Abstract
Protein–protein interactions organize the localization, clustering, signal transduction, and degradation of cellular proteins and are therefore implicated in numerous biological functions. These interactions are mediated by specialized domains able to bind to modified or unmodified peptides present in binding partners. Among the most broadly distributed protein interaction domains, PSD95-disc large-zonula occludens (PDZ) domains are usually able to bind carboxy-terminal sequences of their partners. In an effort to accelerate the discovery of PDZ domain interactions, we have constructed an array displaying 96% of the human PDZ domains that is amenable to rapid two-hybrid screens in yeast. We have demonstrated that this array can efficiently identify interactions using carboxy-terminal sequences of PDZ domain binders such as the E6 oncoviral protein and protein kinases (PDGFRβ, BRSK2, PCTK1, ACVR2B, and HER4); this has been validated via mass spectrometry analysis. Taking advantage of this array, we show that PDZ domains of Scrib and SNX27 bind to the carboxy-terminal region of the planar cell polarity receptor Vangl2. We also have demonstrated the requirement of Scrib for the promigratory function of Vangl2 and described the morphogenetic function of SNX27 in the early Xenopus embryo. The resource presented here is thus adapted for the screen of PDZ interactors and, furthermore, should facilitate the understanding of PDZ-mediated functions.
Beyond enzymatic activities, cellular functions are largely mediated and coordinated by protein–protein interactions. These interactions build genuine protein networks that contribute to the organization of subcellular compartments and allow coordinated cellular functions to occur. Thus, signaling networks employ a broad range of proteins endowed not only with enzymatic activities but also with binding capacities for other proteins or lipids. Deciphering these protein networks is a prerequisite for understanding the principles of physiological and physiopathological cellular responses, but it is a tedious task because of the numerous and specialized interactions in which each protein can be engaged. Protein interactions are usually mediated by specialized domains presenting a spatial organization that defines their binding specificities. In some cases, binding of these domains to peptide sequences can be dependent on post-translational modifications such as phosphorylation (1, 2). More than 70 protein interaction domains are currently known. Among them, SH2 and SH3 domains bind, respectively, to short peptides containing phosphorylated tyrosines and enriched in proline residues. The early identification of the precise binding specificities of these domains has greatly simplified the discovery of numerous SH2 and SH3 partners and has facilitated the study of their roles in cell signaling and related cellular functions (3). Among the protein interaction domains, PSD95-disc large-zonula occludens (PDZ)1 domains are the most widely distributed in genomes (4, 5). PDZ domains can be present in one or several copies in proteins. Some proteins contain PDZ domains only, such as MUPP1, which includes 13 PDZ domains, whereas others exhibit a PDZ domain or domains associated with other functional domains, such as the membrane-associated guanylate kinase (MAGUK) protein family, which associates PDZ, SH3, and guanylate kinase domains (4, 6). PDZ domains are found in vertebrates, Drosophila, Caenorhabditis elegans, and yeast proteomes, emphasizing the conserved functionality of these domains along evolution (6, 7). Protein interactions mediated by PDZ domains are involved in many biological processes, including clustering and targeting receptors to cellular membranes (8), cell signaling, and cell architecture (6). As a classical example, the building, maintenance, and function of all epithelial tissues relies on the organized shaping of cells, the so-called cell polarity process, which engages a large number of PDZ proteins dedicated to the compartmentalization of proteins and lipids. The importance of these highly organized PDZ networks in epithelial homeostasis is demonstrated by the strong defects observed when PDZ functions are disrupted in pathological situations such as infectious diseases or cancers (9–11). It is, for example, now well established that some PDZ proteins are targeted by certain classes of viruses that perturb their cellular functions, contributing to viral spreading and, in some cases, cellular transformation (5, 12–14).
Structurally, PDZ domains consist of 80 to 90 residues forming a packed structure of six β strands and two α helices (15, 16). The initial studies searching for PDZ domain partners revealed their affinity for carboxy-terminal peptide sequences and led to their classification in three main classes (17, 18). Class I PDZ domains were defined for their ability to bind T/S-X-Ψ (where T is threonine, S is serine, X is any amino acid, and Ψ is a hydrophobic residue) motifs. Class II and class III PDZ domains bind to ΨXΨ motifs and D/E-X-Ψ motifs (where D is aspartic acid and E is glutamic acid), respectively. Later on, many studies demonstrated that the mechanisms of PDZ interactions are more complex than initially thought, and some have questioned the early simplistic classification (19–21). Firstly, new models based on refined carboxyl terminus binding preferences classify PDZ domains in more subtle classes (5, 22). Then, adding to their versatility, recent studies have shown that PDZ domains can also bind to internal motifs in their protein partners (23–26), to other PDZ domains (27), and to lipids (28, 29).
We have decided to create a collection of human PDZ domains readily amenable to high-throughput studies of PDZ-mediated interactions. Cloned PDZ domains arranged as an array were used in a straightforward yeast two-hybrid (Y2H) screen to successfully identify known and new interactors for the E6 oncoprotein encoded by human papillomaviruses and for PDGFRβ, BRSK2, PCTK1, ACVR2B, and HER4 protein kinases. These interactions were confirmed by means of peptide pull-down coupled to mass spectrometry identification, demonstrating the strength and robustness of the Y2H approach. We also screened this Y2H array with the carboxy-terminal sequence of Vangl2, a planar cell polarity receptor, and confirmed some of the identified interactions via mass spectrometry. Finally, we reveal the novel role of one of the Vangl2 PDZ partners in cell migration in vitro and in morphogenetic movements in vivo.
EXPERIMENTAL PROCEDURES
Identification of PDZ Domain Sequences in the Human Genome
Domain boundaries were obtained by cross-searching Interpro (V18), PFAM (V23), and SMART version 5.0 (30). Each domain was extended on each side with a 10-amino-acid tail from the original protein to account for variations in border prediction and to ensure the integrity of the structure of the domain. In some cases, the sizes of these tails had to be slightly modified according to the position of the PDZ in protein (extreme end or start) or to ensure correct amplification.
PDZ Domain Cloning
Primers containing Gateway® (Invitrogen) B1 and B2 recombination tails were designed using the OSP program as described elsewhere (31), including a stop codon before the B2 tail (supplemental Table S1). DNA fragments encoding each PDZ domain were amplified via polymerase chain reaction (Platinum HIFI polymerase, Invitrogen) from human cDNA libraries and cloned into a pDONRZeo Entry vector using the Gateway® cloning system as described elsewhere (32, 33), creating a collection of PDZ Entry clones. These clones were sequence-verified using PDONRZeoF primer 5′-GCAATGTAACATCAGAGAT. A pipeline was set up to reassay unsuccessful amplifications using different combinations of cDNA libraries and to obtain wild-type clones from isolated transformed bacterial colonies.
Construction and Screening of a Comprehensive PDZ Domain Y2H Array
PDZ Entry clones were used in a Gateway® LR reaction to transfer the DNA coding for the PDZ domain into the Y2H AD expression vector pACT2. All PDZ domains were transformed into the haploid Y187 yeast strain (MATα, ura3–52, his3–200, ade2–101, leu2–3, 112, gal4Δ, met-, gal80Δ, MEL1, URA3::GAL1UAS -GAL1TATA-lacZ) using a standard procedure. Similarly, E6 and Vangl2 DNA fragments encoding the 15 last residues of the proteins were cloned into the Y2H DB expression vector pGBT9, and the resulting constructs were transformed into the haploid AH109 yeast strain (MATa, trp1–901, leu2–3, 112, ura3–52, his3–200, gal4Δ, gal80Δ, LYS2::GAL1UAS-GAL1TATA-HIS3,GAL2UAS-GAL2TATA-ADE2, URA3::MEL1UAS-MEL1TATA-lacZ, MEL1). Interactions between each PDZ and E6 or Vangl2 were tested through mating of the two yeast strains. Briefly, an overnight culture in a selective medium of the bait and the PDZs expressing yeasts (of the opposite mating type) were grown together in liquid Yeast extract-Peptone-Dextrose (YPAD) supplemented with 10% PEG for 4 h at 30 °C under gentle agitation. After one wash in water, the yeast was spotted on a solid selective medium Tryptophan-Histidine-Leucine (WHL) for phenotypic assay.
