Abstract
Protein modification by small ubiquitin-like modifier (SUMO) modulates the activities of numerous proteins involved in different cellular functions such as gene transcription, cell cycle, and DNA repair. Comprehensive identification of SUMOylated sites is a prerequisite to determine how SUMOylation regulates protein function. However, mapping SUMOylated Lys residues by mass spectrometry (MS) is challenging because of the dynamic nature of this modification, the existence of three functionally distinct human SUMO paralogs, and the large SUMO chain remnant that remains attached to tryptic peptides. To overcome these problems, we created HEK293 cell lines that stably express functional SUMO paralogs with an N-terminal His6-tag and an Arg residue near the C terminus that leave a short five amino acid SUMO remnant upon tryptic digestion. We determined the fragmentation patterns of our short SUMO remnant peptides by collisional activation and electron transfer dissociation using synthetic peptide libraries. Activation using higher energy collisional dissociation on the LTQ-Orbitrap Elite identified SUMO paralog-specific fragment ions and neutral losses of the SUMO remnant with high mass accuracy (< 5 ppm). We exploited these features to detect SUMO modified tryptic peptides in complex cell extracts by correlating mass measurements of precursor and fragment ions using a data independent acquisition method. We also generated bioinformatics tools to retrieve MS/MS spectra containing characteristic fragment ions to the identification of SUMOylated peptide by conventional Mascot database searches. In HEK293 cell extracts, this MS approach uncovered low abundance SUMOylated peptides and 37 SUMO3-modified Lys residues in target proteins, most of which were previously unknown. Interestingly, we identified mixed SUMO-ubiquitin chains with ubiquitylated SUMO proteins (K20 and K32) and SUMOylated ubiquitin (K63), suggesting a complex crosstalk between these two modifications.
SUMOylation is a covalent and reversible post-translational modification that conjugates the small ubiquitin-like modifier (SUMO)1 proteins on Lys residues of target proteins (1). Although this modification is common to all species, the number of SUMO genes expressed vary from a single SUMO gene in lower eukaryotes (e.g. yeast, drosophila, nematodes) to eight different paralogs in plants (e.g. Arabidopsis thaliana) (2). In human, three SUMO genes (SUMO1, SUMO2, and SUMO3) are ubiquitously expressed in all tissues, whereas a forth gene (SUMO4) is uniquely expressed in spleen, lymph nodes, and kidney cells (3). As for other ubiquitin-like proteins (UBLs) such as NEDD8, ISG15, and FAT10, the structure of SUMO proteins share a characteristic three-dimensional fold with ubiquitin, but differ significantly in their sequences.
Protein SUMOylation is an essential cellular process conserved from yeast to mammals, and is associated with many fundamental pathways in both the nucleus and cytoplasm including DNA replication, genome stability, nuclear transport, gene transcription, mitochondrial fission and fusion events (4–6). Despite the low occurrence of SUMOylation compared with ubiquitylation, an increasing number of substrates have been characterized based on the presence of a predicted SUMO consensus motif ψKxE/D, where ψ represents a hydrophobic residue (7). This modification can also take place on Lys residues located on an extended consensus site defined by the phosphorylation-dependent motif (ψKxExxpSP) (8) and the negatively charged amino acid-dependent motif (9). It is noteworthy that the ψKxE/D motif is also found on many proteins that are not SUMO targets, and that many SUMOylation sites are known to be located in non consensus sites.
SUMOylation involves a sequence of enzymatic steps similar to those of ubiquitylation in which SUMO is transferred from the E1 activating enzyme SAE1/SAE2 to the single E2-conjugating enzyme Ubc9 that directly recognize substrates and catalyze the formation of an isopeptide bond between the C-terminal group of SUMO and the ε-NH2 group of Lys residue from the target protein (10). Although Ubc9 is sufficient to promote SUMOylation, substrate recognition can be mediated by one of several E3 ligases including nucleoporin RanBP2, polycomb repressor Pc2, and members of the protein inhibitor of STAT (PIAS) ligase family (PIAS1, PIAS3, PIASxα, PIASxβ, PIASy) that facilitate the conjugation process (11). SUMOylation is reversible and this modification can be cleaved rapidly from its target protein by Sentrin/SUMO-specific proteases (SENPs), a family of conserved proteins that can also process SUMO precursors to expose the di-glycine motif necessary for the conjugation of mature SUMO proteins (12).
The identification of SUMOylation sites in protein substrates by mass spectrometry (MS) represents a sizable analytical challenge because of low abundance and dynamic nature of this modification. In contrast to phosphorylation where affinity media such as TiO2 or immobilized metal affinity chromatography are available to enrich phosphopeptides from complex tryptic digests, the purification of UBLs has been typically performed at the protein level using His6- or TAP-tag of the target UBL proteins (13–15). Although these approaches are effective to identify and quantify UBL-modified proteins, the precise identification of SUMOylation sites is still a daunting task in view of the large remnant SUMO sequence appended to the Lys residues of modified tryptic peptides. The identification of SUMOylated peptides is also complicated by their relatively low abundance and their small proportion in complex protein digests (typically < 1% of all identified peptides). The corresponding cross-linked peptides comprise long SUMO remnant chains up to 32 amino acids for SUMO2 and SUMO3 that complicate the interpretation of the MS/MS spectra, and often lead to misidentification of modified peptides using conventional database search engines tailored to the analysis of linear peptides. Different bioinformatics approaches have been proposed to alleviate these difficulties including an automated recognition pattern tool (SUMmOn) (16) and a database containing “linearized branched” peptides (ChopNSpice) (17).
To facilitate the enrichment and identification of modification sites of distinct SUMO paralogs from in vivo samples, we recently reported a novel proteomics approach using HEK293 cells stably expressing SUMO (1, 2, or 3) mutant proteins with a His6 tag and an Arg residue at the 6th position from the C terminus (18). Also, Gln88 of SUMO3 was mutated to an Asn residue to distinguish this paralog from SUMO2. Our SUMO2 mutant is similar to that reported by Matic et al. (19) except that internal lysines were not replaced by arginines to maintain functional polySUMOylation of protein substrates. Upon tryptic digestion, proteins modified with these SUMO mutants give rise to tryptic peptides with a five amino acid long SUMO remnant that facilitates their identification using conventional database search engines. In the present study, we determined the occurrence and abundance of paralog-specific fragment ions formed by collisional activation (collision induced dissociation, CID; higher energy collision dissociation, HCD) and electron transfer dissociation (ETD) using libraries of synthetic peptides with SUMO1 and SUMO3 remnant chains. We also implemented a data independent acquisition method similar to that of Gillet et al. (20) that repeatedly cycle through consecutive 100-m/z precursor isolation windows to form paralog-specific fragment ions and selectively identify SUMOylated peptides from HEK293 cells expressing our SUMO3 mutant. The combination of HCD and high resolution fragment ion detection on the LTQ-Orbitrap Elite conferred a unique advantage to map SUMO modification sites in endogenous target proteins, and provided complementary identification to traditional data dependent acquisition (DDA).
EXPERIMENTAL PROCEDURES
Peptide Synthesis
The peptides (1) Boc-Glu(OtBu)-Qln(Trt)-Thr(tBu)-Gly-Gly-OH and (2) Boc-Asn(Trt)-Gln(Trt)-Thr(tBu)-Gly-Gly-OH were synthesized manually on a 2-chlorotrityl chloride resin using standard procedures of solid phase peptide synthesis. Cleavage of both fragments from the resin was performed using 25% hexafluoroisopropanol in dichloromethane. To a solution of 400 mg of compound 1 or 2 in 10 ml N,N-dimethylformamide were added 1.2 equivalent N-hydroxybenzotriazole and one equivalent ethyl diisopropylcarbodiimide. The mixture was stirred for 30 min at room temperature followed by addition of one equivalent Fmoc-Lys-OH . HCl and one equivalent N-methylmorpholine. After stirring for 2h at room temperature, the solvent was evaporated and the residue was dissolved in 100 ml ethyl acetate. The solution was then extracted four times with 5% K2HSO3, four times with a saturated aqueous solution of sodium chloride and dried over Na2SO4. After evaporation of the solvent, the building blocks (3) Fmoc-Lys[Boc-Glu(OtBu)-Gln(Trt)-Thr(tBu)-Gly-Gly]-OH (44% yield) and (4) Fmoc-Lys[Boc-Asn(Trt)-Gln(Trt)-Thr(tBu)-Gly-Gly]-OH (77% yield) were precipitated with diethyl ether and dried in vacuo over P2O5. All tryptic peptides with SUMO remnant chains were synthesized by automated SPOT synthesis as described by Frank et al. (21). Acylation of the peptide chain with 3 and 4 was performed by preactivation of the respective building block with 2-(1H-Benzotriazole-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate/N,N-diisopropylethylamine. All synthetic peptides were analyzed by LC-MS on a Agilent 1100 series LC/MSD Trap to confirm purity.
