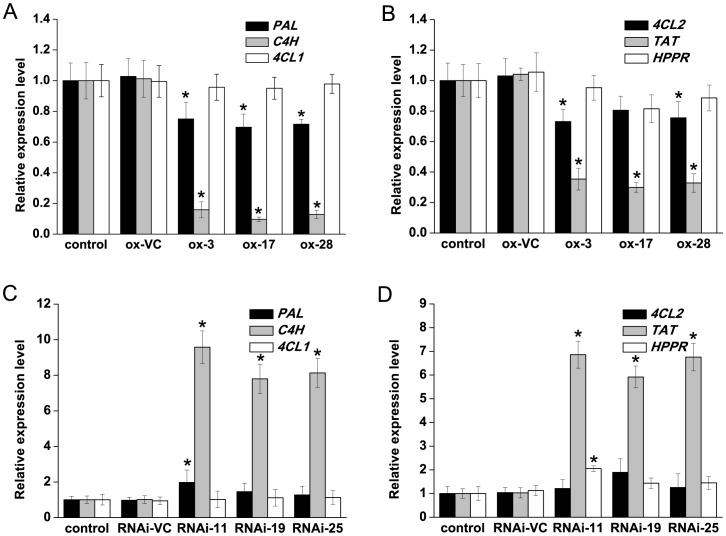
The qRT-PCR analysis of six key enzyme genes (PAL, C4H, 4CL1, 4CL2, TAT, and HPPR) in RA pathway was carried out in transgenic and untransformed plantlets. The results revealed that overexpression of SmMYB39 down-regulated all the six genes at different levels. Compared with the empty-vector control lines, expression levels of C4H were reduced by 84.31% (in ox-3), 90.45% (in ox-17) and 87.36% (in ox-28), respectively, and transcripts of TAT were decreased by 66.01% (in ox-3), 71.28% (in ox-17) and 68.50% (in ox-28), respectively. Yet no cases of ≥2-fold down- regulation were seen in PAL, 4CL1, 4CL2 and HPPR (Fig. 7A, B). In contrast to overexpression of SmMYB39, silencing of this gene up-regulated expression levels of the six key enzyme genes. Relative to the empty-vector control lines, transcripts of C4H reached 9.46- (in RNAi-11), 7.71- (in RNAi-19) and 8.04-fold (in RNAi-25), respectively, and transcripts of TAT reached 6.67- (in RNAi-11), 5.75- (in RNAi-19) and 6.57-fold (in RNAi-25), respectively. Expression levels of PAL, 4CL1, 4CL2 and HPPR were also enhanced, while none of them exhibited a ≥2- fold increase (Fig. 7C, D).
Figure 7. Relative quantitative transcripts analysis of enzyme genes in transgenic lines and controls of S. miltiorrhiza.
The results were analyzed using the comparative Ct method and presented as fold-changes compared with the control sample (untransformed control). The S. miltiorrhiza actin gene was used as an internal control to normalize expression levels. The asterisks indicate statistically significant differences (P<0.05) compared to the empty vector control. control: untransformed plant; ox-VC, RNAi-VC: empty vector controls of SmMYB39-overexpressing lines and SmMYB39-RNAi lines; ox-3, ox-17, ox-28: SmMYB39-overexpressing lines; RNAi-11, RNAi-19, RNAi-25: SmMYB39-RNAi lines; PAL: phenylalanine ammonia-lyase; C4H: cinnamic acid 4-hydroxylase; 4CL: 4-coumaric acid CoA-ligase; TAT: tyrosine aminotransferase; HPPR: 4-hydroxyphenylpyruvate reductase.