Cell Culture
T47D, HEK 293T, Caco-2, and mouse embryonic fibroblast (MEF) cells were grown in accordance with ATCC recommendations. T47D cells were transfected with pEGFP, pEGFP-Vangl2, or pEGFP-Vangl2 mutants using lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen). MEF nucleofection was done according to the manufacturer's protocol (AMAXA, Lonza, Basel/CH).
Antibodies
Goat anti-Scrib (C20) antibody was obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-GFP and rat anti-HA 3F10 were acquired from Roche. Rat anti-Vangl2 clone 2G4 was made in-house (34). Rabbit anti-βPIX antibody was obtained from Chemicon International Millipore/St. Quentin, France. Mouse anti-tubulin was obtained from Sigma. SNX27 antibody was purchased from Abcam, Cambridge, UK. The cDNA of SNX27 was kindly provided by Dr. Philippe Marin (35). Secondary antibodies coupled to horseradish peroxydase were acquired from Dako, Courtaboeuf, France.
Protein Procedure
In pull-down experiments and mass spectrometry, E6 (MSCCRSSRTRRETQL) and Vangl2 (KSHKFVMRLQSETSV) amino-terminal peptides were incubated and analyzed as described elsewhere (36). For immunoprecipitation, after preclearing with agarose beads and incubation with antibodies, protein G-agarose beads were added to the lysates, and bound immune complexes were recovered, washed three times in lysis buffer, and separated on SDS-PAGE for Western blot analysis.
Mass Spectrometry Analysis
Mass spectrometry analyses were performed using a MALDI-TOF Ultraflex instrument (Bruker Daltonics, Inc., Bremen, Germany) with reflector and positive modes, ion acceleration of 25 keV, a 5-Hz laser frequency, and a delay extraction of 110 ns. Six hundred shots were accumulated for each spectrum. Spectra were recalibrated using internal trypsin monoprotonated monoisotopic masses of 842.509, 1045.564, 2211.104, and 2283.180. Raw data were processed using Flex Analysis 3.0 and Biotool 3.1 software (Bruker Daltonics, Inc.). Protein searches were done using an in-house Mascot search engine (MatrixScience Ltd., London, UK) against the SwissProt database 2012_02 version, using “human” for taxonomy (20,247 sequences).
Mascot search results from E6 and Vangl2 pull-down on Caco-2 and/or HEK 293T cell lysates are shown in separate folders. Each folder contains the whole set of protein bands that were analyzed and identified for a peptide pull-down. For each analyzed band, a Mascot search result folder containing a protein summary report, a protein view file, and the Mascot generic file is available in E6 and Vangl2_PullDown.7z files.
Immunofluorescence Experiments
Cells grown on coverslips coated with rat-tail collagen I (Roche) were stimulated with 1 nm heregulin (HRG), fixed in 4% paraformaldehyde in PBS for 20 min, permeabilized in 0.1% Triton X-100 for 5 min, and blocked with 10% fetal calf serum in PBS for 30 min before the addition of antibodies as described elsewhere (37). Images were acquired using a Zeiss Axiovert 200mot microscope linked to a Meta LSM 510 confocal module operated by LSM-FCS software (Carl Zeiss MicroImaging Inc., Jena, Germany) using a 63× oil-immersion objective (plan achromatic lens; numerical aperture = 1.3).
Boyden Chambers Assays
Cell migration was evaluated using 8-μm pore polycarbonate membrane transwell chambers (Corning Costar, Amsterdam, NL). The bottom side of the membrane was coated with 25 μg/ml rat-tail collagen I for T47D and SUM149 cells, or 5 mg/ml human fibronectin for MEF cells. Cells were serum-starved for 16 h and then plated in the top chamber. Medium with or without 1 nm HRG was added to the bottom chamber, and cells were incubated for 12 h. Non-migrated cells were scraped from the top of the membrane. Migrated cells were fixed in 4% formaldehyde and stained with 0.1% crystal violet for counting.
Homogenous Time-resolved Fluorescence Assays
The interaction of E6, βPIX, or Vangl2 C-terminal peptides with PDZ domains was evaluated via homogenous time-resolved fluorescence assay (HTRF). To this end, we produced GST proteins fused to the indicated PDZ domains. Reaction mixtures consisted of GST-PDZ proteins at the indicated concentration in titration experiments or 2.5 × 10−9 m in IC50 experiments, an anti-GST-terbium antibody (1 × 10−9 m), streptavidin-d2 (4 × 10−8 m) (from Cisbio), a biotinylated C-terminal peptide (1.9 × 10−7 m), and the competing non-biotinylated homologous peptide at the indicated concentration in IC50 experiments. Upon excitation of the reaction mixture at 337 nm, a 615-nm fluorescence emission was produced by the donor terbium that in turn excited a 665-nm light emission by the acceptor streptavidin-d2 bound to the biotinylated peptide only if it resided in close vicinity of the donor. The long-lasting emission of the terbium cryptate allowed the fluorescence energy transfer to be measured at a time when all nonspecific light emission that followed the 337-nm illumination had faded.
After the incubation (18 h, 4 °C), the intensity of light emission (A) at 615 and 665 nm was measured in a Polarscan Omega (BMG Labtech, Champigny, France) microplate reader equipped for HTRF. For each condition, the A665/A615 R ratio was obtained and a delta F value (DF) was computed as follows: [(RSample − RNSB)/RNSB] * 100, where RNSB is the light emission produced by the reaction mixture without GST-PDZ protein. IC50 values were determined by measuring the inhibition of the DF0 obtained with the biotinylated peptide by the homologous non-biotinylated peptide (DF) in dose-response and computing the obtained DF/DF0 using Prism software (log inhibitor) versus the response-variable slope (four parameter subprogram). Values with R2 better than 0.99 were considered.
ELISA
Each of the 96 Maxisorb wells (Nunc) was sequentially coated with biotinylated BSA (200 ng), streptavidin (100 ng), biotinylated E6 (2E-7M) in PBS, and finally PBS/BSA 1% w/v. The GST-PDZ domains or GST used as a negative control at the indicated concentration in PBS/BSA were incubated on the coats overnight (4 °C). After rinsing and further incubation for 3 h on ice with peroxidase-conjugated anti-GST antibody 1/5000 (Ab 3416, Abcam), anti-GST amounts bound to the coats were evaluated after rinsing with the tetramethylbenzidine chromogenic HRP substrate. Absorbance (optical density: 450 nm) was determined on an ELISA POLARstar reader.
Xenopus Procedures
Eggs obtained from Xenopus laevis females (NASCO, Fort Atkinson, WI) were fertilized in vitro, dejellied, cultured, and injected as described elsewhere (38). The morpholino-modified antisense oligonucleotide (MO) targeting SNX27 translation initiation (5′-TCCCCCTCCTCGTCCGCCATCTTTT-3′) was purchased from GeneTools LLC (Philomath, OR), resuspended in sterile water to a concentration of 10 mg/ml, and further diluted prior to injection. The MO targeting Xenopus Vangl2 was reported in Refs. 39 and 40. The cDNA encoding Xenopus laevis SNX27 (Image clone 6945202) was purchased from GenomeCube® (Source BioScience, Nottingham, UK). Embryos were processed for whole-mount in situ hybridization with SNX27 and Sox2 digoxygenin-labeled probes (Roche) as described elsewhere (38).