Cell Culture and Isolation of SUMOylated Proteins
HEK293 wild type and HEK293 cells stably expressing His6-SUMO mutants (5 × 106 cells/replicate) were cultured in Dulbecco's modified Eagles medium (DMEM) High Glucose (Hyclone SH30081.02) supplemented with 10% fetal bovine serum (FBS) (Fisher), 1% l-glutamine and 1% Penicillin/Streptomycin (Fisher) (18). Cells were washed with PBS containing 20 mm of N-ethyl maleimide (NEM) and centrifuged at 215 × g for 10 min. NEM is used to stabilize SUMO conjugates by alkylating the sulfhydryl group of the catalytic cysteine on SUMO-specific proteases (SENP). Cells were lysed in hypotonic buffer A (10 mm Tris pH 7.6, 1.5 mm MgCl2, 20 mm NEM, protease inhibitor, Sigma-Aldrich) for 30 min and centrifuged 15 min at 215 × g. Nuclei were washed with buffer A and centrifuged at 215 × g. The nuclei pellet was lysed in denaturing buffer B (6 m Guanidine, 0.1 m NaH2PO4, 10 mm imidazole, 10 mm Tris-HCl pH 8, 20 mm NEM, 10 mm β mercaptoethanol), sonicated and centrifuged at 16000 × g for 15 min. The supernatant was incubated with 200 μl of nickel nitriloacetic acid (NTA) agarose beads (Invitrogen) for 3h at room temperature. Ni-NTA beads were washed once with buffer B and then five times with buffer C (8 m urea, 0.1 m NaH2PO4, 10 mm Tris-HCl pH 6.3, 10 mm β mercaptoethanol, 20 mm imidazole). A portion of the NTA beads were used to determine total protein amounts using a Bradford protein assay. NTA beads were kept frozen until digestion. The enrichment of SUMOylated proteins was verified by immunoblots using rabbit anti-SUMO2/3 and chicken anti-mouse Alexa-Fluor 594-conjugated secondary antibody from Invitrogen, and monoclonal anti-6xHis antibody from Clontech.
Protein Digestion
SUMO-proteins immobilized on NTA beads were solubilized in 4 m urea, reduced in 5 mm tris(2-carboxyethyl)phosphine (TCEP) (Pierce) for 20 min at 37 °C and then alkylated in 5 mm chloroacetamide (Sigma-Aldrich) for 20 min at 37 °C. We used this alkylating agent to differentiate free cysteines (modified by NEM) and residues linked by disulfide-bonds (modified by chloroacetamide). A solution of 5 mm dithiothreitol was added to the protein solution to react with excess chloroacetamide. The solution was diluted to 1 m urea using 50 mm ammonium bicarbonate and digested overnight with modified trypsin (1:50, enzyme:substrate ratio) at 37 °C under high agitation speed. The digest was acidified with trifluoroacetic acid (TFA), desalted using an Oasis HLB cartridge (Waters) and dried using a speed vac prior to MS analyses.
Mass Spectrometry
LC-MS/MS analyses were performed on a nano-LC 2D pump (Eksigent) coupled to a LTQ-Orbitrap Elite hybrid mass spectrometer via a nanoelectrospray ion source (Thermo Fisher Scientific). Peptides were separated on a Optiguard SCX trap column, 5 μm, 300Å, 0.5 ID × 23 mm (Optimize technologies) and eluted on-line to a 360 μm ID × 4 mm, C18 trap column prior to separation on a 150 μm ID x 10 cm nano-LC column (Jupiter C18, 3 μm, 300 Å, Phenomex). Tryptic digests were loaded on the SCX trap and sequentially eluted using salt plugs of 0, 250, 500, 750, 1000, and 2000 mm ammonium acetate, pH 3.5. Peptides were separated on the analytical column using a linear gradient of 5–40% acetonitrile (0.2% formic acid) in 53 min with a flow rate of 600 nL/min.
For the analysis of synthetic peptides, MS/MS spectra were collected in DDA. The conventional MS spectra (survey scan) were acquired in the Orbitrap at a resolution of 60000 for m/z 400 after the accumulation of 106 ions in the linear ion trap. Mass calibration used a lock mass from ambient air [protonated (Si(CH3)2O))6; m/z 445.120029], and provided mass accuracy within 7 ppm for precursor ion mass measurements. The dynamic exclusion of previously acquired precursor ions was enabled (repeat count 1, repeat duration: 15 s; exclusion duration 15 s). MS/MS spectra were acquired in CID, HCD or ETD activation modes using an isolation window of 2 Da. For ETD, up to 12 precursor ions were selected for fragmentation. Precursor cation AGC target was set at 1 × 104, whereas a value of 2 × 105 was used for the fluoranthene anion population and ion/ion reaction duration was fixed at 100 ms. For CID, a normalized collision energy of 35% was used and up to 12 precursor ions were sequentially isolated and accumulated to a target value of 10000 with a maximum injection time of 100 ms. For MS/MS spectra acquired with HCD, a normalized collision energy of 30% was selected. Up to six precursor ions were accumulated to a target value of 50000 with a maximum injection time of 300 ms and fragment ions were transferred to the Orbitrap analyzer operating at a resolution of 30000 at m/z 400.
For the analysis of HEK293 cell extracts, MS/MS spectra were acquired in HCD mode with a normalized energy of 30%. We first used the LTQ-Orbitrap Elite in data dependent acquisition with dynamic exclusion of previously acquired precursor ions (repeat count 1, repeat duration: 30 s; exclusion duration 45 s). In data independent acquisition, the MS instrument cycled through the sequential acquisition of a MS scan and a HCD MS/MS scan to transmit in turn intact peptide ions and fragment ions arising from the dissociation of selected precursor ion windows (up to seven ion trap segments of m/z 100–700). Typically, seven segments of m/z 100 each were transmitted in turn to the HCD cell and the collision energy was scaled according to precursor m/z windows (16–25% for precursor ions m/z 300–1000). An injection time of 50 ms for a target value of 106 counts was used for the HCD MS/MS acquisition. This experiment enabled the generation of an inclusion list of potential SUMO peptides for subsequent targeted MS/MS experiments.
Data Processing and Peptide Identification
MS data were acquired using the Xcalibur software (version 2.1). Peak lists were generated using Mascot distiller (version 2.3.2.0, Matrix science) and MS/MS spectra were searched against a concatenated target/decoy UniProtKB/Swiss-Prot Human containing 37275 forward sequences (released Feb 2013) using Mascot (version 2.3.2, Matrix Science) to achieve a false-positive rate of less than 2% (p < 0.02). MS/MS spectra were searched with a mass tolerance of 7 ppm, for precursor ions and 0.5 Da for fragment ions acquired in CID and ETD modes or 0.02 Da for HCD spectra. The number of allowed missed cleavage sites for trypsin was set to 2 and phosphorylation (STY), oxidation (M), deamidation (NQ), carbamidomethylation (C), ethylmaleimidation (C), ubiquitylation (K), acetylation (K, NH2-term), and SUMOylation (K) (EQTGG: SUMO1 or NQTGG: SUMO3) were selected as variable modifications. A software application was developed to search Mascot generic files (mgf) for specific SUMO fragment ions (e.g. SUMO1: m/z 102.0550, 129.0659, 240.0979, 258.1084, 341.1456, 359.1561; SUMO3: m/z 132.0768, 226.0822, 243.1088, 299.1350, 316.1615, 326.1459, 327.1299, 344.1565, 383.1674, 401.1779; and neutral losses of SUMO remnants) to produce a subset mgf file containing only MS/MS spectra of potential SUMOylated peptide candidates. SUMO fragment ions were removed from the corresponding mgf files and searched using Mascot as indicated above. Manual inspection of all MS/MS spectra for modified peptides was performed to validate assignments.
