Abstract
Next to embryonic stem cell research, adult stem cell research is providing a promising alternative for enhanced tissue regeneration and transplantation. The key biochemical networks controlling the differentiation processes regulating stem cell biology remain largely disputed and or undefined, contributing to a lack of knowledge of the principle phosphoregulatory events propagating signal transduction. To effectively monitor these events relative to adipocyte differentiation, this study utilized a high throughput reverse phase protein microarray platform and characterized adult adipose-derived stem cell (ASC) differentiation through the monitoring of ∼100 phosphospecific endpoints with 33 distinct time points examined across 14 days. This kinetic-based analysis showed time ordered signal transduction ultimately implicating pathways correlated with adipogenic differentiation. To further validate the causal significance of these network activations, pharmacological targeting was implemented to include the chemical inhibitors MAPK inhibitor PD169316, rapamycin, and HNMPA-(AM)3 yielding partial or complete disruption of adipocytic differentiation, as noted by a decrease or lack of lipid formation within the mature adipocytes. Based on this analysis, v-crk sarcoma virus CT10 oncogene homolog (CRKII) and c-abl oncogene 1, non-receptor tyrosine kinase (c-ABL) were implicated as novel key regulators of adipocyte differentiation, with v-akt murine thymoma viral oncogene (AKT), mammalian target of rapamycin (mTOR), and SMAD family member (SMAD) pathways being implicated as secondary regulators. This dynamic molecular profiling provides a novel insight into the signaling architecture of mesenchymal stem cell differentiation and may be useful in the development of therapeutic modulators for clinical applications; in addition to advancing the collective understanding of key cellular processes, ultimately contributing to more confident stem cell manipulation.
Recent breakthroughs in adult stem cell research have increased industry focus on the potency and availability of these cells. Adult stem cells exist in many tissue types, but adult adipose-derived stem cells (ASCs)1 provide a particularly abundant source obtained via through minimally invasive techniques (liposuction) (1, 2, 3). This plastic-adherent, multipotent cell population is also known as adipose-derived stromal cells among other names, with the International Fat Applied Technology Society reaching a consensus to refer to them as ASCs (4). Despite extensive research on the differentiation of stem cells, the underpinning molecular mechanisms remain an enigma. During adipogenesis, terminal differentiation from a mesenchymal stem cell to a mature adipocyte is characterized by the ability to store triglycerides that can be mobilized as fuel for other organs, but the cellular signaling actuating this transformation remains predominately undefined (5, 6). A few well-established factors inducing differentiation are: high concentrations of insulin (resulting in stimulation of the insulin-like growth factor 1 receptor), glucocorticoid agonists, peroxisome proliferator-activated receptor γ (PPARγ) agonist, and agents that elevate cAMP (7, 8).
Primarily research into the process of adipogenesis has focused on gene expression profiling with proteomic technologies being used only more recently. Despite the benefits of gene expression profiling (9, 10), it shows little correlation to protein levels and provides no insight into the timing of protein activation. This simple fact highlights the need for a systems biology approach. More specifically, the monitoring of key post-translational modification events such as phosphorylation is necessary to characterize the signaling architecture regulating differentiation (11, 12). These reversible kinase-driven phosphorylation events alter protein confirmation, ultimately affecting enzymatic activity and protein–protein interactions leading to an array of cellular events from differentiation to gene expression, thus encompassing signal transduction. Characterization of this broad-scale signaling architecture is necessary to provide a more finite depiction of the complex signaling events directing a given cellular phenotype and aid in the understanding of the repercussions of alterations in regulation (13, 14). Presently, phosphoproteomic analysis of cellular differentiation processes such as with mass spectrometry has been performed in a very limited number of independent time points that span the cellular differentiation process, and important dynamic changes in the cellular signaling architecture may have been missed (12, 15, 16). This study however, utilized the reverse phase protein microarray (RPMA) to maximize both the number of time points and phosphoproteins able to be examined simultaneously. RPMAs enable the quantitative interrogation of the phosphorylation state of hundreds of signaling proteins simultaneously for hundreds of cell lysates allowing for broad-scale pathway activation mapping analysis (17). This ultrasensitive platform has demonstrated sensitivity of as little as 1000 molecules per spot with less than 1/10th of a cell equivalent volume analyzed per spot and intraslide and interslide CV between 3–10% (18, 19). RPMAs greatly reduce the required sample size with spot deposits averaging between 0.3–2 nL, and yet show concurrent findings when parallel processing of samples via Western blot is performed (20, 21, 22, 23, 24).
Despite the implication of various markers of adipogenesis, there remains little consensus on the overarching signal transduction governing the differentiation process. The objective of this research was to better characterize this process through utilization of ASCs combined with broad-scale protein pathway activation mapping, using a dynamic experimental design. Lineage signal transduction profiles were established through the monitoring of protein network activation during the course of differentiation into adipocyte, osteoblast, and chondrocyte lineages. This multilineage kinetic experimentation allowed for global examination of signal transduction through the monitoring of ∼100 phosphospecific endpoints, across 33 consecutive time points that spanned a 14 day period to reach terminal differentiation demonstrating time-specific and lineage-specific signaling. This experimental design allowed for isolation of a subset of time-specific endpoints unique to adipogenesis relative to the other lineages that could then be further tested for causal significance using pharmacologic knock-out analysis.
EXPERIMENTAL PROCEDURES
Tissue Culture
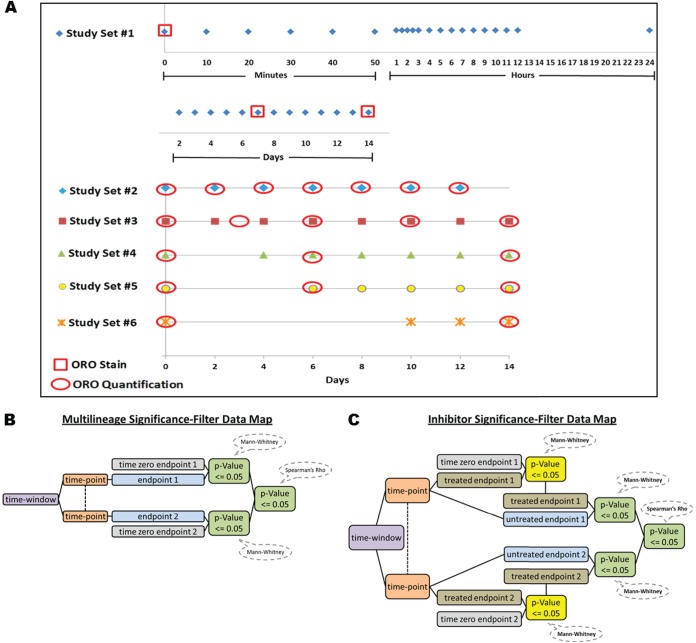
ASCs were purchased from Zen-Bio and then cultured according to the manufacture's protocols. Cells were comprised of a mixed lot of five donors and were shown to be positive for CD29, CD44, and CD 105, while negative for CD14, CD31, CD34, CD45, and CD133 (performed by Zen-bio). Cells were passaged twice in a 75 cm2 flask, then transferred to a 24-well plate, and seeded at a concentration of 39,140 cells/well. 24 h post-transfer, cells had media changed to lineage-specific media per the manufacture specifications, with media being replaced every other day. Samples for the multilineage differentiation time-course (Study Set #1) were performed in triplicate and began with time zero (no differentiation media added), followed by 33 time points for adipocytic and osteoblastic lineages (terminal differentiation reached at day 14), and 39 for the chondrocytic lineage (terminal differentiation reached at 20 days) (Fig. 1). To lyse samples, the media was removed, cells were rinsed three times with 1 × phosphate-buffered saline (PBS), lysed in 70 μl lysis buffer, and stored at −80 °C. The lysis buffer was comprised of 50% T-PER® tissue protein extraction reagent (Thermo Scientific), 47.5% Novex® Tris-glycine SDS sample buffer (Invitrogen, Carlsbad, CA), and 2.5% 2-mercaptoethanol.