RESULTS
Building a Human PDZome Array
We have set up a rational approach to test the potential interactions of a given protein against an array of almost all human PDZ domains. This approach, based on a single Y2H assay, has been shown to be effective in probing the entire set of C. elegans PDZ domains for interacting proteins (23). An initial cross-database search allowed us to identify a total of 256 PDZ domains in 145 distinct proteins, excluding isoforms sharing identical PDZ domains (supplemental Table S1). The DNA fragments coding for 246 PDZ domains were successfully amplified from human cDNA libraries and cloned into the Gateway® Entry vector (32) (supplemental Table S1). Note that the PDZ domains of AHNAK2, ARHGAP23, LIMK1, SHANK3, SHROOM2, SHROOM3, SIPA1, SNTB2, SYNPO2L, and TJP1-PDZ1 are missing from the resource as a result of unsuccessful amplification. In this versatile format, the PDZome resource can rapidly and easily be transferred to a variety of expression vectors, including those used for two Y2H screens.
For the purposes of this study, each PDZ domain was transferred into a Y2H vector and then transformed into haploid Y187 yeast, creating a collection of individual yeast strains each expressing a single PDZ domain. These strains were arrayed in a standard 96-well format, allowing all individual PDZ domains to be probed simultaneously for interaction with a protein of interest through a simple mating procedure. Because of the direct nature of the assay, a single phenotypic readout was chosen (Fig. 1A). This resource is hereinafter referred to as the human PDZome array.
Fig. 1.
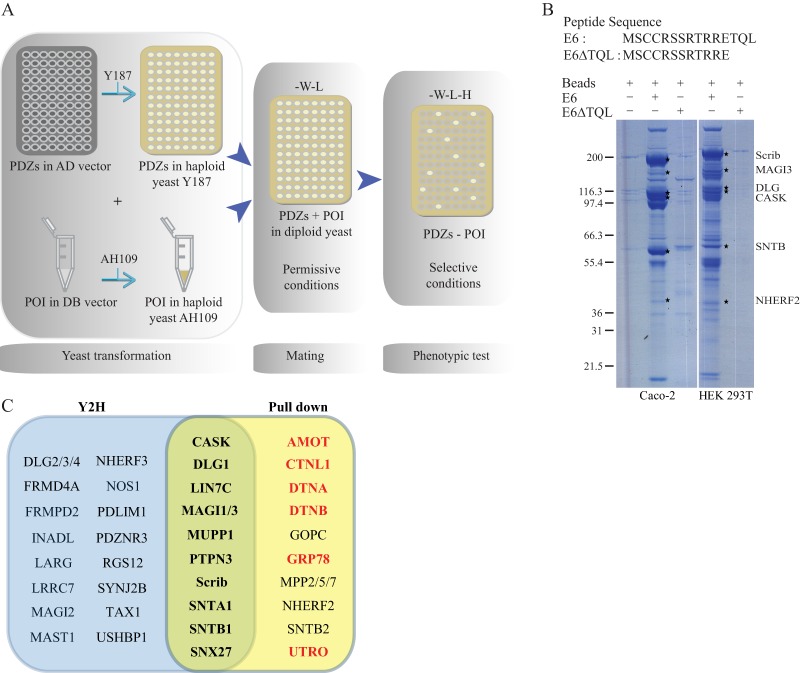
Human PDZome array to detect direct PDZ domain interactors. A, schematic representation of the identification of interactions between the collection of PDZ domains and a protein of interest (POI) using a yeast two-hybrid assay. The PDZ domain collection cloned in the pAD vector is hosted by the yeast Y187 strain. This arrayed collection is probed with the desired POI cloned in the pDB vector and hosted by an AH109 strain. B, Coomassie-stained protein gel after pull-down assay on Caco2 and HEK 293T cell extracts using E6 or E6ΔTQL peptides as affinity reagents. C, graphic representation of Y2H (blue) and mass spectrometry (yellow) screens. Common identified E6 binders are in the green intersection of those two ensembles. In red, binders considered as indirect interactors as explained in the text.
Validation of the Human PDZome Array Using HPV E6 Oncoprotein and Protein Kinase Screens
To validate our PDZome array, we decided to first challenge the resource with the E6 oncoprotein, a well-known PDZ domain interactor encoded by the genome of human papillomavirus (HPV). This oncoprotein involved in the induction of human cervical cancers bears a TQL carboxy-terminal sequence (where T is threonine, Q is glutamic acid, and L is leucine) terminated by a classical class I PDZ binding site and has been studied for its interaction with PDZ domains by many groups (12, 14). We fused the DNA-binding domain of Gal4 to the wild-type E6 carboxy-terminal sequence (DB-E6) or to a mutant version unable to bind PDZ domains (DB-E6ΔTQL) because it lacks the carboxy-terminal TQL PDZ class I motif. These constructs were transfected into the AH109 yeast strain and assayed against the human PDZome array (Fig. 1A). As expected, no interactions were identified with DB-E6ΔTQL. In contrast, screening with DB-E6 gave 37 reproducible interactions through 29 proteins, and the whole set of positive clones is listed in Table I. Among the 37 interactions found, one-third had been described in previous studies referenced in Ref. 12. For example, PDZ3 of Scrib and PDZ1, PDZ2, and PDZ3 of DLG1 have been shown to bind E6 (41, 42). Moreover, our results established the potential of our array not only to identify PDZ binders but also to directly identify which domain in multi-PDZ proteins is responsible for the interaction. For INADL (also called PATJ) comprising 10 PDZ domains, we found that only the PDZ5 domain binds to E6, whereas the interaction was previously shown to occur through a region encompassing PDZ4 and PDZ5 domains (43). Similarly, we confirmed the prevalence of PDZ7 in the 13-PDZ-domain protein MPDZ (also called MUPP1) for binding to the E6 carboxyl terminal peptide (44). Beyond that, these E6 peptide screens allowed us to identify a set of potential new interactions such as CASK and SNX27. To validate the interactions obtained by Y2H screening, we adopted a biochemical approach based on peptide pull-down assay using synthetic C-terminal E6 peptides, wild-type (E6) or lacking the PDZ binding motif (E6ΔTQL), equivalent to those used in Y2H. Those peptides covalently bound to agarose beads were employed as baits to purify partners from HEK 293T and Caco-2 cell extracts. The resulting bound proteins were separated on SDS-PAGE and stained, and protein bands differentially present in the E6 and E6ΔTQL conditions were excised and further identified via mass spectrometry (Fig. 1B). Reproducible results from at least three experiments are compiled in Table II. Fewer interactors were identified in Caco-2 cell extracts than in HEK 293T cell extracts, reflecting potential differential expression for E6 partners in the more differentiated and polarized Caco-2 cell line. Out of 17 PDZ proteins co-purified with the E6 peptide, 11 were also identified as direct E6 interactors by the PDZome Y2H array, among which CASK and SNX27 are newly identified E6 interactors. The results and the overlap between Y2H and biochemical approaches are presented in Fig. 1C.
Table I. List of PDZ domains interacting with E6 as determined by Y2H. This list of Y2H interactors refers to the protein name of each interactor with the total number of PDZ domains per protein (PDZn) and the position of the interacting PDZ domain(s) in each protein (PDZi). Cells in the last column are solid black when the interaction was already described (reviewed in Ref. 12).