For the generation of inclusion lists of potential SUMO peptides, raw LC-MS files that comprised MS and HCD MS/MS data sets were converted into peptide maps using in-house peptide detection and clustering software (22). The HCD MS/MS data sets were searched for specific SUMO fragment ions (e.g. 132.0768, 226.0822, 243.1088 for SUMO3) to identify relevant scans containing SUMO peptide candidates. Multiply charged precursor ions with elution profile overlapping with SUMO fragment ions (± 2 scans) were selected in neighboring MS scans. Only precursor ions with corresponding neutral losses (e.g. 242.1015, 343.1492 for SUMO3, tolerance 10 ppm) identified in the HCD MS/MS scan were retained. Data clustering was then performed to remove redundancy and merge potential precursors from the same peptide. Inclusion lists were created with peptide candidates identified with at least two fragment ions and two neutral losses, and detected in LC-MS/MS runs above an intensity threshold of 5000 counts (supplemental Fig. S1). This script is available at http://www.thibault.iric.ca/ADIA.
RESULTS
Product Ion Spectra of SUMO Mutant Peptides Revealed Paralog-specific Fragment Ions
The identification of SUMOylated peptides by conventional database search engines can be challenging in view of the occurrence of fragment ions from the peptide backbone and the long SUMO remnant chain (up to 32 amino acid long for SUMO2 and SUMO3) that complicates the interpretation of the corresponding MS/MS spectra (16, 17). To facilitate the identification of SUMOylated peptides from digests of cell extracts, we previously generated HEK293 cell lines stably expressing His6-SUMO mutants that comprised an arginine residue strategically located near the C terminus thus leaving a short five amino acid long SUMO remnant on tryptic digestion (18). Although the corresponding SUMOylated peptides were identified using conventional database search engines, we also noted the presence of fragment ions arising from the cleavage of the SUMO remnant chain that could affect the peptide score assignment. To rationalize the fragmentation of these branched peptides under different activation modes, we performed MS analyses on both synthetic peptides containing a SUMO remnant chain and tryptic digests originating from a HEK293 stably expressing SUMO mutant. Synthetic peptides containing either a SUMO1 (EQTGG) or SUMO3 (NQTGG) remnant chain were synthesized on solid phase support and were selected based on sequences predicted from the tryptic digestion of known SUMOylated proteins. Tryptic peptides with SUMO2 (QQTGG) remnant chains were found to fragment in a similar fashion to that of SUMO3 (18), and were not analyzed in the present study. For experiments performed on HEK293 SUMO mutant cells, SUMOylated proteins are first enriched on Ni-NTA columns under denaturing conditions and the corresponding protein extracts are subsequently digested with trypsin before the identification of modified peptides using MS/MS. The overall strategy used for the identification of SUMOylated peptides is presented in Fig. 1.
Fig. 1.
Schematic overview of the work flow used for the analysis of protein SUMOylation using DDA and DIA methods.
Each library of branched tryptic-like peptides contained 96 synthetic peptides from different SUMO substrates, and comprised a modified Lys residue located within a backbone sequence extending up to 20 amino acids in length. Mixtures of SUMO1 or SUMO3 synthetic peptides present at concentrations of 20–50 nm each were analyzed in triplicate by LC-MS/MS on a LTQ-Orbitrap Elite using ETD, CID, and HCD activation modes (Fig. 1). MS/MS spectra were collected in DDA in which up to 12 most abundant multiply charged ions from the survey scan were subjected to ion activation in the ion trap or the HCD collision cell. MS/MS spectra were searched against the UniProtKB/Swiss-Prot human protein database using Mascot (see experimental section). In total, we identified 89 SUMO1 and 93 SUMO3 peptides from the expected synthetic peptide libraries. Manual search of raw LC-MS/MS data indicated that the remaining undetected peptides were either not synthesized efficiently or showed truncated backbone sequences.
A comparison of the number of identified peptides showed that more than 90% of peptides were correctly assigned in all three activation modes though different Mascot scores were obtained (Fig. 2A, supplemental Table S1). Interestingly, more than 90% of all SUMO1 and SUMO3 peptides were identified with higher Mascot scores using CID or HCD activation (Fig. 2B). MS/MS spectra acquired using HCD also showed a larger proportion of fragment ions arising from cleavages of the peptide backbone and the SUMO remnant side chain compared with CID and ETD spectra acquired in the ion trap (supplemental Fig. S2). Also, low mass fragment ions below 1/3 of the precursor ion m/z values are not transmitted for CID and ETD activation modes. This is evidenced in Fig. 3 for a representative example of SUMO1 and SUMO3 synthetic peptides identified in all three activation modes. This peptide sequence corresponds to the heterogeneous nuclear ribonucleoprotein (hnRNP) M with a modified Lys 381 residue. The ETD MS/MS spectra of [M+3H]3+ ions showed prominent c- and z-type fragment ions with limited cleavages of side chain residues (c* and neutral losses, Figs. 3A and 3B). Similarly, the low resolution CID MS/MS spectra of peptide ions displayed a regular distribution of b- and y-type fragment ions with low abundance neutral losses of side chain residues (Figs. 3c and 3d). The successive losses of SUMO remnant residues, and the occurrence of paralog-specific fragment ions (e.g. b2*, b2*-H2O, b3*, b3*-H2O) were more clearly apparent when using HCD fragmentation, presumably because of the higher collision regime and the transmission of low mass ions in the HCD quadrupole cell (Figs. 3A and 3B). Also, HCD spectra acquired in the high resolution Orbitrap mass analyzer enabled the unambiguous assignment of fragment ions via accurate mass measurements.
Fig. 2.
LC-MS/MS analyses of synthetic peptides with SUMO1 and SUMO3 remnant chains using ETD, CID, and HCD activation modes. A, Venn diagram showing the overlap in peptide identification using different activation modes. B, Distribution of Mascot scores for MS/MS of SUMO1 and SUMO3 peptides acquired using all three activation modes.
Fig. 3.
MS/MS spectra of INEILSNALKR with a Lys residue modified with SUMO1 or SUMO3 remnant chains. Product ion spectrum for [M+3H]3+ (m/z 581.651 for SUMO1, m/z 576.652 for SUMO3) acquired with ETD (A, B), CID (C, D) and HCD (E, F). SUMO-specific fragment ions (e.g. c1*, b2*, b2*-H2O, b3*, b3*-H2O) and neutral losses are emphasized in black while other fragment arising from the peptide backbone are shown in gray.
Closer inspection of peptide identification indicated that Mascot scores assigned to [M+2H]2+ precursor ions were generally higher than those observed for other charge states despite the fact that [M+3H]3+ ions from SUMO1 and SUMO3 peptides were clearly more abundant in the corresponding MS spectrum. The propensity to form abundant triply protonated precursor ions was partly attributed to the free amino group on the remnant SUMO chain. The higher difficulty of assigning MS/MS spectra of [M+3H]3+ precursor ions was associated with the larger number and abundance of fragment ions arising from the cleavage of the SUMO remnant chain. This was particularly true for [M+3H]3+ precursor ions with modified lysine residues located closer to the C terminus that gave rise to lower Mascot scores and abundant internal ions with a limited number of y-type fragment ions (supplemental Fig. S3). The higher proportion of nonlinear fragment ions gave rise to complex MS/MS spectra that are generally more difficult to correlate by database search engines.