SmMYB39 inhibits C4H and TAT activities
Activities of C4H and TAT, whose expression were regulated by SmMYB39, were determined in this study. As shown in Fig. 8, activities of these two enzymes were dramatically (P<0.05) reduced in the three SmMYB39-overexpressing lines. Compared with the empty-vector lines, activity of C4H was decreased by 75.31% (in ox-3), 79.79% (in ox-17) and 77.58% (in ox-28), respectively, and activity of TAT was down-regulated by 61.90% (in ox-3), 66.56% (in ox-17) and 63.52% (in ox-28), respectively (Fig. 8A). RNAi-mediated silencing of SmMYB39 obviously (P<0.05) stimulated activities of C4H and TAT. The C4H activity in the SmMYB39-RNAi lines was approximately 4.23- (in RNAi-11), 3.82- (in RNAi-19) and 4.00- fold (in RNAi-25) of that in the empty-vector lines, respectively. TAT activity in these three transgenic lines reached about 3.06- (in RNAi-11), 2.77- (in RNAi-19) and 2.99-fold (in RNAi-25) relative to the empty-vector lines, respectively (Fig. 8B).
Figure 8. Enzyme activities analysis of C4H and TAT in transgenic lines and controls of S. miltiorrhiza.
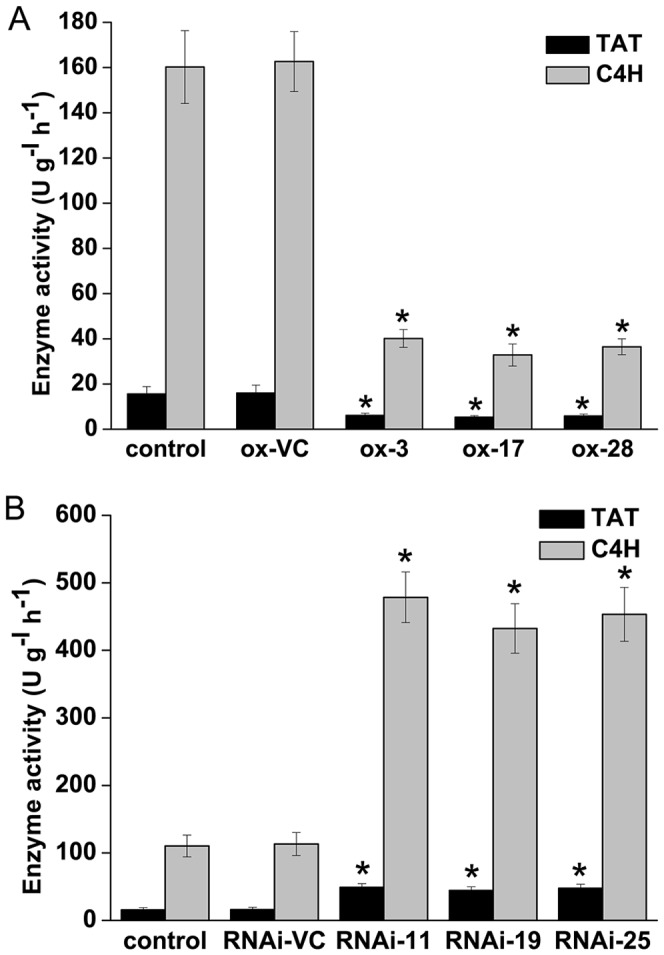
Data presented here are the mean of three replicates with error bars indicating ± SD. The asterisks indicate statistically significant differences (P<0.05) compared to the empty vector control. control: untransformed plant; ox-VC, RNAi-VC: empty vector controls of SmMYB39-overexpressing lines and SmMYB39-RNAi lines; ox-3, ox-17, ox-28: SmMYB39-overexpressing lines; RNAi-11, RNAi-19, RNAi-25: SmMYB39-RNAi lines; C4H: cinnamic acid 4-hydroxylase; TAT: tyrosine aminotransferase.