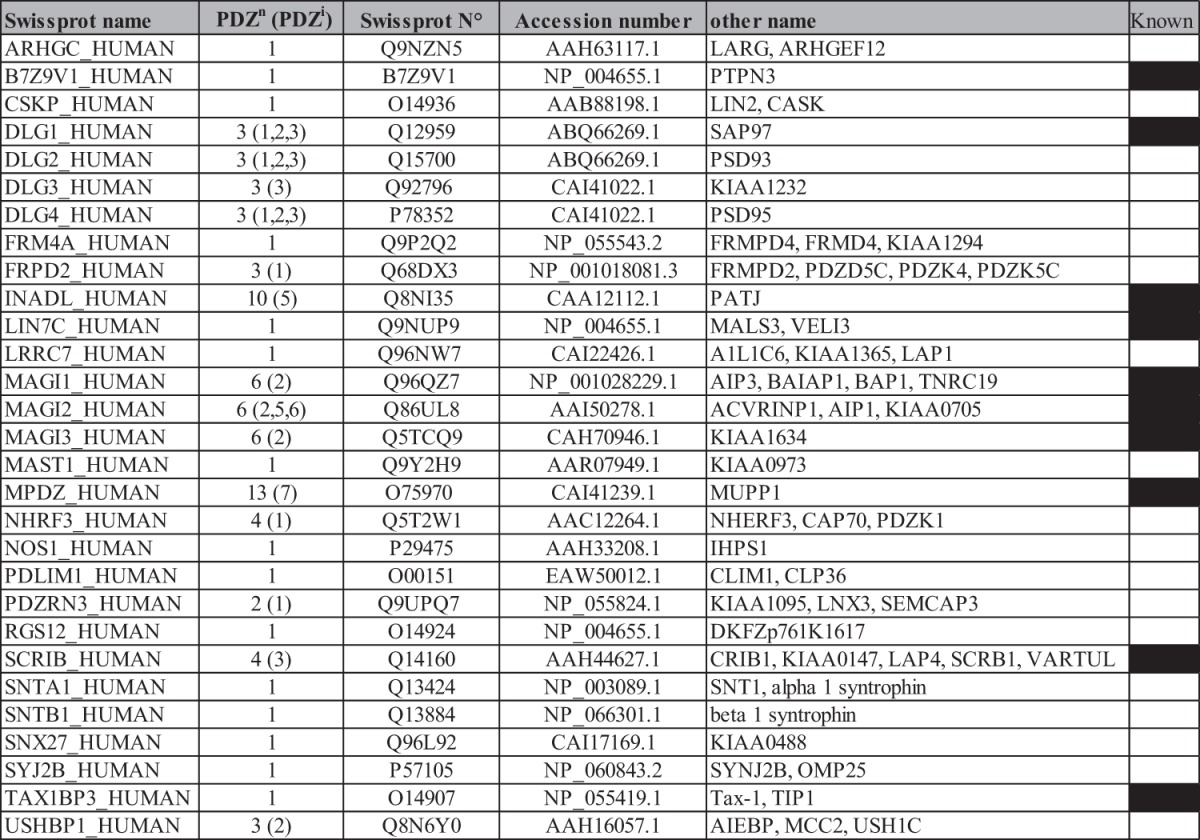
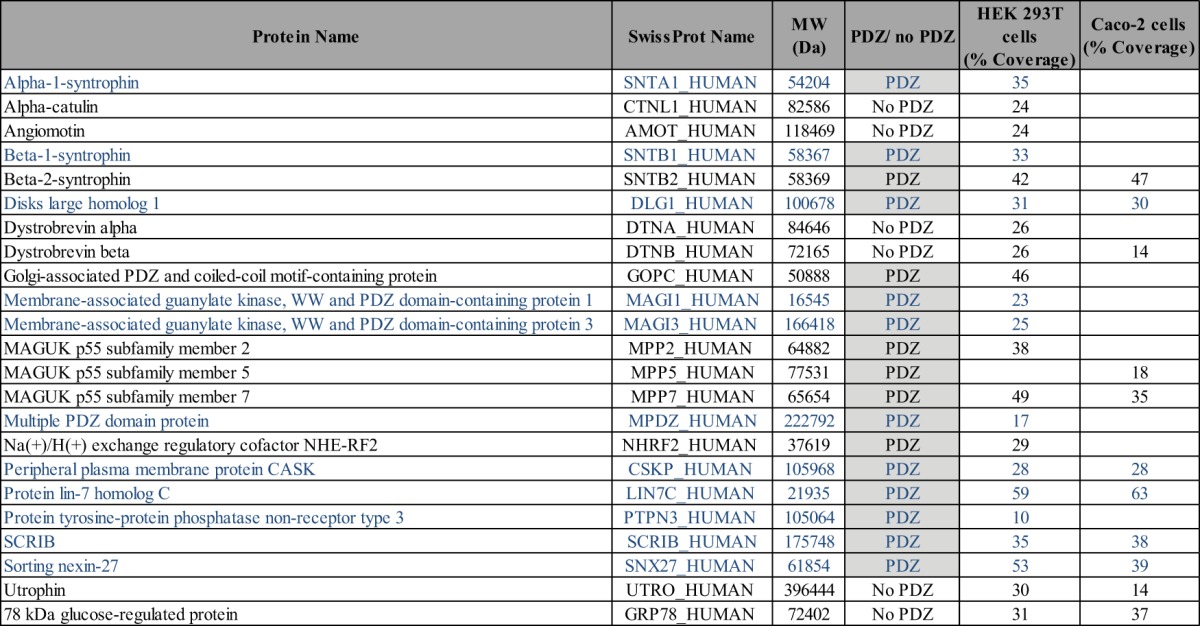
Table II. List of E6 interactors identified via pull-down assay in HEK 293T and Caco-2 cell extracts. This list refers to the protein name and molecular weight of the interactors. The presence of PDZ domains in the protein and percentage of coverage found via mass spectrometry are indicated. PDZ proteins also identified as E6 binders via Y2H screen are presented in blue.
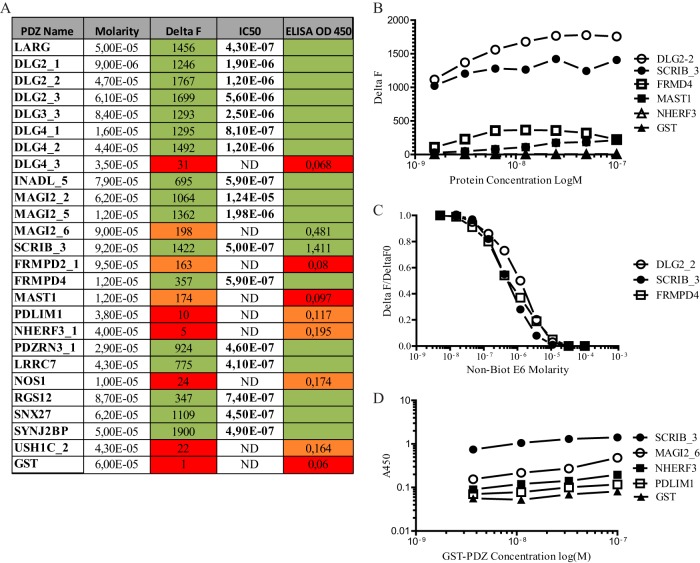
To test our interaction dataset further, we produced some of these PDZ domains as recombinant GST proteins (supplemental Fig. S1) and directly tested their interaction with the synthetic E6 peptide in vitro. In addition, we produced additional proteins: GST alone as a negative control, Scrib_3 and SNX27 as positive controls, and several additional PDZ domains present in DLG2, DLG4, and MAGI2, for a total of 25 PDZ domains. HTRF and ELISA were used to measure the interaction between E6 and the panel of PDZ domains. The two methods detected strong binding of E6 with 17 of these PDZ domains (LARG, DLG2_1, DLG2_2, DLG2_3, DLG3_3, DLG4_1, DLG4_2, INADL_5, MAGI2_2, MAGI2_5, SCRIB_3, FRMPD4, PDZNR3_1, LRCC7, RSG12, SNX27, and SYNJ2B), with apparent affinities ranging from 1E-5 M to 4E-7 M (Figs. 2A–2C). Eight of these PDZ domains (DLG4_3, MAGI2_6, FRMD2_1, MAST1, PDLIM_1, NHERF3_1, NOS1, and USH1C_2) were demonstrated as low or non-binders by HTRF. Compared with GST, four of these domains (PDLIM1, NHERF3_1, NOS1, and USH1C_2) displayed a faint interaction with E6 in ELISA (Figs. 2A, 2D). We thus confirmed that the majority of PDZ interactions identified by two hybrid were confirmed by in vitro binding assays using HTRF or ELISA (17 out of 25) and that the testing PDZ array is robust enough to limit the false positive rate and build a high-confidence dataset. Our results are in accordance with previous work using both high-throughput two-hybrid and affinity purification assays (45). It is commonly admitted now that the incomplete overlap observed with different techniques is mainly due to a high false-negative rate for each of the techniques used, but this overlap allows one to build a high-confidence dataset (46). However, the non-overlapping sets increase the likelihood of finding new partners that can in turn be validated further.