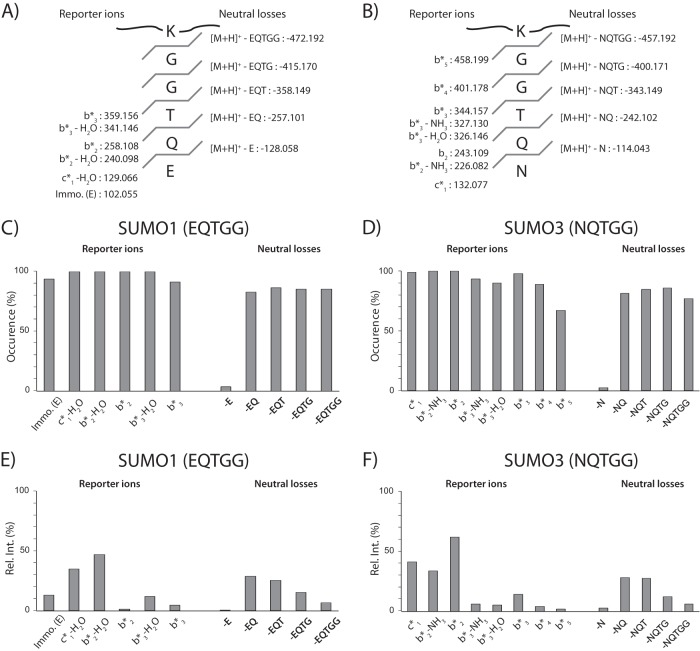
Next, we determined the occurrence and abundance of paralog-specific reporter ions observed in HCD MS/MS spectra of all synthetic peptides (Fig. 4, supplemental Table S2). Typical reporter ions and neutral losses observed for SUMO1 and SUMO3 are presented in Figs. 4A and 4B, respectively. These analyses revealed that cleavages of the last three amino acids of the SUMO remnant side chain were most frequently observed in both paralogs (Figs. 4C and 4D). In particular, dissociation of the amide bonds between Gln-Thr and Thr-Gly residues giving rise to b2* and b3* fragment ions were observed in more than 90% of peptides examined. These ions and their corresponding losses of H2O or NH3 were clearly detected in the HCD MS/MS spectra with intensities above 10% of the base peak (Figs. 4E and 4F). Also, we consistently observed the fragment ion c1* (m/z 132) for SUMO3 and c1*-H20 (m/z 129) for SUMO1 with a relative intensity above 30% of the base peak, though they are not unique to our SUMO peptides and can be found in unmodified peptides. This observation is consistent with earlier reports indicating that c1 fragment ions are prominent in low energy MS/MS spectra of peptide ions containing glutamine as the second amino acid residue from the N terminus (23). A consecutive fragmentation mechanism involving a b2 ion intermediate was proposed to explain the unusual stability of the c1 fragment ion. Interestingly, we also noted the presence of abundant neutral losses corresponding to the cleavage of the amide bond of the SUMO remnant side chain for both paralogs (Figs. 4E and 4F). Although their occurrence is less frequent than paralog-specific reporter ions, they can be readily detected in the HCD MS/MS spectra as discrete peaks of predictable spacing from the precursor ions (Figs. 3E and 3F). The fragmentation features observed for synthetic SUMO peptides in terms of specific diagnostic ions and neutral losses provided characteristic signature ions for identification and confirmatory purposes. The analytical benefits of these features are discussed in a following section.
Fig. 4.
Distribution of intensity for SUMO-specific fragment ions observed using HCD activation. Characteristic reporter ions and neutral losses observed for SUMO1 (A) and SUMO3 (B). Occurrence of SUMO-specific fragment ions and neutral losses for all synthetic peptides of (C) SUMO1 and (D) SUMO3 conjugates. The occurrence indicates the number of times that a SUMO-specific fragment ion is detected in all SUMO1 or SUMO3 conjugates. Relative intensity of SUMO-specific fragment ions observed in the HCD MS/MS of (E) SUMO1 and (F) SUMO3 conjugates.
Increased Identification of SUMO Mutant Peptides Using Conventional Database Search Engines
The formation of multiple fragment ions from branched modified peptides can produce a complex overlapping fragmentation pattern that complicates the interpretation of MS/MS spectra by conventional database search engines. Although SUMOylated peptides containing a pentapeptide remnant chain can be readily identified by Mascot, we surmised that the observation of fragment ions from the modified Lys side chain could compromise peptide identification depending on the length and composition of the peptide backbone, and the presence of other modifications. To determine if fragment ions from the SUMO remnant chain adversely affected their identification by Mascot, we compared the search output obtained from the original MS/MS spectra of synthetic peptides with those from spectra where SUMO-specific reporter ions and neutral losses were removed. This comparison was performed for MS/MS spectra obtained for all activation modes and search parameters were adjusted according to the mass analyzer accuracy (see Experimental section).
Results from Mascot searches using the original MS/MS files and those obtained after the removal of SUMO peaks showed strikingly different results depending on the activation mode used (supplemental Table S3). For ETD MS/MS spectra of both SUMO1 and SUMO3, the removal of SUMO peaks resulted in a 10% reduction of correct assignment with no additional gain in peptide identification. The analyses of CID MS/MS spectra of SUMO1 and SUMO3 peptides revealed that removal of SUMO peaks did not significantly change the outcome of the search results as more than 95% of peptide identifications were shared with those of conventional searches, whereas the average Mascot scores of peptides common to both searches remained unchanged. In contrast, the comparison of search results from HCD MS/MS spectra with and without removal of SUMO peaks indicated a large overlap of peptide identification, though an additional 3 SUMO1 peptides were correctly identified when SUMO peaks were removed. Interestingly, we noted that removal of SUMO peaks led to a 10–30% increase of Mascot scores compared with conventional searches (Figs. 5A and 5B).
Fig. 5.
Enhanced identification and improvement of Mascot scores following the removal of SUMO-specific ions in the HCD MS/MS spectra. Distribution of Mascot scores changes with and without peak removal for (A) SUMO1 and (B) SUMO3 conjugates. MS/MS spectra of [M+2H]2+ at m/z 909.94 (C) without and (D) with peak removal. MS/MS spectra of [M+3H]3+ at m/z 610.33 (E) without and (F) with peak removal. The sequence of the top ranking candidate is shown along with the corresponding Mascot score.
A representative example of a SUMO3 synthetic peptide identified using HCD activation mode is presented in Fig. 5c. This peptide sequence corresponds to that of the peptidyl-prolyl cis-trans isomerase-like 4 with a modified Lys 212 residue. Abundant SUMO3 reporter ions at m/z 132, 226 and 243 along with neutral losses at m/z 1418, 1476 and 1577 were detected in the HCD MS/MS spectrum. Mascot identified the correct peptide sequence and the modified residue with a score of 28. We obtained the same identification with a score of 48 when the search was repeated with the edited MS/MS spectrum where SUMO3 peaks were removed (Fig. 5D). The increase in Mascot score observed here is partly attributed to a shift in weighted average score of sequence-specific fragment ions. We also noted that original MS/MS spectra of SUMOylated peptides could remain unassigned because of the occurrence of internal fragment ions from the cleavage of the SUMO remnant side chain residues. This is illustrated in Fig. 5E for the synthetic peptide GTAGLLEQWLK*R, corresponding to C3orf37 with a modified Lys 339 residue. This peptide was originally unassigned, but was correctly identified with a Mascot score of 7 when the search was repeated following the removal of SUMO3 peaks (Fig. 5F). The observation of internal fragment ions from the SUMO remnant chain confirmed the sequence assignment.
Time and Mass Correlation of Precursor and Fragment Ions Enabled the Identification SUMOylated Peptides from Data Independent Acquisition Experiments
The identification of SUMOylated peptides by LC-MS/MS represents a sizable analytical challenge in view of their relatively low abundance and their small proportion in complex protein digests (typically < 1% of all identified peptides). These considerations, in addition to those associated with the mass spectrometer duty cycle, place significant limitations on the ability to comprehensively identify SUMOylated peptides using DDA. In this context, alternative identification strategies that rely on neither detection nor knowledge of precursor ions to acquire fragment ion spectra, also referred to data independent acquisition (DIA), can be advantageous. In DIA, consecutive survey scans and fragment ion spectra for all the precursors are collected in separate isolation windows of different widths (20, 24–27). Although most applications of DIA were presented using quadrupole time-of-flight instruments, the implementation of this method was recently shown for the Q-Exactive (24) and the LTQ-Orbitrap (28).