Fig. 1.
Timeline for all experimental sets with shapes representing sample lysates and red shape overlays showing when oil red O (ORO) staining or quantification was performed A, Analytical methodology for Study Set #1 significance-filtering (B) and Study Sets #3 - 6 (C) with the Mann-Whitney time point-end point analysis having to show significance for end point consideration.
Inhibitor Treatment
Based on results from inhibitor pilot experiments, three inhibitors were selected that completely or partially disrupted adipocyte differentiation. A time-course using 15 μm of HNMPA-(AM)3 (Calbiochem), 500 nm of mitogen-activated protein kinase (MAPK) inhibitor PD169316 (Calbiochem), and 250 nm of rapamycin (BIOSOURCE) was performed with inhibitors having a final concentration of 0.1% dimethyl sulfoxide (DMSO). Study Set #3 samples were cultured in triplicate, as previously describe, with treatments beginning at the start of differentiation and continuing through day 14 with associated untreated and DMSO controls. Fresh inhibitor was added with each media change and sample lysates were taken every other day through day 14. To further characterize the role of the timing of signaling events, additional inhibitor experiments were performed with inhibitor administered at delayed time-points. Three delayed treatment sets were performed in triplicate; Study Set #4: inhibitor/DMSO treatment was started on day 2 and continued until day 14, Study Set #5: inhibitor/DMSO treatment was started on day 4 and continued until day 14, and Study Set #6: inhibitor/DMSO treatment was started on day 8 and continued until day 14 (Fig. 1A). In all cases, differentiation was monitored through the quantification of Oil Red O (ORO) staining to determine the impact on lipid formation.
Oil Red O Staining and Quantification
At various points in the differentiation process, lipid formation was visualized and quantified utilizing ORO. Briefly, a stock solution of ORO (0.35g ORO and 100 mls isopropanol) was stirred overnight and filtered through a 0.2 μ filter. Before use, a working stock comprising 6 mls of the ORO stock and 4 mls of ddH2O was prepared and filtered (0.2 μ). Cells were rinsed two times with PBS prior to a 10 min rinse with 10% phosphate buffered formalin, followed by a 1 hour incubation period. Following fixation, cells were washed with ddH2O, incubated with 60% isopropanol for 5 min, and dried in an oven at 37 °C. This step was followed by incubation with 500 μl of the working ORO working stock for 10 min, rinsed four times with 2 mls of ddH2O, images taken, and allowed to air dry. The ORO was then eluted using 1 ml of 100% isopropanol and incubated on an orbital shaker for 10 min. An optical density (OD) was then obtained using a spectrophotometer at 500 nm and values normalized to a negative control sample. Lipid based differentiation was established using a time course with lipid quantification performed every other day until day 14 (Study Set #2, Fig. 1A).
RPMA
Samples were heated at 100 °C for 7 min to melt the protein into its primary structure. Sample lysates were loaded into 384-well plates and a two-point dilution curve (neat and 1:4) was generated for each sample. Lysates were printed in duplicate on a customized FAST® nitrocellulose coated slide (Whatman®) using the 2470 Arrayer (Aushon Biosystems) with controls lysates to include jurkat treated with etoposide, jurkat treated with calyculin, HeLa, and HeLa treated with pervanadate printed in a ten point dilution curve (neat, 1:2, 1:4, 1:8, 1:16, 1:32, 1:64, etc.). Slides were stored at −20 °C until further use. Prior to staining, slides were incubated in 1 × Re-Blot (Chemicon) for 14 min on an agitator to relax protein structure. Re-Blot was removed, and slides were washed two times for 5 min in PBS. Slides were then placed into a blocking solution [1g I-block (Tropix, Applied Biosystems), 0.5% Tween-20 in 500 ml PBS] for at least 2 h with agitation before staining.
Protein Microarray Staining
To ensure printing quality and to provide protein quantification, a total protein assay was performed with selected slides using SYPRO® Ruby as previously described and visualized with a NovaRay® (Alpha Innotech) imager (17). Each antibody staining procedure was completed using the Autostainer DAKO Cytomation, which uses a catalyzed signal amplification (CSA) system, according to the manufacturer's protocol. Every antibody used was commercially purchased and specificity was confirmed via Western blot. Antibody staining employed the use of multiple phosphospecific primary antibodys, as well an appropriate secondary antibody (mouse or rabbit), followed by visualization with the fluorophore IR-Dye® 680 streptavidin (Li-Cor® Biosystems). Additionally, every staining run had an associated negative control slide treated with antibody diluent in the place of the primary antibody. Phospho-specific antibodies selected pertained to various areas of cellular functioning, with some being implicated in differentiation and others having no known affiliations. Incubation sequences, reagent dilutions, and incubation times were performed as previously described (17).
Imaging and Analysis
Slides were scanned with the NovaRay for a maximum of 500 milliseconds or until no sample or control saturation was noted. Both antibody stained and SYPRO Ruby stained slides were analyzed using the Microvigene® software package, with local background subtraction and the averaging of duplicated spots resulting in a single data point per sample. Each spot was then normalized to the total protein slides as well as the negative control slide such that the values obtained for any phosphoprotein are independent of the total amount of protein on a per/cell basis. This data was then subjected to unsupervised hierarchical clustering analysis and Spearman's Rho correlation analysis using JMP 5.0 (SAS).
Statistics and Data Filtering
To establish statistical significance, the math library (Apache Foundation Software) was utilized to perform Mann-Whitney U tests. These tests consisted of experiment specific time point and end point (“time point-end point”) comparisons. Statistically significant time point-endpoints were further correlated between endpoints during specific time windows (“time-window-end point”) using a Spearman's Rho analysis, and further paired down to significant at the same time point. During analysis of Study Set #1, two significance-filtering-methods were used. Filter 1) all time point–end point samples for a given lineage were compared with the corresponding untreated time zero end point sample and were required to have a Mann-Whitney p value of ≤ 0.05 and 2) Spearman's Rho time-window-end point correlation samples were required to have a p value of ≤ 0.05 (Fig. 1B). Similarly three significance-filtering-methods were used for Study Sets #3 - 6 with Filter 1) being the same as above. Filter 2) each time point–end point untreated sample was compared with its corresponding time point-end point inhibitor sample and required to have a Mann-Whitney p value of ≤ 0.05, 3) Spearman's Rho time-window-end point correlations of inhibitor samples were required to have a p value of ≤ 0.05 (Fig. 1C).
Data was further visualized using Microsoft Access to create composite data tables and histograms. In Study Set #1, a Mann-Whitney U test table was viewed in histogram format showing the frequency of significant time-points for a given end point within each lineage for a specific time-window. Endpoints that were unique to a treatment or those having a frequency minimally 2 higher than other inhibitor treatments were highlighted for further examination. Similarly for Study Sets #3–6, a table was created linking the Mann-Whitney U table for the time zero sample compared with an inhibitor treated sample and the Mann-Whitney U table for the untreated to treated comparison, with each linked by time point, inhibitor treatment, and end point. Spearman's Rho correlations were also taken into consideration for each experimental set, with experimental sets ultimately being compared with each other to isolate endpoints unique to adipogenesis. Alterations in signaling for inhibitor treated experimental sets were further examined using CScape pathway maps to reflect signaling pathway changes relative to the related untreated samples (25). These maps examined alterations in the early and late signaling events by analysis of day 4 and day 8 data. The intensity values were normalized with unaltered endpoints displaying a value of 1, decreased phosphorylation displaying a value closer to 0 and increased phosphorylation showing a value closer to 2.