Fig. 2.
Interaction between E6 and selected PDZ domains measured by HTRF and ELISA. A, shown in the table are the PDZ name, the molarity of the GST-PDZ fusion protein in purification eluates, Delta F (FRET intensity obtained with GST-PDZs, equal to 1E-8 M, determined as shown in B), IC50 (the apparent affinity determined via homologous competition assay as shown in C for representative PDZ domains (ND: not done)), and the binding intensity (A450) of the GST proteins (1E-7M) to solid phase E6 measured via ELISA, as shown in D (note the logarithmic scale of the y-axis). Green: high interaction; orange: low interaction; red: no interaction.
Three known E6 PDZ partners (GIPC, PTPN13, and GOPC) were missed by our Y2H screen. For GIPC, the original published interaction was described for experiments using E6 from HPV-18 E6 as a bait, the C-terminal sequence of which is ETQV (47). Our screen was done with E6 from HPV-16, which contains a slightly different C-terminal sequence (ETQL). For PTPN13, the original interaction, considered as a weak one by the authors and obtained via GST-E6 pull-down experiments, was not described as a direct interaction (13). Note that the GIPC and PTPN13 PDZ domains were efficiently selected by Vangl2 (Fig. 4D), ruling out that these preys were not functional. Given this, we do not consider GIPC and PTPN13 as false negative hits. As for GOPC (also called CAL), Jeong et al. recovered the interaction via pull-down using a recombinant GST-E6 protein and further confirmed the interaction using in vitro translated proteins (48). Although GOPC was not selected by our Y2H screen, we did isolate this PDZ protein in the E6 peptide pull-down assay (Fig. 1D), emphasizing the usefulness of combining Y2H and pull-down/mass spectrometry approaches to decrease the false negative rate.
Fig. 4.
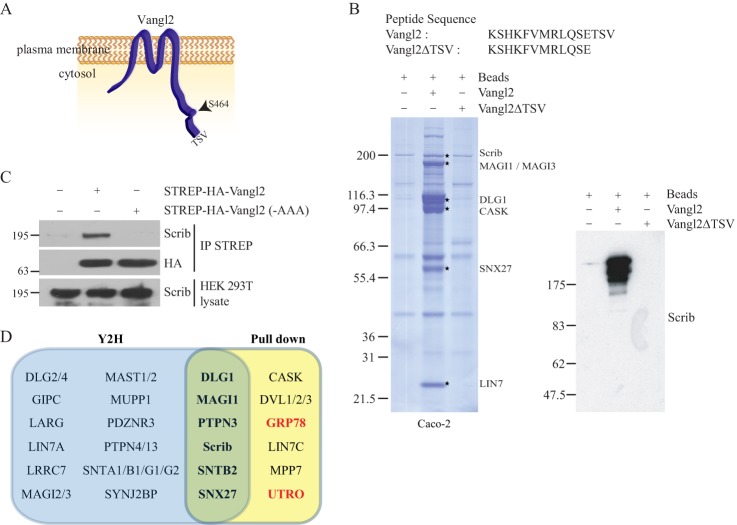
Identification of PDZ partners of the Vangl2 carboxy-terminal region. A, schematic representation of Vangl2 receptor. B, Coomassie-stained protein gel after pull-down assay on Caco-2 cell extracts using Vangl2 or Vangl2ΔTSV peptides as affinity reagents. Each star corresponds to PDZ interactors of the Vangl2 carboxy-terminal peptide found via mass spectrometry. In the right panel, an immunoblot from the same experiment using anti-Scrib antibody confirms the Vangl2–Scrib interaction. C, Scrib interacts with Vangl2 through its PDZ binding motif (PDZBM) sequence. HEK 293T cells were transfected with epitope-tagged Vangl2 or Vangl2 mutated at its PDZBM (last three amino acids mutated in alanine). Vangl2 proteins were purified using streptavidin beads, and the presence of Scrib was assessed by means of co-affinity purification and Western blotting. 15 μg of lysate were loaded in total lysate, and 400 μg were used for affinity purification. D, graphic representation of Y2H (blue) and mass spectrometry (yellow) screens. Common identified Vangl2 binders are in the green intersection of those two ensembles. In red are the binders considered as indirect interactors (see text).
Furthermore, PDZ proteins isolated by pull-down assays but not by Y2H screening with E6 are likely indirect E6 partners. This was the case for three members of the MAGUK p55 subfamily (MPP2, -5, and -7) in this study (Table II, Fig. 1D). Interestingly, these proteins are known to strongly interact with the well-characterized DLG-CASK-LIN7 protein complex (49), suggesting co-purification with the E6 peptide through this complex. Other proteins isolated by pull-down assay that do not contain a PDZ domain (e.g., utrophin, angiomotin, dystrobrevin α and β, α-catulin) belong to known PDZ complexes and are also probably indirectly associated with E6. Indeed, utrophin, α-catulin, and dystrobrevins form a dystrophin-associated protein complex that also contains syntrophin (SNTB1) (50), a direct partner for E6 (Fig. 1D, Table I). Angiomotin, a cell surface receptor containing a PDZ-binding site, binds to PDZ proteins, especially to the PDZ1 domain of MUPP1 (51), whereas E6 interacts with the MUPP1 PDZ_7 domain (Table I). We thus believe that angiomotin is an indirect E6 partner.