In the present study, we investigated the capability of the LTQ-Orbitrap Elite to cycle through the repeated acquisition of survey scans and fragment ion spectra of precursor ions from preselected m/z windows. We first optimized precursor windows and accumulation time in the ion trap to avoid space-charge issues that could result in overfilling the Orbitrap analyzer with fragment ions generated in the HCD cell. LC-MS/MS experiments were performed using a set of six synthetic SUMO3 peptides spiked at 100 fmoles each in a tryptic digest of HEK293 cell extracts. These peptides were previously reported to be SUMOylated on PML (K380, K400, and K490), RanGAP (K524), SAFB2 (K525), and E2–25K (K14) (18). The distribution of relative intensity of SUMO-specific fragment ions for different precursor windows (m/z 300–1000) and accumulation periods (5–100 ms) is presented in supplemental Fig. S4. These experiments indicated that optimal fragment ion transmission and signal to noise ratio were obtained using an accumulation time of 50 ms and 7 segments of 100 m/z units which corresponded to an overall duty cycle of 6.6 s. Additional LC-MS/MS experiments were performed using a library of 96 synthetic SUMO3 peptides to select optimal HCD fragmentation conditions for the generation of neutral loss fragment ions and a collision energy scaled according to precursor m/z windows was varied from 16–25% for all LC-MS/MS experiments (supplemental Fig. S5).
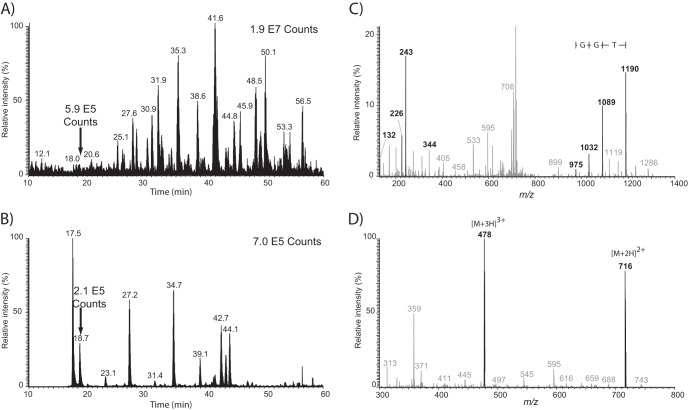
An example of a LC-MS/MS experiment performed under optimized conditions is shown in Fig. 6 for the analysis of 250 ng of a HEK293 tryptic digest spiked with six synthetic SUMO3 peptides (100 fmoles each). The base peak chromatogram (BPC) presented in Fig. 6A shows a complex pattern of peaks. However, the presence of SUMO3 peptides was clearly observed from the extracted ion chromatogram (XIC) of the characteristic fragment ion b2* at m/z 243.1088 for peaks eluting at 17.5, 18.7, 27.2, 34.6, 41.7, and 42.2 min (Fig. 6B). Other peaks identified in Fig. 6B corresponded to tryptic peptides from HEK293 cells with a N terminus NQ undistinguishable from the characteristic b2* SUMO3 fragment ion. For example, the peak at 42.7 min was subsequently identified as the tryptic peptide NQVAMNPTNTVFDAK from the Isoform 1 of Heat shock cognate 71 kDa protein. Obviously, the co-elution of additional SUMO3-specific fragment ions can be used to rule out potential interfering peaks. Interestingly, we noted that the human proteome contains 3334 putative tryptic peptides of mass greater than 600 Da with a N terminus NQ, 206 with NQT, 5 and 4 with NQTG and NQTN and 0 with NQTGG. The relative proportion of peptides beginning with NQ and NQT compared with the in silico digestion of the human proteome (1630481 theoretical tryptic peptides) corresponds to 0.01 and 0.2%, and provided a realistic estimate of false positive identification expected from HEK293 cell extracts. However, parallel precursor fragmentation can generate fragment ions that could be falsely assigned as neutral losses from SUMO peptides, thereby contributing to additional non-SUMO identification.
Fig. 6.
LC-MS/MS analysis (DIA) of SUMOylated peptides spiked in a tryptic digest of HEK293 cells. A, BPC, B, XIC for m/z 243.109, C, extracted fragment ion spectrum (precursor isolation window of m/z 750 ± 50) and D, survey scan for peak shown at 18.7 min. Peptide standard eluting at 18.7 min correspond to the synthetic SAFB2 peptide TVIKK(NQTGG)EEK. Peptide standards were spiked at a level of 100 fmols each in 250 ng of tryptic digest.
Data obtained from fragment ions of large precursor windows enabled the detection of additional peaks that could be used to identify potential SUMO3 remnant tryptic peptides. This is illustrated in Fig. 6C for fragment ions generated from the selected precursor window of m/z 750 ± 50 of the peak eluting at 18.7 min in Fig. 6B. Characteristic SUMO3-specific fragment ions (e.g. c1*: 132.078, b2*-NH3: 226.083, b2*: 243.109, b3*: 344.157) were within 5 ppm of the expected mass and confirmed the identification of the synthetic SAFB2 peptide TVIKK(NQTGG)EEK. Importantly, neutral losses of SUMO3 side chain ([M+H] + - NQ: 1189.679, [M+H] + - NQT: 1088.631, [M+H] + - NQTG: 1031.610, [M+H] + - NQTGG: 974.588) were clearly observed in Fig. 6C and enabled the calculation of the tryptic peptide mass at 1430.772 Da. The multiply-charged ions of the corresponding peptide were clearly evidenced in the survey scan (Fig. 6D) at m/z 477.931 ([M+3H]3+) and m/z 716.394 ([M+2H]2+).
The observation of SUMO3-specific fragment ions generated from large precursor windows and the possibility of determining the mass of potential SUMO tryptic peptides for correlation in the survey scan, prompted us to develop a script to compute the mass of putative SUMO3 peptides for targeted MS/MS identification using inclusion lists (see experimental section). Next, we compared the analytical merits of the DIA method with that of the conventional DDA for the identification of SUMO3 peptides from a NTA-enriched nuclear extract from HEK293-SUMO3 cells (Fig. 1). In each case, triplicate injections of 2 μg (5 × 106 cells) of tryptic digest were analyzed by on-line 2D-LC-MS/MS on a LTQ-Orbitrap Elite using HCD fragmentation.
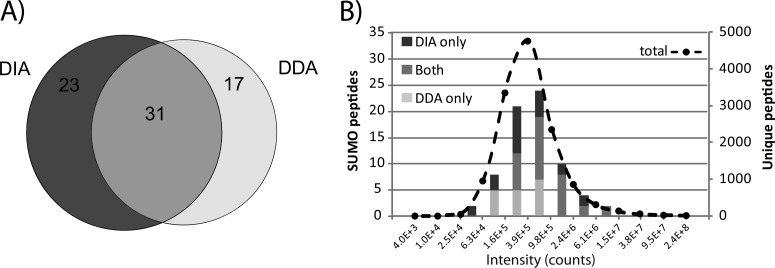
We identified a total of 12626 unique peptides corresponding to 2099 proteins (FDR: 1%) in all DDA experiments. Subsequent data analysis performed after filtering out SUMO fragment ions from the original MS/MS spectra led to the assignment of 115 potential candidates, of which 48 SUMO3 peptides were confirmed by manual validation. In comparison, the targeted approach enabled the identification of a total of 7741 unique peptides corresponding to 1849 proteins (FDR: 1%). We identified 125 potential SUMO3 peptides from Mascot searches performed on MS/MS spectra where SUMO3 fragment ions were removed. Manual validation of these assignments confirmed the identification of 54 SUMO3 unique peptides, of which 23 were not identified by DDA (Fig. 7A). A total of 31 SUMO3 peptides were identified in both DDA and DIA whereas 17 SUMO3 peptides were unique to the DDA approach and corresponded to large tryptic peptides that gave rise to neutral losses of low abundance or fragment ions outside of the scanning range. The list of identification is provided in supplemental Table S4 and MS/MS spectra of modified peptides are presented in supplemental Fig. S6. It is noteworthy that editing MS/MS spectra to remove SUMO fragment ions provided a 30% increase in Mascot score with a 15% gain in new identification compared with the traditional search, consistent with that observed previously for synthetic peptides (supplemental Table S5). These results suggest that DIA provided complementary identification to those of obtained by DDA, and that both methods enabled the identification of low abundance SUMO peptides (Fig. 7B).