Discussion
Although many subgroup 4 R2R3 MYB transcription factors have been identified as transcriptional repressors, the mechanism of repression is still not totally understood. As one of first R2R3 MYB transcription factors suggested as transcriptional repressors of phenylpropanoid pathway, AtMYB4 was reported to act both by directly repressing and by competing with activators on binding motifs located on the promoters of target genes [25]. Most of C-terminus of subgroup 4 R2R3 MYB protein sequences contain three typical protein motifs ‘LlsrGIDPxT/SHRxI/L’, ‘pdLNLD/ELxiG/S’ and‘CX1–2CX7–12CX2C’, which were termed as C1, C2 and Zf motif, respectively [22], [26], [45]. Amongst them the C2 motif was proposed to play key roles in repression activity [23], [25], [44]. All the three motifs were present in the C-terminus of SmMYB39 and its highly homologous protein sequences that have been reported to act as repressors of phenylpropanoid pathway (Fig. 2) [27], [44]–[46]. MYB proteins from different plant species with a similar sequence structure were suggested to regulate similar pathways and possess same type of regulation (activation or repression) [28]. The overall structural homology between SmMYB39 and MYB proteins mentioned above suggests that SmMYB39 function as a true repressor of RA pathway in S. miltiorrhiza.
R2R3 MYB transcription factors have been demonstrated to regulate different branches of phenylpropanoid metabolism in plants, such as biosynthesis of lignin [26], anthocyanin and flavonols [28]. Yet there are only a few reports about whether anther branch of phenylpropanoid metabolism, the RA biosynthetic pathway, can be regulated by R2R3 MYB transcription factors. For example, Zhang et al. (2010) revealed that ectopic expression of AtPAP1 from Arabidopsis strikingly induced the accumulation of SAB in transgenic S. miltiorrhiza [47]. In this study, Overexpresison/RNAi-mediated silencing of SmMYB39 in S. miltiorrhiza plantlets dramatically (P<0.05) altered accumulations of 4-coumaric acid, rosmarinic acid, salvianolic acid B and salvianolic acid A, indicating that the RA pathway could be regulated by the R2R3 MYB transcription factor. Besides, content of t-cinnamic acid remained consistent in all the tested lines, implying that this precursor was not the bottleneck of RA biosynthesis.
A single R2R3 MYB transcription factor usually has several target genes and regulates multiple steps of the pathway that it is involved in [48]. This enables the regulator to control the metabolic flux more efficiently. In this study, we identified two key enzyme genes, C4H and TAT, whose transcripts and enzyme activities were all regulated by SmMYB39 as the target genes of this transcription factor. C4H (EC 1.14.13.11) is the second key enzyme in the phenylpropanoid pathway and catalyzes the hydroxylation of t-cinnamic acid to 4-coumaric acid [49]. C4H gene has been identified as the target gene of subgroup 4 R2R3 MYB transcription factors in many plant species, such as AmMYB308 and AmMYB330 from Antirrhinum majus [24], AtMYB4 from Arabidopsis [25], PhMYB4 from Petunia × hybrida [46] and VvMYB3 from grapevine [50]. By down-regulating of C4H, these transcription factors reduced biosynthesis of various phenylpropanoid metabolites. TAT (EC 2.6.1.5) is the first key enzyme of the tyrosine-derived pathway and transaminates tyrosine to 4-hydroxyphenylpyruvic acid [13]. TAT is involved in biosynthesis of several metabolites including rosmarinic acid, tocopherols and plastoquinones [13], [51], [52], while little is known about transcription factors that regulate this enzyme gene. C4H and TAT belong to the two parallel tributaries of RA pathway, phenylpropanoid pathway and tyrosine-derived pathway, respectively. With the co-ordinate control of the two key enzyme genes, SmMYB39 effectively regulated the accumulations of phenolic acids in S. miltiorrhiza.
Transcription factors usually active or repress gene transcripts by binding to cis-acting elements contained within promoters of target genes [53]. R2R3 MYB proteins from subgroup 4 have been reported to repress phenylpropanes metabolism by binding to AC elements [27], [44], [45], [54]. In this work, promoters of C4H gene (Genbank accession number: GQ896332.1) and TAT gene (Genbank accession number: EF192320.1) were analyzed with PlantCARE program (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). Results showed that both of them contained one AC-I elements with sequence ACCTACC (approximately 228 bp upstream from the transcription initiation site of C4H promoter and 776 bp upstream from the transcription initiation site of TAT promoter). SmMYB39 was likely to bind these AC elements and regulated the two parallel pathways of RA pathway simultaneously.