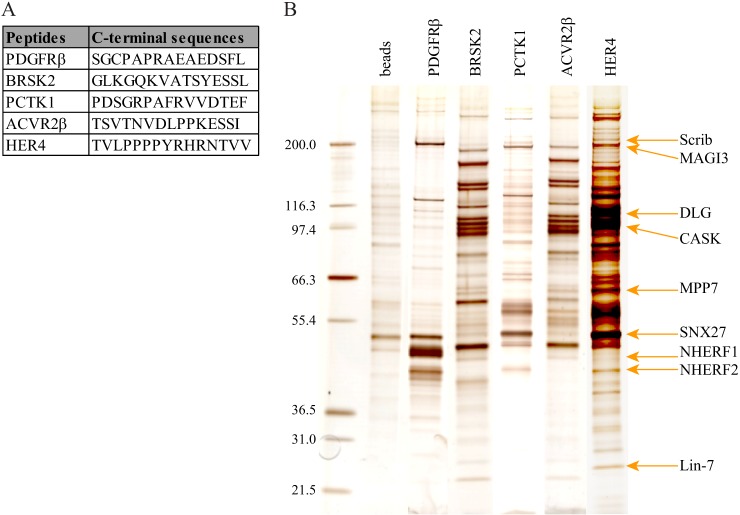
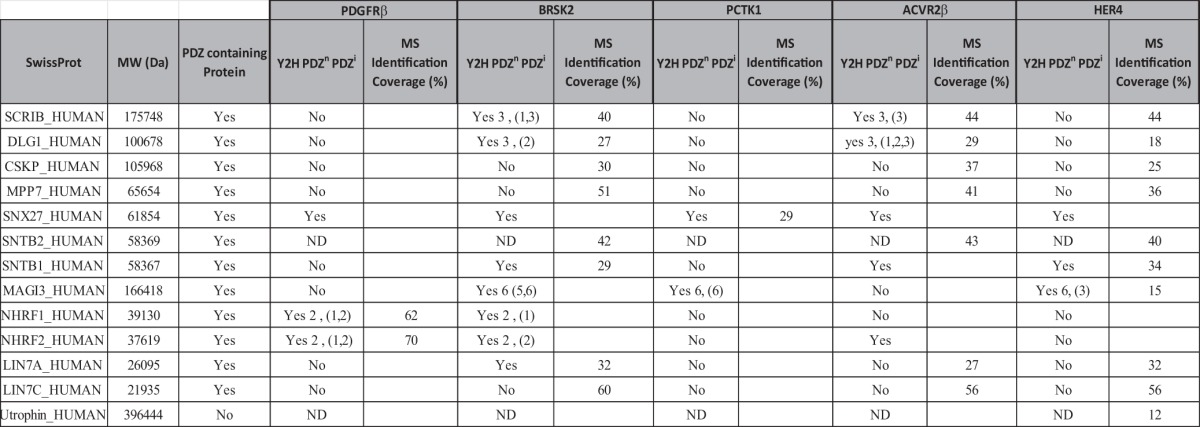
In an effort to challenge the PDZome with other potential PDZ binding motifs, we screened with carboxy-terminal peptides of several human protein kinases: PDGFRβ, BRSK2, PCTK1, ACVR2B, and HER4 (Fig. 3A). PDGFRβ and HER4 are known to bind members of the DLG and NHERF families, respectively (52, 53), and indeed we recovered these interactions via Y2H and, for some of them, pull-down assays (Fig. 3B, Table III). We confirmed that NHERF proteins (NHERF1 and NHERF2) are strong and specific PDGFRβ partners, easily isolated via both methods. As for HER4, DLG1 was efficiently precipitated by this peptide but was not selected by Y2H (Table III). Nevertheless, we found that the very similar PDZ domains of DLG2 (DLG2_1, DLG2_2), DLG3 (DLG3_2), and DLG4 (DLG4_2) interacted with HER4 in the PDZome array (data not shown). In addition, we isolated novel interactions of interest for HER4, in particular with SNTB1 and MAGI3, which are positive in Y2H and pull-down assays, as well as for BRSK2, PCTK1, and ACVR2B, for which no PDZ interactions have been, to our knowledge, described previously. Altogether, these data demonstrate that the human PDZome Y2H array is suitable for the rapid screening of direct PDZ interactions, the refinement of already described interactions, and, in combination with affinity purification approaches, the identification of novel PDZ complexes.
Fig. 3.
Screening of the PDZome with the PDZ binding site of protein kinases. A, carboxy-terminal sequences of selected protein kinases. B, silver-stained protein gel after pull-down assay on HEK 293T cell extracts using peptides from A as affinity reagents.
Table III. List of PDZ proteins interacting with protein kinases as determined via Y2H and pull-down assays. This list refers to the protein name and molecular weight of the interactors. The presence of PDZ domains in the protein and the percentage of coverage found via mass spectrometry are indicated as in Table I.
The Vangl2 Carboxy-terminal Region Binds to a Set of PDZ Proteins
In a more prospective study, we used our PDZome resource to identify the PDZ interactome of Vangl2, an evolutionarily conserved receptor originally identified in Drosophila melanogaster for its role in planar cell polarity (PCP) (54, 55). In mice, a spontaneous Vangl2 loss of function also leads to PCP defects characterized by failure of neural tube closure and stereociliary bundle misorientation in the cochlea (56, 57). Vangl2 is a four-transmembrane receptor bearing amino- and carboxy-terminal cytoplasmic sequences and two limited extracellular loops (Fig. 4A) (58). Vangl2 possesses two types of PDZ binding motifs: a typical class I PDZ binding motif (TSV) in its carboxy-terminal region, and an unconventional internal PDZ binding motif containing the serine 464 residue known to bind Dischevelled (Fig. 4A) (59). The typical class I PDZ motif is known to bind PDZ proteins such as Scrib, Discs Large, and MAGI3 proteins, but its functional role remains uncertain (60). Mutation of serine 464 to asparagine (mutant Vangl2Lp) leads to the failure of Vangl2 to bind to Dishevelled and to PCP defects (58).
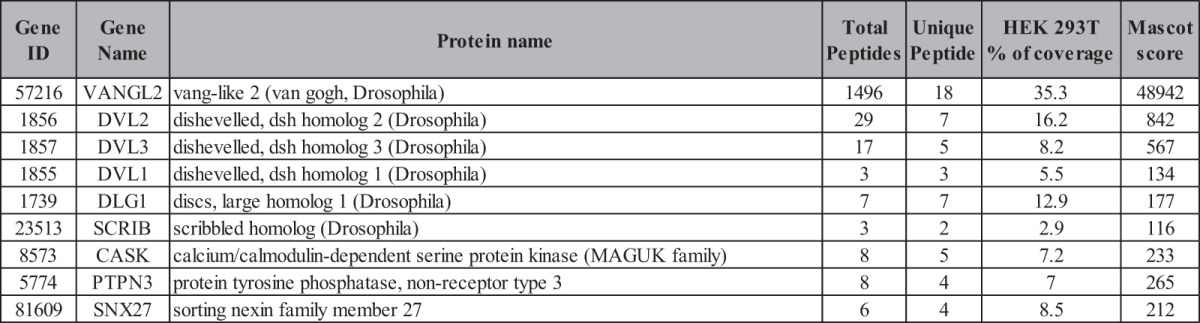
In order to identify the whole set of PDZ domains able to interact with the Vangl2 PDZ type I motif, we used the same strategy that we applied to E6 and protein kinase C-terminal sequences. First, we screened the PDZome with a Vangl2 peptide encompassing the 15 last residues including or not including the TSV PDZ binding site as bait (Vangl2 and Vangl2ΔTSV, respectively). No interaction was recovered in the absence of the TSV motif (not shown), whereas a screen with the wild-type bait selected 36 PDZ domains in 24 proteins (supplemental Table S2). In addition to already characterized Vangl2 PDZ partners such as Scrib, DLG1, MAGI1, and MAGI3 (60, 61), we identified potential new binders. Among them, we noticed an overrepresentation of proteins involved in traffic such as SNX27 (62, 63), PDZNR3, and GIPC1, the latter being a G-protein regulator thought to control vesicular trafficking (64). In parallel, we carried out reciprocal Vangl2 peptide pull-down assays on Caco-2 cell extracts, followed by mass spectrometry analysis. These biochemical experiments confirmed known PDZ protein partners such as Scrib, DLG1 (60), and MAGI3 (61), or they identified previously uncharacterized partners such as CASK and SNX27 (Fig. 4B, supplemental Table S3). Thanks to the Y2H data, we could identify which PDZ domain is implicated in the interaction with Vangl2 in each multi-PDZ protein (supplemental Table S2). For example, the Scrib PDZ3 domain is a potent interactor for Vangl2, as it was for E6 (Table I). Non-PDZ proteins such as utrophin and GRP78 were also isolated via pull-down assay, indicating that they are likely indirect Vangl2 binders (supplemental Table S3). In order to confirm that our screen based on the last Vangl2 carboxy-terminal residues was relevant in the context of the full-length protein, we chose to use the tandem affinity purification (TAP) protocol and further identified associated proteins by means of mass spectrometry (65, 66). We stably overexpressed in HEK 293T cells a TAP-tagged version of Vangl2, and we confirmed the presence of Scrib in the Vangl2 complex via proteomic analysis and Western blot (Fig. 4C and Table IV). A mutant version of Vangl2 in which the TSV motif was replaced by an AAA motif was used as a control. This mutant was unable to co-immunoprecipitate with Scrib. The results obtained corroborated our peptide-based screens, as we identified Scrib and DLG1 as well as potential new Vangl2 interactors CASK and SNX27. Moreover, Dishevelled proteins (DVL1, DVL2, DVL3) were also identified through this technique, whereas we did not find them using the PDZome Y2H array using the carboxy-terminal region of Vangl2. This indicates that the Dishevelled PDZ domain interacts with Vangl2 through the unconventional internal motif as previously reported (58). Overall, the combination of mass spectrometry analysis from peptide and full-length Vangl2 and our PDZome Y2H array approach allows us to define a network of PDZ proteins associated with Vangl2 (Fig. 4D).