Fig. 7.
Identification of SUMO3peptides from 2D-LC-MS/MS analyses of a tryptic digest of HEK293-SUMO3 cells using DDA and DIA methods. A, Venn diagram showing the overlap in peptide identification using both methods. B, Distribution of intensities for SUMO peptides and all other tryptic peptides identified using both methods.
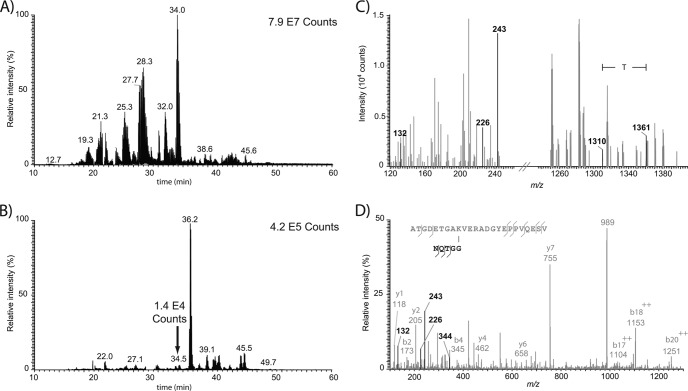
Fig. 8 shows an example of a SUMO3 peptide uniquely identified in the DIA method. The 2D-LC-MS/MS analysis of the tryptic digest from a NTA-purified protein extract of HEK293-SUMO3 peptide showed discrete peaks for the XIC at m/z 243.109 (Fig. 8B) that could not be readily identified in the BPC of the corresponding analysis (Fig. 8A). The fragment ion spectrum from the peak eluting at 34.5 min in Fig. 8B showed characteristic SUMO3 neutral losses from which the mass was calculated to be 2961.36 Da (Fig. 8C). This candidate of low abundance was identified in a subsequent targeted 2D-LC-MS/MS analysis as a tryptic peptide from the 40S ribosomal protein S3a with a K249 SUMOylated residue, a site previously reported to be acetylated (29) (Fig. 8D).
Fig. 8.
2D-LC-MS/MS analysis of tryptic digest of HEK293-SUMO3 cells (2 μg inj.) using DIA and targeted identification with an inclusion list. A, BPC, B, XIC for m/z 243.109, C, extracted fragment ion spectrum (precursor isolation window of m/z 950 ± 50) from which the mass of the SUMO3 peptide candidate was calculated to be 2961.36 Da based on the observation of the [M+3H]3+ ion at m/z 988.1254 for the peak eluting at 34.5 min. D, MS/MS spectrum (30 V collision energy) from the targeted analysis of [M+3H]3+ at m/z 988.1234 confirming the sequence of the SUMO3 peptide as ATGDETGAK(NQTGG)VERADGYEPPVQESV corresponding to 40S ribosomal protein S3a with a modified K249 residue.
Altogether, these analyses enabled the identification of 71 unique SUMO3 peptides corresponding to 44 different SUMOylation sites on 32 protein substrates (Table I). Approximately half of the sites identified are novel, and are mostly represented by the consensus ψKxE/D SUMO motif. However, we noted that 37% of SUMO sites harbored nonconsensus sites including an inverted SUMO motif E/DxK previously reported by Matic et al. (19). (supplemental Fig. S7). We also identified branched SUMO3 or SUMO2 chains at residues K7, K11, and K41, confirming the presence of polySUMOylation on protein substrates. Only residue K11 bear a SUMO motif in the unstructured N-terminal region, although SUMOylation at other nonconsensus sites were described earlier by different groups (16, 18, 30, 31). Interestingly, we observed the SUMOylation of ubiquitin at K63 and the ubiquitylation of SUMO3 at K20 and K32. These results suggest that protein substrates could be modified by mixed SUMO-ubiquitin chains highlighting a complex crosstalk between these two modifications. Further confirmation of the SUMOylated ubiquitin at K63 residue was obtained using a synthetic peptide (supplemental Fig. S8).
Table I. Examples of identified SUMOylated peptides from LC-MS/MS analyses of HEK293 His6-SUMO3 mutant cells.
| Gene name | Proteins | Site | Knowna | DDA | DIA | Function |
|---|---|---|---|---|---|---|
| BCLAF1 | Bcl-2-associated transcription factor 1 | 831 | S | X | Death-promoting transcriptional repressor | |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | 17 | S | X | Pre-mRNA binding protein in vivo, binds avidly to poly(G) and poly(U) RNA homopolymers in vitro | |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | 145 | S, U | X | X | |
| HNRNPM | Heterogeneous nuclear ribonucleoprotein M | 667 | U, A | X | ||
| NPM1 | Nucleophosmin | 215 | A | X | X | Ribosome biogenesis, centrosome duplication, protein chaperoning, histone assembly, cell proliferation, and regulation of tumor suppressors p53/TP53 and ARF |
| NPM1 | Nucleophosmin | 248 | U | X | X | |
| PARP1 | Poly [ADP-ribose] polymerase 1 | 203 | S | X | Involved in the base excision repair (BER) pathway | |
| RPL3 | 60S ribosomal protein L3 | 399 | X | Constituent of ribosome | ||
| RPS5 | 40S ribosomal protein S5 | 18 | U | X | Constituent of ribosome | |
| RPS6 | 40S ribosomal protein S6 | 243 | X | Constituent of ribosome | ||
| RSL1D1 | Ribosomal L1 domain-containing protein 1 | 435 | X | Constituent of ribosome | ||
| NHP2 | H/ACA ribonucleoprotein complex subunit 2 | 5 | S | X | Constituent of ribosome | |
| SAFB2 | Scaffold attachment factor B2 | 293 | S, U, A | X | X | Estrogen receptor corepressor and can inhibit cell proliferation |
| SAFB2 | Scaffold attachment factor B2 | 517 | S | X | X | |
| SAFB2 | Scaffold attachment factor B2 | 524 | S | X | X | |
| UBE2I | SUMO-conjugating enzyme UBC9 | 49 | S, U | X | E2 enzyme of the SUMOylation cascade | |
| UTP14A | U3 small nucleolar RNA-assoc. protein 14 | 733 | A | X | Biogenesis of ribosomes |
a S, U and A refer to known SUMOylated, ubiquitylated and acetylated sites, respectively.
These analyses confirmed previously identified substrates (18) including isoform 1 of transcription intermediary factor 1-beta, TIF-1β (K750 and K779), nucleolar protein 5 (K467) and Ran GTPase-activating protein 1 (K524) using both DDA and DIA. SUMOylated proteins uniquely identified by DIA included H/ACA ribonucleoprotein complex subunit 2 (K5), Bcl-2-associated transcription factor 1 (K831), Poly [ADP-ribose] polymerase 1 (K203), probable U3 small nucleolar RNA-associated protein 11 (K235), Ubiquitin (K63), TSPYL protein (K156), and transcriptional repressor protein YY1 (K203). Several of these sites were previously reported to be acetylated and/or ubiquitylated (Table I). Bioinformatics analyses of our data sets using Ingenuity Pathway Analysis (supplemental Fig. S9) revealed that a number of identified SUMOylated proteins were enriched in GO terms associated with RNA processing, p value: 5.50E-07 (NOP58,NPM1,RPL26,RPS6) or double-strand DNA break repair, p value: 4.01E-04 (NPM1, PARP1, SUMO1, TRIM28). These analyses confirmed the involvement of protein SUMOylation in ribosomal biogenesis and in the regulation of DNA repair (1, 4, 5).