RESULTS
Multilineage Time Course (Study Set#1)
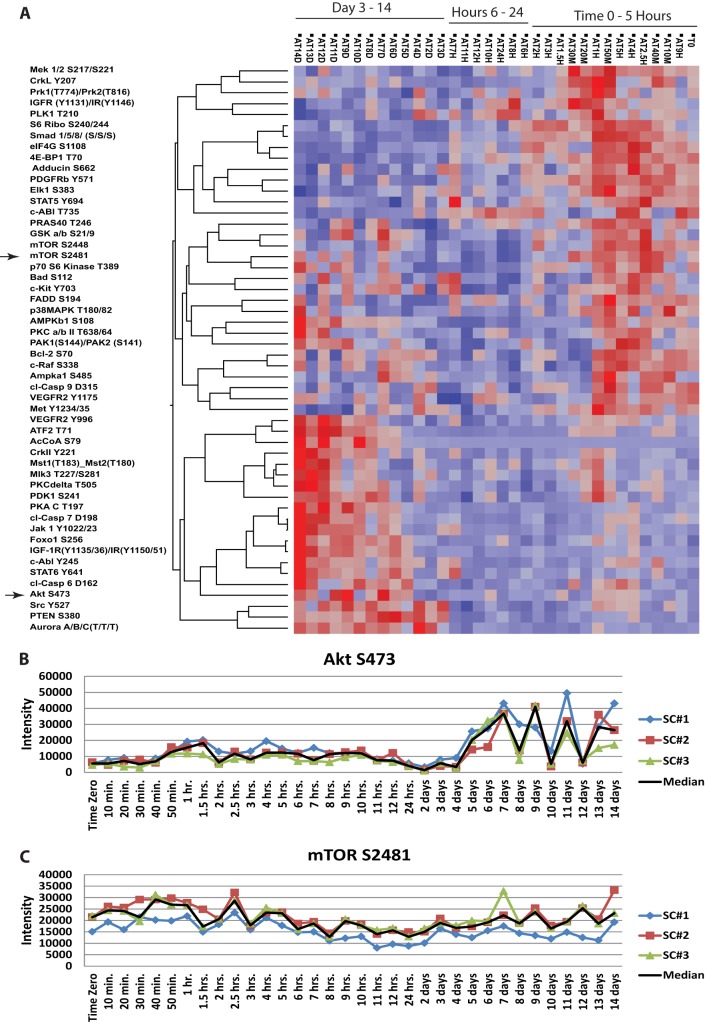
Dynamic time course RPMA data from each lineage was visualized via unsupervised hierarchical clustering, which facilitated end point clustering and established time windows used for histogram based analysis. The analysis displayed time ordered signal transduction (Fig. 2A) that revealed a near-perfect ordering of signaling dynamics that produced a distinct series of unique dynamic up-and-down signaling activation patterns following a defined time series. The reproducibility of the signaling dynamics was seen through the independent analysis of differentiated independent triplicate samples. This portrayed kinetic activation portraits that were cleanly reproducible across all three samples, including even subtle and time-dependent changes (Fig. 2B and 2C) and interesting oscillations in phosphorylation levels seen for AKT at the later time points (Fig. 2B). The cause and/or consequence of these reproducible phosphorylation level oscillations is unknown and could be because of cell cycling feedback and further investigation is warranted. The unsupervised clustering revealed activation of the insulin-like growth factor receptor (IGFR)/IR, SMAD, and mTOR pathways within the first few hours, with AKT pathway activation a few days post-differentiation. Time-windows were visualized with histograms to isolate lineage unique endpoints per time-window (Supplemental Data Table ST1). This analysis showed extracellular-signal-regulated kinases (ERK), phosphatase and tensin homolog (PTEN) and c-ABL signaling activation to be unique to adipogenesis in the early time points, with SMAD and CRK activation unique in later time points. This outcome suggested lineage-time point-specificity and enabled the implication of potential pathways to be targeted pharmacologically for further detailed molecular analysis on adipocyte differentiation and the associated signaling network activation correlates.
Fig. 2.
Hierarchical clustering of adipocytic samples for Study Set #1 time course in triplicate through terminal differentiation A, Plots of raw triplicate data pertaining to adipocytic differentiation for a 14 day period, to include AKT S473 (B) and mTOR S2481 (C).
ORO Quantification (Study Set #2)
Adipocyte differentiation was monitored through the quantification of ORO to validate increased lipid formation throughout differentiation and to generate a quantifiable numeric measurement of functional biology (lipid accumulation). To ensure methodological validity, ASCs were differentiated until day 12, with ORO quantification and cellular imaging performed (supplemental Fig. S1). This showed ORO staining levels to correspond to visual lipid accumulations enabling a quantifiable inhibitor time course to be pursued.
Inhibitor Studies (Study Sets #3–6)
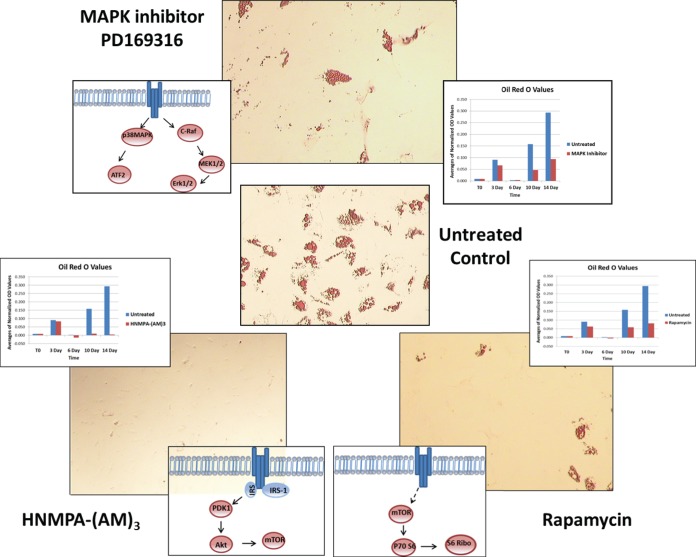
Inhibitor screening was performed with inhibitor added at the start of differentiation and maintained until day 14, when cultures were imaged (Fig. 3). The results showed that the HNMPA-(AM)3 inhibitor (15 μm concentration) completely inhibited lipid formation, while rapamycin (250 nm concentration) and the MAPK inhibitor PD169316 (500 nm concentration) partially inhibited lipid formation. To evaluate inhibitor affect for Study Set #3, specific endpoints that should have been targeted by the inhibitor treatment were examined. In the case of the HNMPA-(AM)3 (IR inhibitor), two insulin receptor autophosphorylation sites were examined to validate inhibitor targeting. The end point type I insulin-like growth factor receptor/IR [IGF-1R (Y1135/36)/IR (Y1150/51)] showed inhibition relative to the untreated sample, whereas the end point IGF-1R (Y113)/IR (Y1146) did not (supplemental Fig. S2). ORO values displayed a gradual increase in lipid formation in untreated cells, with HNMPA-(AM)3, treated cells displaying lipid detection levels close to that of the negative control (Fig. 3C). For rapamycin, the end point mTOR S2481, an autophosporylation site, showed a significant decrease in activation relative to the untreated; whereas the end point mTOR S2448, phosphorylated by AKT, was not as significantly altered relative to the untreated sample (SF2). A downstream target of mTOR, ribosomal protein S6 kinase (RPS6) S244/40, showed a significant decrease in activation relative to the untreated samples, and lipid accumulation was approximately half that of the untreated control samples (Fig. 3D). Last, the same examination was performed for the MAPK inhibitor PD169316. The endpoints p38 MAPK, ERK1/2, and MAP kinase kinase 1/2 (MEK1/2) were examined as potential direct targets in addition to downstream targets such as activating transcription factor 2 (ATF2) (supplemental figure S2).
Fig. 3.