Table IV. List of Vangl2 interactors identified by ectopic expression of tagged Vangl2 in HEK 293T. List of proteins found in the Vangl2 complex by means of LC-MS/MS. The peptides and resulting proteins were identified using an integrated version of SEQUEST on a Sorcerer platform (SageN) and were analyzed with Peptide and Protein Prophet software (ISB, Seattle, WA). The “Unique Peptides” column represents the total number of unique peptides successfully identified. “Total Peptides” correspond to the total number of peptides matching a given protein identification. The percentage (%) of coverage is the percentage of the protein sequence covered by the peptide sequences identified.
The Vangl2–Scrib Complex Is Required for Cell Migration
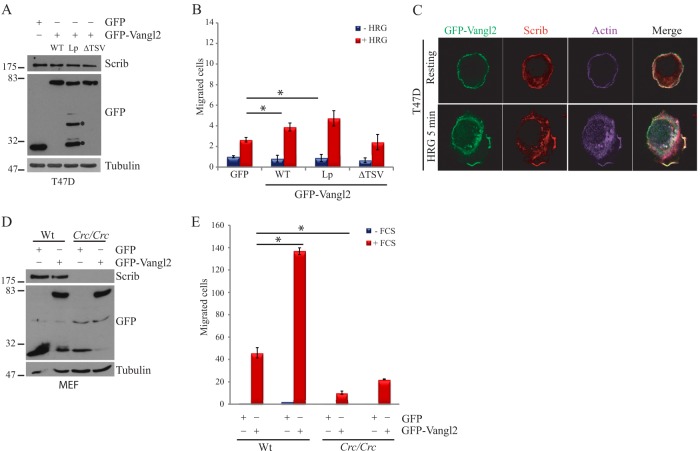
In an attempt to investigate the functional contribution of the two PDZ binding motifs of Vangl2 in a cultured cell model, we overexpressed GFP, GFP-Vangl2, and its mutant forms (Vangl2Lp and Vangl2ΔTSV) in a breast cancer T47D cell line that does not express endogenous Vangl2 (data not shown) and tested their effect on cell migration induced by stimulation with HRG, a ligand for EGF receptors (37). Although all constructs were expressed equally well (Fig. 5A), we observed increased cell migration in Boyden chamber assays when GFP-Vangl2, but not GFP, was expressed (Fig. 5B). Disruption of the Vangl2 unconventional internal PDZ binding site (GFP-Vangl2Lp) did not affect this increased cell migration, in contrast to the deletion of the classical PDZ binding motif (GFP-Vangl2ΔTSV), as shown in Fig. 5B. These results clearly demonstrate that Vangl2 exerts promigratory properties in induced cell migration assays, and that this function requires its carboxy-terminal PDZ type I motif but not its internal PDZ binding site. Although Scrib was initially described in Drosophila as a gene implicated in apico-basal polarity (67), we and others have previously established that it plays a role in cell migration (37, 68). In the mouse, Scrib and Vangl2 genetically interact during embryonic development to regulate PCP (56, 69). Based on these results, we next tried to determine whether Scrib is required for the promigratory properties of Vangl2. As expected, in pull-down assays using a GST fused to the four Scrib PDZ domains, we demonstrated the efficient binding of this recombinant protein to GFP-Vangl2 and GFP-Vangl2Lp, but not GFP-Vangl2ΔTSV (supplemental Fig. S2A). Similar experiments using the individual Scrib PDZ domains demonstrated that the Scrib PDZ3 domain appears to be the most efficient for precipitating Vangl2, although some weak binding also occurred with the Scrib PDZ4 (supplemental Fig. S2B). Previous studies have demonstrated that Scrib controls cell migration through binding to βPIX, a Rac/Cdc42 GEF that interacts with Scrib PDZ1 and PDZ3 domains (36, 37). We recovered this binding by reprobing the GST-Scrib PDZ membrane with anti-βPIX antibody (supplemental Fig. S2B), as well as via reconstitution of the interactions using the HTRF method (supplemental Figs. S2C–S2F). Although Scrib has the capacity to bind βPIX independently of Vangl2 by interacting with its PDZ1 domain, it may also potentially compete for binding to its PDZ3 domain. We used HTRF-based measurements to evaluate the relative affinity of soluble βPIX, Vangl2, and E6 carboxy-terminal peptides for the four independent Scrib PDZ domains. As expected, PDZ3 was able to bind the three peptides, whereas PDZ1 strongly bound to βPIX and E6 but not to Vangl2. We could not recover a detectable interaction between PDZ4 and Vangl2, suggesting a weak interaction in HTRF (supplemental Figs. S2C–S2E). We also defined the EC50 of each peptide for PDZ1 and PDZ3 domains and found a higher affinity for E6 (0.4 μm) than βPIX (8 μm) toward PDZ3, whereas Vangl2 displayed the lowest affinity (13 μm) for the PDZ3 domain (supplemental Fig. S2F). In order to further define Scrib's contribution to Vangl2-induced cell migration, we tested their potential co-localization during this process via immunofluorescence using T47D cells expressing GFP-Vangl2 stimulated by HRG. As shown in Fig. 5C, GFP-Vangl2 and Scrib are present at the plasma membrane in unstimulated conditions. After 5 min of HRG stimulation, T47D cells spread a large lamellipodium at the edge of migrating cells that is enriched in actin, Scrib (as previously described (37, 68)), and GFP-Vangl2, demonstrating their co-localization in the protruding membrane. Furthermore, we evaluated the Scrib requirement in cell migration by overexpressing GFP-Vangl2 in MEFs deficient for Scrib (MEFCrc/Crc) relative to wild-type MEFs (37). Using Boyden chamber assays and serum-induced cell migration, we showed that increased cell migration behavior is retrieved by overexpressing GFP-Vangl2 in wild-type MEFs, but not in MEFCrc/Crc, where the cell migration rate is not significantly affected relative to control conditions (Figs. 5C, 5D). Altogether, these data demonstrate a requirement of the PDZ partner Scrib in Vangl2 promigratory activity.
Fig. 5.
Involvement of Scrib in Vangl2 promigratory function. A, Western blot analysis of protein extracts of T47D cells stably transfected with GFP, GFP-Vangl2, or GFP-Vangl2Lp and GFP-Vangl2ΔTSV mutants. Degradation products of GFP-Vangl2Lp are marked by black points. B, each T47D cell population was submitted to transmigration assays in Boyden chambers without stimulation (blue bars) or with 1 nm HRG (red bars). Data are represented as fold changes and are the sum of three independent experiments. C, GFP-Vangl2 T47D cells either unstimulated or stimulated for 5 min with HRG treatment were fixed and labeled with anti-Scrib and Actin staining by TRITC-label and phalloidin, respectively. Confocal acquisition shows the localization of GFP-Vangl2 and Scrib at the leading edge under HRG stimulation. D, wild-type (Wt) MEF cells or circletail homozygous cells (Crc/Crc) were transiently transfected with GFP or GFP-Vangl2. The expression of proteins was evaluated with the mentioned antibodies. E, Wt and Crc/Crc MEF cells transiently transfected with GFP or GFP-Vangl2 were submitted to Boyden chamber assays using serum (FCS) as a chemoattractant. Filters were coated with fibronectin. Asterisks represent significant differences relative to the control (* p < 0.05 by Mann–Whitney U test). Data are represented as fold changes.