In summary, this work isolated and characterized a subgroup 4 R2R3 MYB transcription factor that regulated biosynthesis of phenolic acids in S. miltiorrhiza. It was located in nucleus and acted as a repressor by suppressing transcripts of C4H and TAT. Our results will be useful to better understand the regulating mechanism of phenolic acids production in S. miltiorrhiza and provide a train of thought to improve contents of bioactive compounds in this traditional herbal.
Supporting Information
The expression plasmids used in transformation and molecular analyses of transgenic S. miltiorrhiza plantlets. (A) Schematic representation of transgene expression plasmids. PNOS, nopaline synthase promoter; NPTII, neomycin phosphotransferase II gene; TNOS, nopaline synthase terminator; P35S, CaMV 35S promoter; F-259bp, R-259bp, forward 259 bp and reverse 259 bp fragments of SmMYB39; TOCS, octopine synthase terminator; LB, T-DNA left border; RB, T-DNA right border; NPTII-1F, NPTII-1R, Sm35S1F, SmMYB39-11R, NPTII-2F, NPTII-2R, Sm35S2F, SmMYB39-22R, primers used for identification of transgenic lines; Restriction sites are marked. (B) PCR analyses for SmMYB39-overexpression and SmMYB39-RNAi S. miltiorrhiza plantlets. M, DNA marker (100–1200 bp); P, the corresponding engineered plasmids (positive control); N, the wild-type S. miltiorrhiza plantlet (negative control); C, no template control.
(TIF)
Schematic representation of gene structure, predicted secondary structure and 3-D structure of SmMYB39. (A) Structure of the SmMYB39 genomic sequence. The exons are shown as blocks and the intron as line. The R2 and R3 repeats that constitute the MYB domain are shown as green and yellow shaded boxes, respectively. Numbers refer to position relative to the first nucleotide of the start codon. (B) Predicted secondary structure of SmMYB39 by SOPMA. Alpha helices, extended strands, beta turns and random coils are indicated by the longest, the second longest, the second shortest and the shortest vertical lines, respectively. (C) Predicted 3-D structure model of SmMYB39 MYB domain by SWISS-MODEL program. The structural model was based on the known crystal structure of a c-MYB from Mus musculus (PDB ID: 1H88, Chain ID: C) and shown using protein solid ribbon.
(TIF)
The prediction of secondary structure and 3-D structural model of SmMYB39.
(DOCX)
Acknowledgments
We thank Dr. Fei Yu and Dr. Xiayan Liu of Northwest A & F University who provided vectors used in this study.
Funding Statement
The work has been supported by the National Natural Science Foundation of China (Grant No. 81373908; http://www.nsfc.gov.cn/Portal0/default152.htm) and the Ph.D. Programs Foundation of Ministry of Education of China (Grant No. 20110204110028; http://www.cutech.edu.cn/cn/kyjj/gdxxbsdkyjj/A010301index_1.htm). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Xu H, Zhang L, Zhou CC, Xiao JB, Liao P, et al. (2010) Metabolic regulation and genetic engineering of pharmaceutical component tanshinone biosynthesis in Salvia miltiorrhiza . J Med Plants Res 4: 2591–2597. [Google Scholar]