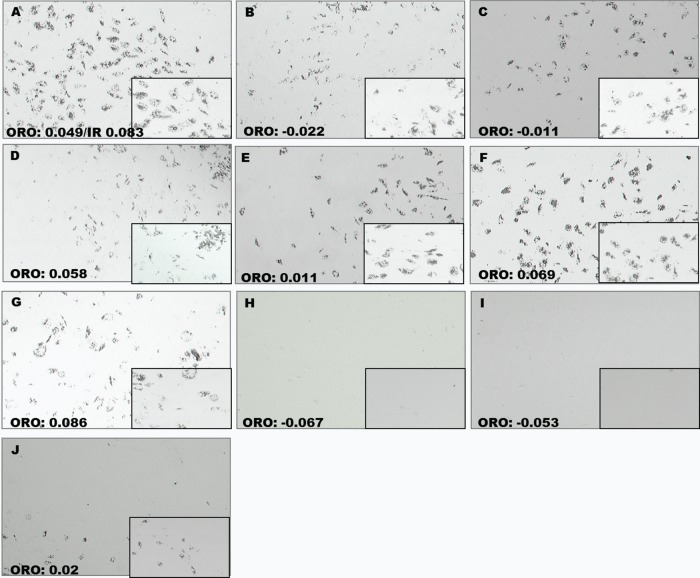
ORO images of inhibitor treated cells at day 14, to include time lapsed graphs of ORO quantifications related to untreated cultures. Samples include the untreated control (B), MAPK inhibitor PD169316 (A), HNMPA-(AM)3 (C), and rapamycin (D).
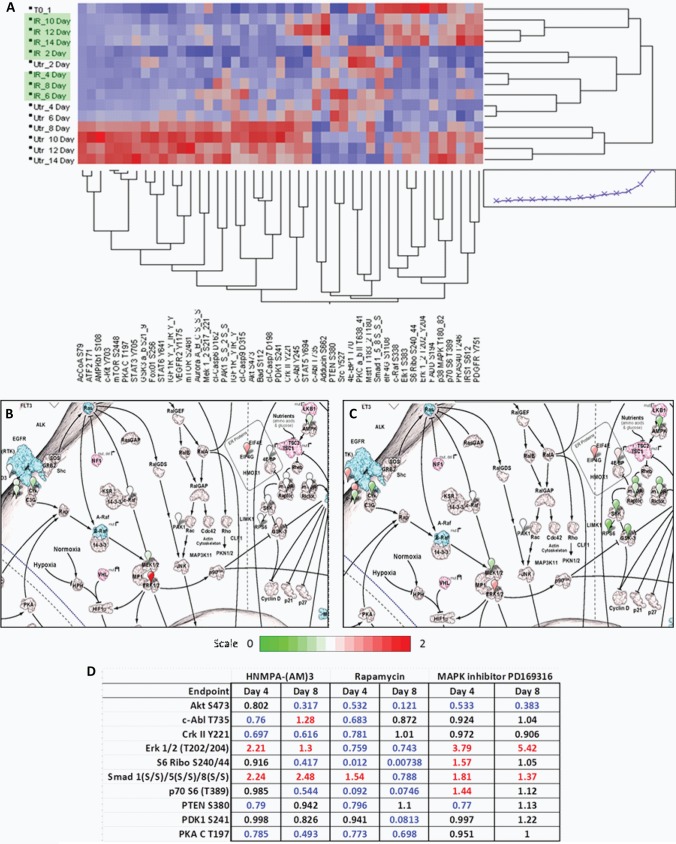
Following analysis, unsupervised hierarchical clustering analysis was performed to generate heat-maps. Heat-map data further corroborated time-ordered-signaling and captured the disruption in signal architecture as a result of inhibitor treatment. HNMPA-(AM)3 inhibitor showed the highest degree of individualized clustering from the untreated experimental set (Fig. 4A). To further delineate signaling pathway differences between inhibitor treatments, early (day 4) and late (day 8) pathway maps were constructed as described above using CScape (Fig. 4B and 4C), in addition to a summary table displaying values of treated samples normalized to their untreated matched sample (Fig. 4D). When examining the HNMPA-(AM)3 inhibitor samples on day 4, an increased activation was noted in SMAD and ERK1/2 pathways, with a reduction of activation seen in some proteins involved in the AKT signaling pathway. On day 8, the SMAD pathway was still highly phosphorylated, with ERK 1/2 activated to a lesser degree. When comparing selected endpoints potentially promoting differentiation, it was noted that on day 4 c-ABL T735 and CRKII Y221 activity was lower in HNMPA-(AM)3 and rapamycin treated samples, but unaffected in MAPK treated samples. On day 8 in HNMPA-(AM)3 treated samples, c-ABL T735 exhibited increased activation whereas CRKII Y221 retained a decreased activation; however, in rapamycin and MAPK treated samples, c-ABL T735 and CRKII Y221 activation levels returned to that of the untreated samples. These profiles were then compared across treatments in an attempt to isolate endpoints showing a distinct profile in the HNMPA-(AM)3 inhibitor relative to the other two treatments.
Fig. 4.
Samples treated with HNMPA-(AM)3 and untreated control samples showed a time-ordered mechanism of differentiation, with an alteration of signaling noted in the presence of inhibitor A, CScape image of HNMPA-(AM)3 inhibitor treatment on day 4 (B) and day 8 (C), with summary table (D).
To more fully understand the kinetic nature of signaling pathway activation and the cellular commitment processes during differentiation, delayed inhibitor treatments (Study Sets #4–6) were used. Again, the IR inhibitor HNMPA-(AM)3 displayed a strong disruption of differentiation, with a loss of lipid formation being noted when inhibitor was added on day 8 and maintained until day 14. Rapamycin treatment disrupted lipid formation in Study Sets #4 and 5, but showed a lesser effect in Study Set #6. The MAPK inhibitor PD169316 only showed a notable disruption of lipid formation in Study Set #4 (Fig. 5). These findings led to only inhibitor sets with inhibitor started on day 2 being used for further analysis.
Fig. 5.
ORO images of day 14 for Study Sets #4–6. Samples include untreated (A); rapamycin started on day 2 (B), day 4 (C), and day 8 (D); MAPK started on day 2 (E), day 4 (F), and day 8 (G); HNMPA-(AM)3 started on day 2 (H), day 4 (I), and day 8 (J).
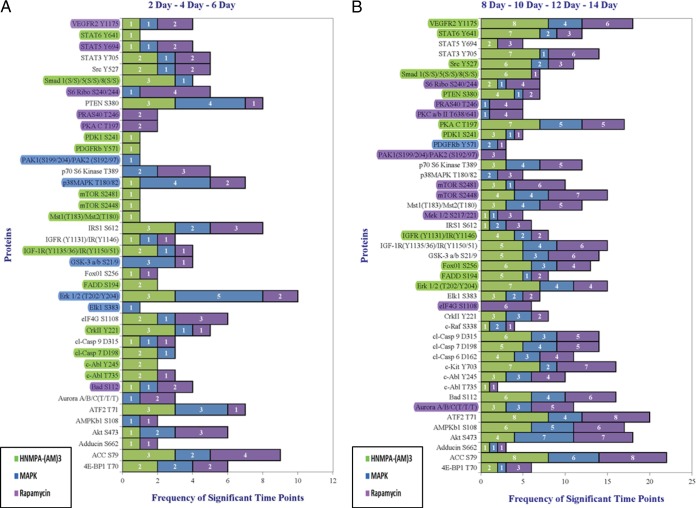
In an effort to identify endpoints promoting the differentiation process, data was condensed into an early (days 2–6) and late (days 8–14) time-window with a tabulation of significance per endpoint being represented in a histogram format (Figure 6). Again, based on ORO results, only Study Sets #3 and 4 were utilized in keeping with the focus on samples showing a disruption in differentiation.