DISCUSSION
Detailed analyses of MS/MS spectra from synthetic peptides with SUMO1 and SUMO3 remnant chains using CID, HCD and ETD activation modes revealed important structural features specific to each paralog. While most synthetic peptides were identified using each activation mode, HCD fragmentation generally provided more confident identification with higher Mascot scores and accurate mass measurements. Notably, HCD fragmentation favored the observation of paralog-specific reporter ions corresponding to the SUMO remnant side chain (e.g. c1*, b2*, b2*-H2O, b3*, b3*-H2O) together with neutral losses that can be used for confirmation purposes. These characteristic structural features facilitated the identification of low abundance SUMOylated peptides from tryptic digests of cell extracts that typically represent less than 0.3% of all identified peptides.
However, branched peptides typically yielded additional side chain fragment ions that further complicated the interpretation of MS/MS spectra and their assignments using conventional database search engines. The presence of fragment ions from the SUMO remnant and peptide backbone can adversely affect the database search outcome by reducing the corresponding Mascot scores and/or giving rise to false identification. Interestingly, we found that the location of the crosslink site and the charge state of the precursor ions can influence significantly the database search outcome. In contrast to doubly protonated precursor ions, triply protonated SUMO peptide ions typically generate a higher number of internal fragment ions arising from the cleavage of peptide bond from the SUMO remnant chain. The propensity to generate abundant internal fragment ions was correlated with the location of the modified lysine residue where intense ions were observed for sites closer to the C terminus. This observation has important implications for the development of database search engines tailored to the analysis of crosslinked peptides.
In this context, the development of a script that removed SUMO-specific fragment ions from the original MS/MS spectra before database search provided important advantages to improve both identification rates and Mascot scores. When compared with database searches using the original data sets, we obtained a 30% increase in Mascot scores with a 15% gain in new identifications for both synthetic peptides and tryptic digests from HEK293-SUMO3 cells. This approach could also be beneficial for the analysis of MS/MS spectra from other branched peptides or protein modifications to improve the identification of target peptide sequences using conventional search engines.
Peptide sequencing using DDA becomes progressively more challenging with decreasing ion abundance and increasing occurrence of mixed MS/MS spectra from the coselection of neighboring precursor ions. This is particularly true for trace-level SUMOylated peptides that represent a very small proportion of the entire pool of cell digests. These limitations were alleviated using a DIA identification strategy based on the detection of SUMO-specific reporter ions and neutral losses that could be correlated to potential candidates. The detection of the appropriate neutral losses from high resolution MS/MS spectra containing these reporter ions enabled the calculation and thus identification of the correct precursor masses. Confirmation of SUMOylated peptides was achieved in subsequent targeted LC-MS/MS experiments using inclusion lists of potential candidates. The use of this DIA method provided a 30% gain in new assignments compared with DDA alone, resulting in the identification of 71 SUMOylated peptides using both methods. A main limitation of the current approach is the requirement for conducting two LC-MS/MS analyses on the same sample to determine SUMO candidates first by DIA and then for targeted identification with an inclusion list. However, we anticipate that future improvements in MS duty cycle and on-board computing analysis will enable the detection of SUMO candidates and their identification in a single LC-MS/MS run.
The present study enabled the identification of 71 SUMOylated peptides with 44 distinct SUMOylation sites on 32 protein substrates. This was achieved using only 20 μg of NTA-enriched protein extracts (5 × 107 HEK293-SUMO3 cells) where SUMOylated peptides represent a very small subset of all identified peptides. Although these results are relatively modest in comparison to the number of known SUMOylated substrates, they highlight important biological insights on protein SUMOylation. In addition to the observation of polySUMO chains (K7, K11, K41), our study provided the first direct evidence of the formation of mixed SUMO-ubiquitin chains on protein substrates where reciprocal modifications on branched UBLs were observed. Indeed, we identified SUMOylation of ubiquitin (K63) and the ubiquitylation of SUMO3 (K20 and K32), suggesting that significant crosstalk exists between these modifications. Although SUMOylation of ubiquitin at K63 was observed, we cannot rule out the possibility that ubiquitin is an innocent bystander present at the site of physiologically relevant SUMOylation reactions. However, previous studies highlighted that conjugates of SUMO2 and ubiquitin copurified together, and that proteasomal inhibition led to the accumulation of SUMO2 and/or SUMO3 conjugates (14). Several proteins were found to be SUMOylated on multiple sites or formed polySUMO chains before subsequent polyubiquitylation and targeted proteasomal degradation, although no clear evidence was shown to confirm that SUMO and ubiquitin were linked to one another in the same chain. The modification of substrates by heterologous modification was previously shown for the promyelocytic leukemia (PML) protein whereby SUMOylation led to the recruitment of the ubiquitin ligase really interesting new gene finger protein 4 (RNF4) resulting in its polyubiquitylation and degradation by the proteasome (32, 33). The biological significance of mixed SUMO-ubiquitin chains is still unknown, and further studies are required to determine if SUMOylation directly affects proteasomal recognition or is necessary for the recruitment of polyubiquitin machinery, and whether substrates modified by mixed chains can be degraded.
As part of this study we also uncovered new SUMOylated substrates including several ribosomal proteins (e.g. RPL26, RPL3, RPS3A, RPS3A, RPS5, RPS6), and proteins involved in ribosome biogenesis (e.g.NPM1, NOP58), suggesting a potential role for protein SUMOylation in the regulation of protein synthesis. Several proteins involved in chromatin structure (e.g. H3, H4) or DNA double-strand DNA break repair (NPM1, PARP1) were also found to be SUMOylated, and support previous reports describing the significance of protein SUMOylation in DNA damage response (34, 35). We also noted that several SUMOylation sites were previously reported to be modified by other PTMs where the target Lys residue can be acetylated or ubiquitylated (Table I). In the case of RanGAP1, a GTPase activator for the nuclear Ras-related regulatory protein Ran, its SUMOylation (K524) stabilizes the interaction with RANBB2/NUP58 and favors its translocation to the nuclear pore complex (36). The interplay between protein modifications provides another layer of regulation that affects protein functions and protein interactions. MS-based proteomics approaches such those presented in this report are poised to play an increasing role in unveiling the complex regulation of protein SUMOylation and its crosstalk with other protein modifications.
Supplementary Material
Acknowledgments
We thank Alain Verreault, Antoine Zieger (Université de Montréal) and Mounira Chelbi-Alix (Université Paris-Descartes) for technical assistance and valuable suggestions during the course of this study.
Footnotes
* This work was supported by the Natural Sciences and Engineering Research Council (NSERC). FL was supported by a scholarship from the faculty of graduate studies of the Université de Montreal. The Institute for Research in Immunology and Cancer (IRIC) receives infrastructure support from the Canadian Center of Excellence in Commercialization and Research, the Canadian Foundation for Innovation, and the Fonds de recherche du Québec - Santé (FRQS).
 This article contains supplemental Figs. S1 to S9 and Tables S1 to S5.
This article contains supplemental Figs. S1 to S9 and Tables S1 to S5.
1 The abbreviations used are::
- SUMO
- Small Ubiquitin-related Modifier
- BPC
- base peak chromatogram
- CID
- collision induced dissociation
- DDA
- data dependent acquisition
- DIA
- data independent acquisition
- E2–25K
- E2 ubiquitin ligase – 25 kiloDalton
- ETD
- electron transfer dissociation
- FDR
- false discovery rate
- HCD
- higher energy collisional dissociation
- HEK
- human embryonary kidney
- hnRNP
- Heterogeneous nuclear ribonucleoprotein
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- MS/MS
- tandem mass spectrometry
- NEM
- N-Ethylmaleimide
- NTA
- Nickel nitriloacetic acid
- PIAS
- protein inhibitor of activated STAT
- RanBP2
- Ran Binding Protein 2
- RanGAP1
- Ran GTPase activating Protein 1
- RNF4
- RING finger 4 protein
- SAE
- SUMO activating Enzyme
- SAFB2
- Scaffold Assembly factor B2
- SENP
- Sentrin/SUMO-specific protease
- TBS
- Tris buffered saline
- TIF1β
- (TRIM28) Transcription intermediary factor 1-beta (TRIM28)
- Ubc9
- E2 SUMO conjugating enzyme
- UBLs
- Ubiquitin-like proteins
- XIC
- Extracted ion chromatogram.