- 2. Wu JY, Shi M (2008) Ultrahigh diterpenoid tanshinone production through repeated osmotic stress and elicitor stimulation in fed-batch culture of Salvia miltiorrhiza hairy roots. Appl Microbiol Biotechnol 78: 441–448. [DOI] [PubMed] [Google Scholar]
- 3. Zhou LM, Zuo Z, Chow MSS (2005) Danshen: An overview of its chemistry, pharmacology, pharmacokinetics, and clinical use. J Clin Pharmacol 45: 1345–1359. [DOI] [PubMed] [Google Scholar]
- 4. Nizamutdinova IT, Lee GW, Son KH, Jeon SJ, Kang SS, et al. (2008) Tanshinone I effectively induces apoptosis in estrogen receptor-positive (MCF-7) and estrogen receptor-negative (MDA-MB-231) breast cancer cells. Int J Oncol 33: 485–491. [PubMed] [Google Scholar]
- 5. Song YH, Liu Q, Lv ZP, Chen YY, Zhou YC, et al. (2008) Protection of a polysaccharide from Salvia miltiorrhiza, a Chinese medicinal herb, against immunological liver injury in mice. Int J Biol Macromol 43: 170–175. [DOI] [PubMed] [Google Scholar]
- 6. Wan JMF, Sit WH, Lee CL, Fu KHM, Chan DKO (2006) Protection of lethal toxicity of endotoxin by Salvia miltiorrhiza BUNGE is via reduction in tumor necrosis factor alpha release and liver injury. Int Immunopharmacol 6: 750–758. [DOI] [PubMed] [Google Scholar]
- 7. Chen H, Chen F, Chiu FCK, Lo CMY (2001) The effect of yeast elicitor on the growth and secondary metabolism of hairy root cultures of Salvia miltiorrhiza . Enzyme Microb Technol 28: 100–105. [DOI] [PubMed] [Google Scholar]
- 8. Li LN (1998) Biologically active components from traditionaI Chinese medicines. Pure Appl Chem 70: 547–554. [Google Scholar]
- 9. Liu AH, Li L, Xu M, Lin YH, Guo HZ, et al. (2006) Simultaneous quantification of six major phenolic acids in the roots of Salvia miltiorrhiza and four related traditional Chinese medicinal preparations by HPLC-DAD method. J Pharm Biomed Anal 41: 48–56. [DOI] [PubMed] [Google Scholar]
- 10. Petersen M, Simmonds MSJ (2003) Molecules of interest - Rosmarinic acid. Phytochemistry 62: 121–125. [DOI] [PubMed] [Google Scholar]
- 11. Don MJ, Shen CC, Syu WJ, Ding YH, Sun CM (2006) Cytotoxic and aromatic constituents from Salvia miltiorrhiza . Phytochemistry 67: 497–503. [DOI] [PubMed] [Google Scholar]
- 12. Shi Z, He J, Yao T, Chang W, Zhao M (2005) Simultaneous determination of cryptotanshinone, tanshinone I and tanshinone IIA in traditional Chinese medicinal preparations containing Radix salvia miltiorrhiza by HPLC. J Pharm Biomed Anal 37: 481–486. [DOI] [PubMed] [Google Scholar]
- 13. Petersen M, Häusler E, Karwatzki B, Meinhard J (1993) Proposed biosynthetic pathway for rosmarinic acid in cell cultures of Coleus blumei Benth. Planta 189: 10–14. [Google Scholar]
- 14. Ellis B, Towers G (1970) Biogenesis of rosmarinic acid in Mentha . Biochem J 118: 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.China Pharmacopoeia Committee (2010) Chinese Pharmacopoeia of People's Republic of China. Beijing: Chemical Industry. 71.