Fig. 6.
Compilation of all inhibitor treatments, with treatment initiated at the start of differentiation, started on day 2, and started on day 4 complied into a single data set displayed in two time windows: days 2–6 (A) and days 8–14 (B). Endpoints showing at least two points higher or exclusive inhibitor representation highlighted.
Data Compilation and Analysis
Because of the complete inhibition seen in HNMPA-(AM)3 treated sets, treatment unique endpoints were isolated as potentially responsible for the complete disruption of lipid formation noted in this treatment. The list of endpoints unique to the HNMPA-(AM)3 was related back to endpoints found to be unique to adipocytic differentiation in Study Set #1 (Table IC). This revealed a substantial number of endpoints in common between the two studies, thus implicating them as potential key regulators of differentiation. To identify possible end point interactions, a Spearman's Rho correlation was performed in accordance with the previously described data filtering methods (Table IA and IB). This displayed predominantly unique correlations when comparing inhibitor treatments, with some treatments lacking a correlation with a given end point entirely.
Table I. Spearman's Rho correlations and endpoints unique to the HNMPA-(AM)3 treatment.
|
A Treatment started at Time 0: Early-Time Window (2–6 Days)a | ||||
|---|---|---|---|---|
| HNMPA-(AM)3 |
Rapamyacin |
MAPK |
||
| Abbreviation | Phosphoprotein | Abbreviation | Phosphoprotein | Phosphoprotein |
| ACC S79 | Acetyl-CoA Carboxylase (Ser79) | BAD S112 | BCL2-associated agonist of cell death (Ser112) | No significant correlations |
| c-ABL Y245 | c-Abl, non-receptor tyrosine kinase (Tyr245) | CRKII Y221 | v-crk sarcoma virus CT10 oncogene homolog (Tyr221) | |
| CRKII Y221 | v-crk sarcoma virus CT10 oncogene homolog (Tyr221) | IRS1 S612 | Insulin receptor substrate 1 (Ser612) | |
| PTEN S380 | Phosphatase and tensin homolog (Ser380) | p38MAPK T180/Y182 | p38 Mitogen Activated Protein Kinase (Thr180/Tyr182) | |
| SRC Y527 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (Thr527) | VEGFR2 Y1175 | Kinase insert domain receptor (a type III receptor tyrosine kinase) (Tyr1175) | |
| STAT5 Y694 | Signal transducer and activator of transcription 5 (Tyr694) | |||
|
B Treatment started on Day 2: Early-Time Window (2–6 Days)a | ||||
|---|---|---|---|---|
| HNMPA-(AM)3 |
MAPK |
Rapamycin |
||
| Abbreviation | Phosphoprotein | Abbreviation | Phosphoprotein | Phosphoprotein |
| ACC S79 | Acetyl-CoA Carboxylase (Ser79) | ATF2 T71 | Activating transcription factor 2 (Thr71) | No significant correlations |
| AKT S473 | v-akt murine thymoma viral oncogene (Ser473) | BAD S112 | BCL2-associated agonist of cell death (Ser112) | |
| BAD S112 | BCL2-associated agonist of cell death (Ser112) | CRKII Y221 | v-crk sarcoma virus CT10 oncogene homolog (Tyr221) | |
| c-ABL Y245 | c-Abl, non-receptor tyrosine kinase (Tyr245) | ERK 1/2 (T202/Y204) | Mitogen-activated protein kinase 1/2 (T202/Y204) | |
| CC7 D198 | Caspase-7, cleaved (Asp198) | p38MAPK T180/Y182 | p38 Mitogen Activated Protein Kinase (Thr180/Tyr182) | |
| CC9 D315 | Caspase-9, cleaved (Asp315) | PTEN S380 | Phosphatase and tensin homolog (Ser380) | |
| CRKII Y221 | v-crk sarcoma virus CT10 oncogene homolog (Tyr221) | STAT5 Y694 | Signal transducer and activator of transcription 5 (Tyr694) | |
| ERK 1/2 (T202/Y204) | Mitogen-activated protein kinase 1/2 (T202/Y204) | |||
| FOX01 S256 | FoxO1 forkhead box O1 (Ser256) | |||
| IGF-1R (Y1131)/IR(Y1146) | Insulin-like growth factor 1 receptor (Tyr1131)/Insulin Receptor (Tyr1146) | |||
| IGF-1R(Y1135/36)/IR(Y1150/51) | Insulin-like growth factor 1 receptor (Try1135/36)/Insulin Receptor (Try1150/51) | |||
| IRS1 S612 | Insulin receptor substrate 1 (Ser612) | |||
| MST1(T183)/MST2(T180) | Macrophage stimulating 1 (hepatocyte growth factor-like) (Thr183)/Mst2 (Thr180) | |||
| mTOR S2481 | Mechanistic target of rapamycin (Ser2481) | |||
| PDK1 S241 | Pyruvate dehydrogenase kinase, isozyme 1 (Ser241) | |||
| PTEN S380 | Phosphatase and tensin homolog (Ser380) | |||
| SRC Y527 | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (Thr527) | |||
| STAT3 Y705 | Signal transducer and activator of transcription 3 (Y705) | |||
| STAT6 Y641 | Signal transducer and activator of transcription 6 (Try641) | |||
|
C Early and late endpoints that are unique to the HNMPA-(AM)3 treatments | |||
|---|---|---|---|
| Early Endpoints |
Late Endpoints |
||
| Abbreviation | Phosphoprotein | Abbreviation | Phosphoprotein |
| c-ABL T735b | c-Abl, non-receptor tyrosine kinase (Thr735) | ERK 1/2 (T202/Y204) | Mitogen-activated protein kinase 1/2 (T202/Y204) |
| c-ABL Y245 | c-Abl, non-receptor tyrosine kinase (Tyr245) | FADD S194 | Fas (TNFRSF6)-associated via death domain (Ser194) |
| CC7 D198 | Caspase-7, cleaved (Asp198) | FOX01 S256 | FoxO1 forkhead box O1 (Ser256) |
| CRKII Y221b | v-crk sarcoma virus CT10 oncogene homolog (Tyr221) | IGF-1R(Y1131)/IR (Y1146) | insulin-like growth factor 1 receptor (Tyr1131)/Insulin Receptor (Tyr1146) |
| FADD S194 | Fas (TNFRSF6)-associated via death domain (Ser194) | PDK1 S241b | pyruvate dehydrogenase kinase, isozyme 1 (Ser241) |
| IGF-1R(Y1135/36)/IR (Y1150/51) | Insulin-like growth factor 1 receptor (Try1135/36)/Insulin Receptor (Try1150/51) | PKA C T19b 7 | Protein kinase, cAMP-dependent, catalytic (Thr197) |
| MST1(T183)/MST2(T180) | Macrophage stimulating 1 (hepatocyte growth factor-like) (Thr183)/Mst2 (Thr180) | PTEN S380b | Phosphatase and tensin homolog (Ser380) |
| mTOR S2448b | Mechanistic target of rapamycin (Ser2448) | SMAD 1(S/S)/5(S/S)/8(S/S)b | Smad family member 1 (Ser463/465)/Smad family 5 (Ser463/465)/Smad family member 8 (Ser426/428) |
| mTOR S2481b | Mechanistic target of rapamycin (Ser2481) | SRC Y527b | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (Thr527) |
| PDGFRb Y571 | platelet-derived growth factor receptor beta (Try751) | STAT6 Y641 | Signal transducer and activator of transcription 6 (Try641) |
| PDK1 S241 | Pyruvate dehydrogenase kinase, isozyme 1 (Ser241) | VEGFR2 Y1175 | Kinase insert domain receptor (a type III receptor tyrosine kinase) (Tyr1175) |
| SMAD 1(S/S)/5(S/S)/8(S/S)b | Smad family member 1 (Ser463/465)/Smad family 5 (Ser463/465)/Smad family member 8 (Ser426/428) | ||
| STAT6 Y641 | Signal transducer and activator of transcription 6 (Try641) | ||
a Spearman's Rho correlations for treatments started at time 0 and day 2 for the early time window.
b Endpoints in common in between Study Set #1 and HNMPA-(AM)3 treated samples.