REFERENCES
- 1. Hay R. T. (2005) SUMO: a history of modification. Mol Cell 18, 1–12 [DOI] [PubMed] [Google Scholar]
- 2. Lois L. M. (2010) Diversity of the SUMOylation machinery in plants. Biochem. Soc Trans 38, 60–64 [DOI] [PubMed] [Google Scholar]
- 3. Guo D., Li M., Zhang Y., Yang P., Eckenrode S., Hopkins D., Zheng W., Purohit S., Podolsky R. H., Muir A., Wang J., Dong Z., Brusko T., Atkinson M., Pozzilli P., Zeidler A., Raffel L. J., Jacob C. O., Park Y., Serrano-Rios M., Larrad M. T., Zhang Z., Garchon H. J., Bach J. F., Rotter J. I., She J. X., Wang C. Y. (2004) A functional variant of SUMO4, a new I kappa B alpha modifier, is associated with type 1 diabetes. Nat. Genet. 36, 837–841 [DOI] [PubMed] [Google Scholar]
- 4. Bossis G., Melchior F. (2006) SUMO: regulating the regulator. Cell Div 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Meulmeester E., Melchior F. (2008) Cell biology: SUMO. Nature 452, 709–711 [DOI] [PubMed] [Google Scholar]
- 6. Seeler J. S., Dejean A. (2003) Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4, 690–699 [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez M. S., Dargemont C., Hay R. T. (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 276, 12654–12659 [DOI] [PubMed] [Google Scholar]
- 8. Hietakangas V., Anckar J., Blomster H. A., Fujimoto M., Palvimo J. J., Nakai A., Sistonen L. (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. U.S.A. 103, 45–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang S. H., Galanis A., Witty J., Sharrocks A. D. (2006) An extended consensus motif enhances the specificity of substrate modification by SUMO. EMBO J. 25, 5083–5093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 [DOI] [PubMed] [Google Scholar]
- 11. Melchior F., Schergaut M., Pichler A. (2003) SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci 28, 612–618 [DOI] [PubMed] [Google Scholar]
- 12. Mukhopadhyay D., Dasso M. (2007) Modification in reverse: the SUMO proteases. Trends Biochem. Sci 32, 286–295 [DOI] [PubMed] [Google Scholar]
- 13. Golebiowski F., Matic I., Tatham M. H., Cole C., Yin Y., Nakamura A., Cox J., Barton G. J., Mann M., Hay R. T. (2009) System-wide changes to SUMO modifications in response to heat shock. Sci Signal 2, ra24. [DOI] [PubMed] [Google Scholar]
- 14. Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., Mann M., Andersen J. S., Vertegaal A. C. (2008) The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol Cell Proteomics 7, 2107–2122 [DOI] [PubMed] [Google Scholar]
- 15. Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., Lamond A. I. (2006) Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol Cell Proteomics 5, 2298–2310 [DOI] [PubMed] [Google Scholar]
- 16. Pedrioli P. G., Raught B., Zhang X. D., Rogers R., Aitchison J., Matunis M., Aebersold R. (2006) Automated identification of SUMOylation sites using mass spectrometry and SUMmOn pattern recognition software. Nat Methods 3, 533–539 [DOI] [PubMed] [Google Scholar]
- 17. Hsiao H. H., Meulmeester E., Frank B. T., Melchior F., Urlaub H. (2009) “ChopNSpice,” a mass spectrometric approach that allows identification of endogenous small ubiquitin-like modifier-conjugated peptides. Mol Cell Proteomics 8, 2664–2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Galisson F., Mahrouche L., Courcelles M., Bonneil E., Meloche S., Chelbi-Alix M. K., Thibault P. (2011) A novel proteomics approach to identify SUMOylated proteins and their modification sites in human cells. Mol Cell Proteomics 10, M110.004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Matic I., Schimmel J., Hendriks I. A., van Santen M. A., van de Rijke F., van Dam H., Gnad F., Mann M., Vertegaal A. C. (2010) Site-Specific Identification of SUMO-2 Targets in Cells Reveals an Inverted SUMOylation Motif and a Hydrophobic Cluster SUMOylation Motif. Mol. Cell 39, 641–652 [DOI] [PubMed] [Google Scholar]
- 20. Gillet L. C., Navarro P., Tate S., Röst H., Selevsek N., Reiter L., Bonner R., Aebersold R. (2012) Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank F. (1992) Spot-synthesis: an easy technique for the positionally addressable, parallel chemical synthesis on a membrane support. Tetrahedron 48, 9217–9232 [Google Scholar]
- 22. Marcantonio M., Trost M., Courcelles M., Desjardins M., Thibault P. (2008) Combined enzymatic and data mining approaches for comprehensive phosphoproteome analyses: application to cell signaling events of interferon-gamma-stimulated macrophages. Mol. Cell. Proteomics 7, 645–660 [DOI] [PubMed] [Google Scholar]
- 23. Lee Y. J., Lee Y. M. (2004) Formation of c(1) fragment ions in collision-induced dissociation of glutamine-containing peptide ions: a tip for de novo sequencing. Rapid Commun. Mass Spectrom. 18, 2069–2076 [DOI] [PubMed] [Google Scholar]
- 24. Geiger T., Cox J., Mann M. (2010) Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Purvine S., Eppel J. T., Yi E. C., Goodlett D. R. (2003) Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics 3, 847–850 [DOI] [PubMed] [Google Scholar]
- 26. Silva J. C., Denny R., Dorschel C., Gorenstein M. V., Li G. Z., Richardson K., Wall D., Geromanos S. J. (2006) Simultaneous qualitative and quantitative analysis of the Escherichia coli proteome: a sweet tale. Mol. Cell. Proteomics 5, 589–607 [DOI] [PubMed] [Google Scholar]
- 27. Venable J. D., Dong M. Q., Wohlschlegel J., Dillin A., Yates J. R. (2004) Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 [DOI] [PubMed] [Google Scholar]
- 28. Panchaud A., Scherl A., Shaffer S. A., von Haller P. D., Kulasekara H. D., Miller S. I., Goodlett D. R. (2009) Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal. Chem. 81, 6481–6488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 30. Cooper H. J., Tatham M. H., Jaffray E., Heath J. K., Lam T. T., Marshall A. G., Hay R. T. (2005) Fourier transform ion cyclotron resonance mass spectrometry for the analysis of small ubiquitin-like modifier (SUMO) modification: identification of lysines in RanBP2 and SUMO targeted for modification during the E3 autoSUMOylation reaction. Anal. Chem. 77, 6310–6319 [DOI] [PubMed] [Google Scholar]
- 31. Matic I., van Hagen M., Schimmel J., Macek B., Ogg S. C., Tatham M. H., Hay R. T., Lamond A. I., Mann M., Vertegaal A. C. (2008) In vivo identification of human small ubiquitin-like modifier polymerization sites by high accuracy mass spectrometry and an in vitro to in vivo strategy. Mol. Cell. Proteomics 7, 132–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) RNF4 is a poly-SUMO-specific E3 ubiquitin ligase required for arsenic-induced PML degradation. Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 33. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Arsenic degrades PML or PML-RARalpha through a SUMO-triggered RNF4/ubiquitin-mediated pathway. Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 34. Morris J. R. (2010) SUMO in the mammalian response to DNA damage. Biochem. Soc. Trans. 38, 92–97 [DOI] [PubMed] [Google Scholar]
- 35. Dou H., Huang C., Van Nguyen T., Lu L. S., Yeh E. T. (2011) SUMOylation and de-SUMOylation in response to DNA damage. FEBS Lett. 585, 2891–2896 [DOI] [PubMed] [Google Scholar]
- 36. Swaminathan S., Kiendl F., Körner R., Lupetti R., Hengst L., Melchior F. (2004) RanGAP1*SUMO1 is phosphorylated at the onset of mitosis and remains associated with RanBP2 upon NPC disassembly. J. Cell Biol. 164, 965–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.