- 16. Xiao Y, Gao SH, Di P, Chen JF, Chen WS, et al. (2010) Lithospermic acid B is more responsive to silver ions (Ag+) than rosmarinic acid in Salvia miltiorrhiza hairy root cultures. Biosci Rep 30: 33–40. [DOI] [PubMed] [Google Scholar]
- 17. Petersen M, Abdullah Y, Benner J, Eberle D, Gehlen K, et al. (2009) Evolution of rosmarinic acid biosynthesis. Phytochemistry 70: 1663–1679. [DOI] [PubMed] [Google Scholar]
- 18. Whetten R, Sederoff R (1995) Lignin biosynthesis. Plant Cell 7: 1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feller A, Machemer K, Braun EL, Grotewold E (2011) Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J 66: 94–116. [DOI] [PubMed] [Google Scholar]
- 20. Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, et al. (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis . BMC Genomics 13: 544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jin HL, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41: 577–585. [DOI] [PubMed] [Google Scholar]
- 22. Stracke R, Werber M, Weisshaar B (2001) The R2R3-MYB gene family in Arabidopsis thaliana . Curr Opin Plant Biol 4: 447–456. [DOI] [PubMed] [Google Scholar]
- 23. Kranz HD, Denekamp M, Greco R, Jin H, Leyva A, et al. (1998) Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana . Plant J 16: 263–276. [DOI] [PubMed] [Google Scholar]
- 24. Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, et al. (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10: 135–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin HL, Cominelli E, Bailey P, Parr A, Mehrtens F, et al. (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis . EMBO J 19: 6150–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fornale S, Sonbol FM, Maes T, Capellades M, Puigdomenech P, et al. (2006) Down-regulation of the maize and Arabidopsis thaliana caffeic acid O-methyl-transferase genes by two new maize R2R3-MYB transcription factors. Plant Mol Biol 62: 809–823. [DOI] [PubMed] [Google Scholar]
- 27. Fornale S, Shi XH, Chai CL, Encina A, Irar S, et al. (2010) ZmMYB31 directly represses maize lignin genes and redirects the phenylpropanoid metabolic flux. Plant J 64: 633–644. [DOI] [PubMed] [Google Scholar]
- 28. Aharoni A, De Vos CHR, Wein M, Sun ZK, Greco R, et al. (2001) The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J 28: 319–332. [DOI] [PubMed] [Google Scholar]
- 29. Gao JJ, Shen XF, Zhang Z, Peng RH, Xiong AS, et al. (2011) The myb transcription factor MdMYB6 suppresses anthocyanin biosynthesis in transgenic Arabidopsis . Plant Cell Tissue Organ Cult 106: 235–242. [Google Scholar]
- 30. Yan YP, Wang ZZ (2007) Genetic transformation of the medicinal plant Salvia miltiorrhiza by Agrobacterium tumefaciens-mediated method. Plant Cell Tissue Organ Cult 88: 175–184. [Google Scholar]
- 31. Murashige T, Skoog F (1962) A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol Plant 15: 473–497. [Google Scholar]
- 32. Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 33. Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18: 2714–2723. [DOI] [PubMed] [Google Scholar]
- 35. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465–W469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liu XY, Rodermel SR, Yu F (2010) A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876. [DOI] [PubMed] [Google Scholar]
- 38. Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, et al. (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590. [DOI] [PubMed] [Google Scholar]
- 39. Gleave AP (1992) A versatile binary vector system with a T-DNA organizational-structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol Biol 20: 1203–1207. [DOI] [PubMed] [Google Scholar]
- 40. Song J, Wang ZZ (2011) RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J Plant Res 124: 183–192. [DOI] [PubMed] [Google Scholar]
- 41. Liang ZS, Yang DF, Liang X, Zhang YJ, Liu Y, et al. (2012) Roles of reactive oxygen species in methyl jasmonate and nitric oxide-induced tanshinone production in Salvia miltiorrhiza hairy roots. Plant Cell Rep 31: 873–883. [DOI] [PubMed] [Google Scholar]
- 42. Yan Q, Shi M, Ng J, Wu JY (2006) Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci 170: 853–858. [Google Scholar]
- 43. Lamb Cj, Rubery PH (1975) A spectrophotometric assay for Trans-cinnamic acid 4-Hydroxylase activity. Anal Biochem 68: 554–561. [DOI] [PubMed] [Google Scholar]
- 44. Shen H, He XZ, Poovaiah CR, Wuddineh WA, Ma JY, et al. (2012) Functional characterization of the switchgrass (Panicum virgatum) R2R3-MYB transcription factor PvMYB4 for improvement of lignocellulosic feedstocks. New Phytol 193: 121–136. [DOI] [PubMed] [Google Scholar]
- 45. Legay S, Lacombe E, Goicoechea M, Briere C, Seguin A, et al. (2007) Molecular characterization of EgMYB1, a putative transcriptional repressor of the lignin biosynthetic pathway. Plant Sci 173: 542–549. [Google Scholar]
- 46. Colquhoun TA, Kim JY, Wedde AE, Levin LA, Schmitt KC, et al. (2011) PhMYB4 fine-tunes the floral volatile signature of Petunia × hybrida through PhC4H . J Exp Bot 62: 1133–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang YA, Yan YP, Wang ZZ (2010) The Arabidopsis PAP1 transcription factor plays an important role in the enrichment of phenolic acids in Salvia miltiorrhiza . J Agric Food Chem 58: 12168–12175. [DOI] [PubMed] [Google Scholar]
- 48. Laitinen RAE, Ainasoja M, Broholm SK, Teeri TH, Elomaa P (2008) Identification of target genes for a MYB-type anthocyanin regulator in Gerbera hybrida . J Exp Bot 59: 3691–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Russell DW (1971) The metabolism of aromatic compounds in higher plants. J Biol Chem 246: 3870–3878. [PubMed] [Google Scholar]
- 50.Carra A, Schubert A, Martin C (2005) Isolation and preliminary characterization of VvMYB3, a novel MYB transcription factor from grape. In: Williams LE, Proceedings of the Seventh International Symposium on Grapevine Physiology and Biotechnology. 529–531.