DISCUSSION
Detailed characterization of stem cell differentiation will contribute to confident stem cell manipulation for tissue reconstruction as well as enhance disease understanding relating to obesity, diabetes, and some cancers (26, 27). Although the current study is substantial in scale, it only provides a starting point for characterization of the signaling architecture regulating ASC differentiation. Because of total protein levels fluctuating over the course of cell cycling and differentiation, this study focused analysis on the phosphorylation status of key signaling “hubs” relating to underpinning biological processes, with only a small pool of a given kinase substrate being phosphorylated at any given time. Further interrogation of the total levels of proteins that were found to be differentially activated/phosphorylated in our study will be performed in future studies. Additionally, only a subset of the kinome was examined, leaving an inherent potential for key signaling proteins to be overlooked. This can be expanded with newly validated antibodies and the incorporation of a more global mass spectrometry (MS)-based phosphoprotein analysis to provide the necessary depth of coverage to more fully elucidate the signaling changes occurring. The use of the RPMA technology, which can quantitatively measure large numbers of analytes with extremely low cell number requirements, enabled the survey of a broad number of time points to provide a starting basis to isolate time windows that could be explored more fully with MS approaches that provide more broad scale analysis yet require larger cellular input.
Despite these facts, results from Study Set #1 displayed a time-ordered signal transduction profile and directed the pharmacological targeting of likely key regulators of adipocytic differentiation via kinase inhibitors that completely or partially inhibited adipocytic differentiation. This screening identified HNMPA-(AM)3, rapamycin, and MAPK inhibitors to most effectively disrupt differentiation and warrant further exploration via Study Sets #3–6 (Fig. 3). Although HNMPA-(AM)3 and rapamycin showed a reduction in phosphorylation levels in expected targets, the MAPK inhibitor showed an increased phosphorylation in the endpoints p38MAPK, MEK ½, and ERK1/2 (SF2). This hyperphosphorylation may have been attributed to cellular compensation, yet despite this finding, all the inhibitors showed a clearly definable disruption in lipid formation relative to untreated samples. Speanman's Rho analysis of endpoints unique to HNMPA-(AM)3 showed CRKII and c-ABL Y245, in addition to other endpoints, to be uniquely correlated in the HNMPA-(AM)3 treated set relative to the other treatments. Data set congruency was further demonstrated with the same time-ordered signaling activation noted across all studies in addition to c-ABL T735 having no correlations with the endpoints examined, just as was noted in Study Set #1. Ultimately, CRKII and c-ABL were implicated as primary regulators of adipocyte commitment with AKT, mTOR, and SMAD pathways implicated secondarily.
The current study ensured protein activation analysis was based on clinically derived human primary ASCs and while primary cultures are becoming more readily available, mouse embryonic cell lines 3T3-L1 and C3H10T1/2 are still the most commonly studied relating to adipocyte biology. These cell lines are useful as human analogs although true parity remains to be validated (28). Rapamycin has been previously cited for complete disruption of differentiation or more minimally leading to a substantial decrease in lipid formation, in addition to a decreased expression of adipocytic markers (29). One study utilizing human primary preadipocytes, with a comparable amount of inhibitor applied, noted a complete disruption of differentiation (13). An additional study examining 3T3-L1 cells treated with rapamycin demonstrated a substantial decrease in lipid formation and moreover, a reduction in the gene expression of the adipocytic markers PPARγ, adipsin, adducin 1/sterol regulatory element binding transcription factor 1 (ADD1/SREBP1c), and TNF receptor superfamily, member 6 (FAS) (30). Although both studies relied on insulin as the main stimulator of differentiation, the secondary media components differed, which may explain the noted variation in lipid disruption. mTOR itself inactivates eukaryotic translation initiation factor 4E-binding protein 1 (4E-BP1) and activates p70 S6 kinase (p70S6K) and subsequently RPS6 to regulate protein synthesis. One study examining mesenchymal stem cells found p70S6K and subsequent RPS6 phosphorylation levels to be almost undetectable, while 4E-BP1 was not impacted equally. This study implicated the AKT pathway and subsequently mTOR to be necessary drivers of differentiation (31). In agreement, this study noted the same trends in the magnitude of inhibition in the downstream targets of mTOR and a possible relation to AKT. However, this pathway was not deemed a necessary component of differentiation in the current study.
The MAPK inhibitor PD169316 has widely been held to be specific to p38MAPK, but this belief is coming into question. The current study has found p38MAPK, ERK 1/2, and MEK 1/2 all to show increased phosphorylation levels which indicates that the inhibitor was targeting MAPKs in general. One study using this inhibitor with the 3T3-L1 cell line suggested that it disrupted differentiation, drawing as evidence a substantial decrease in lipid formation as well as a decrease of CCAAT/enhancer binding protein beta (C/EBPβ) phosphorylation, a protein implicated in adipocytic commitment. Additionally, this same laboratory found that mouse and human derived preadipocytes responded differently to the PD169316 inhibitor, with promoted differentiation and increased phosphorylation of C/EBPβ noted in the human preadipocytes (32, 33). Another study examining the specificity of PD169316 found that when used at concentrations of 5 μm or higher the inhibitor also targeted transforming growth factor beta (TGFβ) hence inhibiting SMAD signaling (34). Although previous studies implicate p38MAPK as a driver of differentiation, it is important to note that samples were treated with 10 μm of inhibitor, whereas the current study treated with 500 nm which could have contributed to the difference of magnitude noted. A study targeting p38MAPK with RNAi knockdown in C3H10T1/2 stem cells noted only a partial inhibition to stem cell commitment, which would be in agreement with the current study (35). Moreover, MEK 1/2 mediated activation of ERK 1/2 has also been implicated in the disruption of differentiation. One study examining embryonic stem cells found the inhibition of ERK 1/2 early in differentiation disrupted adipocyte formation (36). Although another study found ERK inhibition to block osteogenic differentiation and instead promote adipogenesis, as noted by the expression of adipose-specific mRNA (37). Another laboratory expanded on this finding by examining MEK/ERK activation via preadipocyte factor 1 (PREF-1) and found ERK 1/2 activation to inhibit differentiation. This study also noted that a rapid burst of ERK activation was necessary following the addition of differentiation stimuli (38). These findings substantiate a portion of the trends seen in the present study, which demonstrated an increase in ERK 1/2 phosphorylation to correlate with a disruption in differentiation, but without the same magnitude of disruption being noted.
Although no other studies were found to relate the usage of HNMPA-(AM)3 to the disruption of adipogenesis, other effective demonstrations of the inhibitor were examined. One study explored the role of insulin signaling in umbilical endothelial cells. Various inhibitors were used to include a phosphatidylinositol 3-kinases (PI3-K) inhibitor (LY294002), IR/IGFR inhibitor (tyrphostin 23), and HNMPA-(AM)3 inhibitor. All of these showed a decrease in the proliferative effects of insulin suggesting insulin mediated signaling through IGFR/IR followed by PI3-K and subsequent AKT activation, thus confirming the reproducibility of the effects seen through the HNMPA-(AM)3 inhibitor (39). Another study examined the effects of visfatin, an adipocytokine, on osteoblasts. The HNMPA-(AM)3 inhibitor was used in this study to inhibit both insulin and visfatin signaling, because of the ability of visfatin to act through the insulin receptor. When cells were stimulated with visfatin or insulin in the presence of inhibitor, IR, IRS-1, and IRS-2 phosphorylation were inhibited in addition to other cellular properties, thus showing the consistent effects of the HNMPA-(AM)3 inhibitor (40).
CRKII and c-ABL, which were implicated as potential drivers of differentiation in the current study, have not been previously implicated in the differentiation of adipocytes, thus appearing to be a novel finding. CRKII has been predominantly studied in the context of cancer research and implicated in cellular functions such as proliferation, differentiation, and migration (41, 42, 43). One study examining synovial sarcoma cells discovered a novel signaling mechanism with CRKII activating v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (SRC) and subsequently p38MAPK, with CRKII suppression leading to decreased proliferation (14). The current study found a similar relationship with CRKII and SRC, although p38MAPK was not implicated because of a lack of correlation with CRKII. Furthermore, one study employing the L6C6 cell line, rat skeletal muscle, found CRKII overexpression led to enhanced activation of ERK in an insulin-dependent manor (44). Although the current study found ERK to correlate with CRKII in HNMPA-(AM)3 treatments, the relation was dissimilar due to CRKII activity being repressed whereas ERK activity was enhanced. c-ABL has also been studied predominantly in the context of cancer research. c-ABL Y245 has been found to be phosphorylated by other SRC members and platelet derived growth factor receptor (PDGFR), whereas c-ABL T735 has been implicated in cytoplasmic localization and is phosphorylated by CDC-like kinase 1 (CLK1), CLK4, macrophage stimulating 1 (MST1) and MST2. Moreover, c-ABL has been found to associate with CRKII and effect signal transduction and cytoskeletal remodeling (1). This suggests that these two proteins may function in cellular remodeling to aid in the morphological changes necessary to achieve an adipocytic fate.
Although CRKII and c-ABL have been implicated as key regulators of adipocytic differentiation for the first time in this study, further examination is required to validate and characterize causal roles and pathway interactions. If CRKII and c-ABL are validated as key regulators of differentiation, their roles can be further characterized in an in vivo system to provide a complete picture of specific functioning. The implication of CRKII and c-ABL as key regulators is only a starting point, these findings have potential implications in obesity research as a means to regulate and limit the transformation of stem cells into new adipose cells. The global nature of the current study has provided further insight into the time-specific phosphoregualtory events to broaden our understanding of the signal transduction events directing adipocyte differentiation. Advances such as these, may lead to more precise tuning of end lineage dynamics, answer fundamental questions concerning stem cell biology, and perhaps aid in the identification of new therapeutic targets of adipogenesis and metabolic derangements during fat metabolism.
Footnotes
* Financial support provided by the Henry and Alice Greenwald Foundation and the George Mason University College of Science.
 This article contains supplemental Figs. S1 and S2.
This article contains supplemental Figs. S1 and S2.
1 The abbreviations used are::
- ASC
- Adipose-derived stromal/stem cell
- AKT
- v-akt murine thymoma viral oncogene
- c-ABL
- and c-abl oncogene 1 non-receptor tyrosine kinase
- CRKII
- v-crk sarcoma virus CT10 oncogene homolog
- ERK
- extracellular-signal-regulated kinases
- IGFR
- insulin-like growth factor receptor
- IR
- insulin receptor
- mTOR
- mammalian target of rapamycin
- ORO
- Oil red O
- PTEN
- phosphatase and tensin homolog
- RPMA
- Reverse phase protein microarray
- SMAD
- SMAD family member.
REFERENCES
- 1. Colicelli J. (2010) Abl tyrosine kinases: evolution of function, regulation, and specificity. Sci. Signal 3, 1–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Katz A., Tholpady A., Tholpady S. S., Shang H., Ogle R. (2005) Cell surface and transcriptional characterization of human adipose-derived adherent stromal (hADAS) Cells. Stem Cells 23, 412–423 [DOI] [PubMed] [Google Scholar]
- 3. Wu L., Wang T., Ge Y., Cai X., Wang J., Lin Y. (2012) Secreted factors from adipose tissue increase adipogenic differentiation of mesenchymal stem cells. Cell Prolif. 45, 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gimble J., Katz A., Bunnell B. (2007) Adipose-derived stem cells for regenerative medicine. Circ. Res. 100, 1249–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bost F., Aouadi M., Caron L., Binétruy B. (2004) The role of MAPKs in adipocyte differentiation and obesity. Biochimie 87, 51–56 [DOI] [PubMed] [Google Scholar]
- 6. Gonzalez F. (2005) Getting fat: Two new players in molecular adipogenesis. Cell Metab. 1, 85–86 [DOI] [PubMed] [Google Scholar]
- 7. Liu J., DeYoung S. M., Zhang M., Zhang M., Cheng A., Saltiel A. R. (2005) Changes in integrin expression during adipocyte differentiation. Cell. Metab. 2, 165–176 [DOI] [PubMed] [Google Scholar]
- 8. Rodriguez A., Elabd C., Delteil F., Astier J., Vernochet C., Saint-Marc P., Guesnet J., Guezennec A., Amri E. Z., Dani C., Ailhaud G. (2004) Adipocyte differentiation of multipotent cells established from human adipose tissue. Bio. Biophys. Res. Commun 315, 255–263 [DOI] [PubMed] [Google Scholar]
- 9. Menssen A., Häup T., Sittinger M., Delorme B., Charbord P., Ringe J. (2011) Differential gene expression profiling of human bone marrow-derived mesenchymal stem cells during adipogenic development. BMC Genomics 12, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sibov T., Severino P., Marti L., Pavon L. F., Oliveira D. M., Tobo P. R., Campos A. H., Paes A.T., Amaro E., Jr, Gamarra L., Moreira-Filho C. A. (2012) Mesenchymal stem cells from umbilical cord blood: parameters for isolation, characterization and adipogenic differentiation. Cytotechnology 64, 511–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Waters K. M., Pounds J. G., Thrall B. D. (2006) Data merging for integrated microarray and proteomic analysis. Brief. Funct. Genomic. Proteomic 5, 261–272 [DOI] [PubMed] [Google Scholar]
- 12. Williamson A. J., Whetton A. D. (2011) The requirement for proteomics to unravel stem cell regulatory mechanisms. J. Cell. Physiol 226, 2478–2483 [DOI] [PubMed] [Google Scholar]
- 13. Arsenault R., Griebel P., Napper S. (2011) Peptide arrays for kinome analysis: New opportunities and remaining challenges. Proteomics 11, 4595–4609 [DOI] [PubMed] [Google Scholar]
- 14. Tichy A., Salovskaa B., Rehulkab P., Klimentova J., Vavrova J., Stulik J., Hernychova L. (2011) Phosphoproteomics: Searching for a needle in a haystack. J. Proteomics 74, 2786–2797 [DOI] [PubMed] [Google Scholar]
- 15. Brill L. M., Xiong W., Lee K. B., Ficarro S., Crain A., Xu Y., Terskikh A., Snyder E., Ding S. (2009) Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell 5, 204–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Hoof D., Munoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 17. Gallager R. I., Silvestri A., Petricoin E. F., 3rd, Liotta L. A., Espina V. (2011) Reverse phase protein microarrays: fluorometric and colorimetric detection. Methods Mol. Biol 723, 275–301 [DOI] [PubMed] [Google Scholar]