- 51. Sandorf I, Hollander-Czytko H (2002) Jasmonate is involved in the induction of tyrosine aminotransferase and tocopherol biosynthesis in Arabidopsis thaliana . Planta 216: 173–179. [DOI] [PubMed] [Google Scholar]
- 52.Douce R, Joyard J (1996) Biosynthesis of thylakoid membrane lipids. In: Ort DR, Yocum CF, Advances in photosynthesis/oxygenic photosynthesis: the light reactions Dordrecht: Kluwer. 69–101.
- 53. Liu LS, White MJ, MacRae TH (1999) Transcription factors and their genes in higher plants - Functional domains, evolution and regulation. Eur J Biochem 262: 247–257. [DOI] [PubMed] [Google Scholar]
- 54. Zhao JF, Zhang WH, Zhao Y, Gong XM, Guo L, et al. (2007) SAD2, an importin beta-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. Plant Cell 19: 3805–3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The expression plasmids used in transformation and molecular analyses of transgenic S. miltiorrhiza plantlets. (A) Schematic representation of transgene expression plasmids. PNOS, nopaline synthase promoter; NPTII, neomycin phosphotransferase II gene; TNOS, nopaline synthase terminator; P35S, CaMV 35S promoter; F-259bp, R-259bp, forward 259 bp and reverse 259 bp fragments of SmMYB39; TOCS, octopine synthase terminator; LB, T-DNA left border; RB, T-DNA right border; NPTII-1F, NPTII-1R, Sm35S1F, SmMYB39-11R, NPTII-2F, NPTII-2R, Sm35S2F, SmMYB39-22R, primers used for identification of transgenic lines; Restriction sites are marked. (B) PCR analyses for SmMYB39-overexpression and SmMYB39-RNAi S. miltiorrhiza plantlets. M, DNA marker (100–1200 bp); P, the corresponding engineered plasmids (positive control); N, the wild-type S. miltiorrhiza plantlet (negative control); C, no template control.
(TIF)
Schematic representation of gene structure, predicted secondary structure and 3-D structure of SmMYB39. (A) Structure of the SmMYB39 genomic sequence. The exons are shown as blocks and the intron as line. The R2 and R3 repeats that constitute the MYB domain are shown as green and yellow shaded boxes, respectively. Numbers refer to position relative to the first nucleotide of the start codon. (B) Predicted secondary structure of SmMYB39 by SOPMA. Alpha helices, extended strands, beta turns and random coils are indicated by the longest, the second longest, the second shortest and the shortest vertical lines, respectively. (C) Predicted 3-D structure model of SmMYB39 MYB domain by SWISS-MODEL program. The structural model was based on the known crystal structure of a c-MYB from Mus musculus (PDB ID: 1H88, Chain ID: C) and shown using protein solid ribbon.
(TIF)
The prediction of secondary structure and 3-D structural model of SmMYB39.
(DOCX)