- 18. Liotta L. A., Espina V., Mehta A. I., Calvert V., Rosenblatt K., Geho D., Munson P. J., Young L., Wulfkuhle J., Petricoin E. F., 3rd. (2003) Protein microarrays: meeting analytical challenges for clinical applications. Cancer Cell 3, 317–325 [DOI] [PubMed] [Google Scholar]
- 19. Rapkiewicz A., Espina V., Zujewski J. A., Lebowitz P. F., Filie A., Wulfkuhle J., Camphausen K., Petricoin E. F., 3rd, Liotta L. A., Abati A. (2007) The needle in the haystack: application of breast fine-needle aspirate samples to quantitative protein microarray technology. Cancer 111, 173–184 [DOI] [PubMed] [Google Scholar]
- 20. Espina V., Mehta A. I., Winters M. E., Calvert V., Wulfkuhle J., Petricoin E.F., 3rd, Liotta L.A. (2003) Protein microarrays: molecular profiling technologies for clinical specimens. Proteomics 3, 2091–100 [DOI] [PubMed] [Google Scholar]
- 21. Gulmann C., Sheehan K. M., Conroy R. M., Wulfkuhle J. D., Espina V., Mullarkey M. J., Kay E. W., Liotta L. A., Petricoin E. F., 3rd. (2009) Quantitative cell signalling analysis reveals down-regulation of MAPK pathway activation in colorectal cancer. J. Pathol. 218, 514–519 [DOI] [PubMed] [Google Scholar]
- 22. Herrmann P. C., Gillespie J. W., Charboneau L., Bichsel V. E., Paweletz C. P., Calvert V. S., Kohn E. C., Emmert-Buck M. R., Liotta L. A., Petricoin E. F., 3rd. (2003) Mitochondrial proteome: altered cytochrome c oxidase subunit levels in prostate cancer. Proteomics 3,1801–1810 [DOI] [PubMed] [Google Scholar]
- 23. Frederick M. J., VanMeter A. J., Gadhikar M. A., Henderson Y. C., Yao H., Pickering C. C., Williams M. D., El-Naggar A. K., Sandulache V., Tarco E., Myers J. N., Clayman G. L., Liotta L. A., Petricoin E. F., 3rd, Calvert V. S., Fodale V., Wang J., Weber R. S. (2011) Phosphoproteomic analysis of signaling pathways in head and neck squamous cell carcinoma patient samples. Am. J. Pathol. 178, 548–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silvestri A., Calvert V., Belluco C., Lipsky M., De Maria R., Deng J., Colombatti A., De Marchi F., Nitti D., Mammano E., Liotta L., Petricoin E., Pierobon M. (2013) Protein pathway activation mapping of colorectal metastatic progression reveals metastasis-specific network alterations. Clin, Exp, Metastasis 30, 309–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Einspahr J. G., Calvert V., Alberts D. S., Curiel-Lewandrowski C., Warneke J., Krouse R., Stratton S. P., Liotta L., Longo C., Pellacani G., Prasad A., Sagerman P., Bermudez Y., Deng J., Bowden G. T., Petricoin E. F., 3rd. (2012) Functional protein pathway activation mapping of the progression of normal skin to squamous cell carcinoma. Cancer Prev. Res. 5, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cristancho A. G., Lazar M. A. (2011) Forming functional fat: a growing understanding of adipocyte differentiation. Nat. Rev. Mol. Cell. Biol. 12, 722–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Donzelli E., Lucchini C., Ballarini E., Scuteri A., Carini F., Tredici G., Miloso M. (2011) ERK1 and ERK2 are involved in recruitment and maturation of human mesenchymal stem cells induced to adipogenic differentiation. J. Mol. Cell. Biol. 3, 123–131 [DOI] [PubMed] [Google Scholar]
- 28. Qian S. W., Li X., Zhang Y., Huang H., Liu Y., Sun X., Tang Q. (2010) Characterization of adipocyte differentiation from human mesenchymal stem cells in bone marrow. BMC Dev. Biol 10, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bell A., Grunder L., Sorisky A. (2000) Rapamycin Inhibits Human Adipocyte Differentiation in Primary Culture. Obes. Res. 8, 249–254 [DOI] [PubMed] [Google Scholar]
- 30. Cho H., Park J., Lee H. W., Lee Y. S., Kim J. B. (2004) Regulation of adipocyte differentiation and insulin action with rapamycin. Biochem. Biophys. Res. Commun. 321, 942–948 [DOI] [PubMed] [Google Scholar]
- 31. Xiang X., Zhao J., Xu G., Li Y., Zhang W. (2011) mTOR and the differentiation of mesenchymal stem cells. Acta. Biochim. Biophys. Sin. 43, 501–510 [DOI] [PubMed] [Google Scholar]
- 32. Aouadi M., Jager J., Laurent K., Gonzalez T., Cormont M., Binétruy B., Le Marchand-Brustel Y., Tanti J. F., Bost F. (2007) p38MAP Kinase activity is required for human primary adipocyte differentiation. FEBS Lett. 581, 5591–5596 [DOI] [PubMed] [Google Scholar]
- 33. Aouadi M., Laurent K., Prot M., Le Marchand-Brustel Y., Binétruy B., Bost F. (2006) Inhibition of p38MAPK increases adipogenesis from embryonic to adult stages. Diabetes 55, 281–289 [DOI] [PubMed] [Google Scholar]
- 34. Fu Y., O'Connor L. M., Shepherd T. G., Nachtigal M. W. (2003) The p38 MAPK inhibitor, PD169316, inhibits transforming growth factor b-induced Smad signaling in human ovarian cancer cells. Biochem. Biophys. Res. Commun. 310, 391–397 [DOI] [PubMed] [Google Scholar]
- 35. Huanga H., Songa T. J., Li X., Hu L., He Q., Liu M., Lane M. D., Tang Q. (2009) BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. PNAS 106, 12670–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bost F., Caron L., Marchetti I., Dani C., Le Marchand-Brustel Y., Binétruy B. (2002) Retinoic acid activation of the ERK pathway is required for embryonic stem cell commitment into the adipocyte lineage. Biochem. J. 361, 62–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jaiswal R., Jaiswal N., Bruder S., Mbalaviele G., Marshak D. R., Pittenger M. F. (2000) Adult human mesenchymal stem cell differentiation to the osteogenic or adipogenic lineage is regulated by mitogen-activated protein kinase. J. Biol. Chem. 275, 9645–9652 [DOI] [PubMed] [Google Scholar]
- 38. Kim K., Kim J. H., Wang Y., Sul H. S. (2007) Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol. Cell. Biol. 27, 2294–2308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shrader C., Bailey K. M., Konat G. W., Cilento E. V., Reilly F.D. (2009) Insulin enhances proliferation and viability of human umbilical vein endothelial cells. Arch. Dermatol. Res. 301, 159–16 [DOI] [PubMed] [Google Scholar]
- 40. Xie H., Tang S. Y., Luo X. H., Huang J., Cui R. R., Yuan L. Q., Zhou H.D., Wu X. P., Liao E. Y. (2007) Insulin-like effects of visfatin on human osteoblasts. Calcif. Tissue Int. 80, 201–210 [DOI] [PubMed] [Google Scholar]
- 41. Isakov N. (2008) A new twist to adaptor proteins contributes to regulation of lymphocyte cell signaling. Trends Immunol. 29, 388–396 [DOI] [PubMed] [Google Scholar]
- 42. Watanabe T., Tsuda M., Tanaka S., Ohba Y., Kawaguchi H., Majima T., Sawa H., Minami A. (2009) Adaptor protein Crk induces Src-dependent activation of p38MAPK in regulation of synovial Sarcoma cell proliferation. Mol. Cancer Res. 7, 1582–1592 [DOI] [PubMed] [Google Scholar]
- 43. Yamada S., Yanamoto S., Kawasaki G., Rokutanda S., Yonezawa H., Kawakita A., Nemoto T. K. (2011) Overexpression of CRKII increases migration and invasive potential in oral squamous cell carcinoma. Cancer Lett. 303, 84–91 [DOI] [PubMed] [Google Scholar]
- 44. Prosser S., Sorokina E., Pratt P., Sorokin A. (2003) CrkIII: a novel and biologically distinct member of the Crk family of adaptor proteins. Oncogene 22, 4799–480 [DOI] [PubMed] [Google Scholar]