SNX27, a Novel Partner of Vangl2, Is Implicated in Morphogenetic Movements
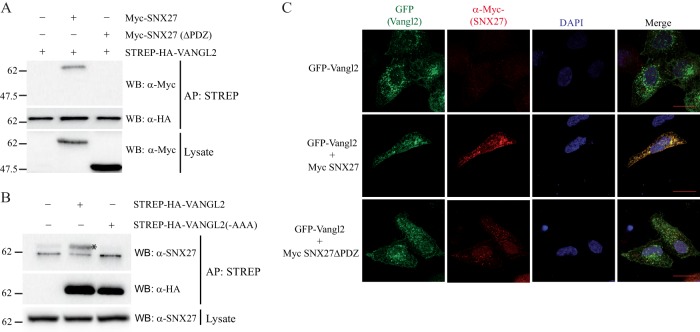
Of the novel PDZ domain proteins interacting with Vangl2, we next focused our attention on SNX27. The Vangl2–SNX27 interaction is obviously robust, as it was recovered by Y2H, peptide pull-down, and co-immunoprecipitation (Figs. 4B and 4D, Table IV, and supplemental Tables S2 and S3). SNX27 has been shown to regulate the membrane localization and intracellular trafficking of several membrane proteins (35, 63). To confirm the PDZ domain-mediated interaction, we first expressed HA-tagged Vangl2 with either Myc-tagged SNX27 or Myc-tagged SNX27 deleted of its PDZ domain. We confirmed that SNX27 interacts with Vangl2 through its PDZ domain, as the deletion of the domain abrogated the interaction (Fig. 6A). Conversely, the PDZBM of Vangl2 is required for the interaction, as replacement of the motif with three alanine residues disrupted the formation of the Vangl2–SNX27 complex (Fig. 6B). Through immunofluorescence and confocal analysis, we observed that GFP-Vangl2 colocalized with Myc-SNX27 in intracellular compartments (Fig. 6C). Deletion of the SNX27 PDZ domain strongly decreased colocalization, suggesting again that interaction of the two proteins, which occurs in living cells, is mediated by the PDZ domain.
Fig. 6.
SNX27 interacts with Vangl2 through its PDZ domain. A, Western blot analysis of protein extracts of HEK 293T cells expressing STREP-HA-Vangl2 and either Myc-SNX27 or Myc-SNX27 deleted of its PDZ domain. Vangl2 proteins were purified using streptavidin beads, and the presence of SNX27 was assessed by means of co-affinity purification and Western blotting using anti-Myc antibody. 15 μg of lysate were loaded in total lysate, and 400 μg were used for affinity purification in A and B. B, SNX27 interacts with Vangl2 through its PDZBM sequence. HEK 293T cells were transfected with Vangl2 or Vangl2 mutated at its PDZBM (last three amino acids mutated in alanine). Vangl2 proteins were purified using streptavidin beads, and the presence of SNX27 (as indicated by an asterisk on the blot) was assessed by means of co-affinity purification and Western blotting. C, co-localization of Vangl2 with Myc-SNX27 but not Myc-SNX27ΔPDZ in transfected HEK 293T cells.
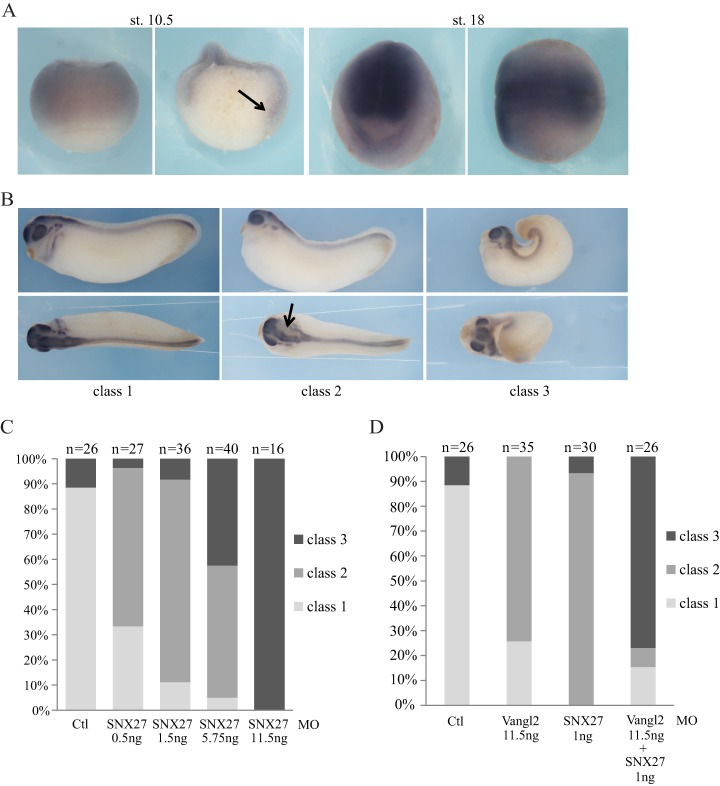
To further assess the functional interaction between Vangl2 and SNX27, we turned to the Xenopus embryo, which offers a simple, integrated model with which to evaluate the role of PCP components in morphogenetic movements such as convergence-extension and neural tube closure (70). Previous work showed that in Xenopus, Vangl2 contributes to convergence-extension (39). We found that SNX27 transcripts are present in tissues undergoing convergent extension, such as the mesoderm and the neural ectoderm at gastrula to neurula stages (Fig. 7A). When endogenous SNX27 function was inhibited through injection of an MO, neural tube closure was strongly impaired in a dose-dependent manner (Figs. 7B and 7C). This phenotype is typical of defective PCP signaling (71). Furthermore, co-injection of sub-optimal amounts of SNX27 and Vangl2 MOs yielded stronger neural tube closure phenotypes than separate injections (Fig. 7D), suggesting that these two factors cooperate to achieve proper morphogenesis. Together with the above biochemical evidence, these in vivo data confirm the importance of the newly detected interaction between Vangl2 and SNX27 and illustrate the power of our Y2H array to uncover novel interactions and associated biological functions.
Fig. 7.
SNX27 knockdown impairs morphogenetic movements in Xenopus embryos. A, in situ hybridization with SNX27 antisense digoxygenin-labeled riboprobe revealed expression in early gastrula mesoderm (st. 10.5) and neural plate (st. 18). The st. 10.5 embryo on the right was bisected following staining to reveal expression in the internal mesodermal mantle (arrow). The st. 18 embryo is shown in front view (left) and dorsal view (right). Sense probe did not produce any signal. B, embryos were injected in dorsal blastomeres at 4- to 8-cell stage with SNX27 or Vangl2 morpholino-oligonucleotides (MOs) and stained for the neural marker Sox2 at the tailbud stage. Embryos representative of the three classes observed are shown. Class 1 embryos are wild-type, class 2 embryos display anterior neural tube opening (arrow), and class 3 embryos exhibit severe neural tube opening and a kinked body typical of deficient convergent extension. C, graph showing the phenotypic distribution of embryos injected with increasing amounts of SNX27 MOs. D, graph showing the synergistic effect of Vangl2 and SNX27 MOs.
DISCUSSION
PDZ domains are one of the most widely distributed protein interaction domains encoded in genomes. Protein interactions mediated by these domains are involved in a wide range of cellular processes, including cell signaling, protein trafficking, and cell polarity (4). Cell polarity requires a number of PDZ proteins to create networks that organize apico-basal and planar cell polarities and build functional epithelial tissues (6, 72). Similar PDZ-derived mechanisms are required for the polarity of non-epithelial cells such as endothelial and neuronal cells. In addition, PDZ-mediated interaction networks are involved in directed cell migration, which relies on a profound polarized reorganization of the cell body and its organelles following stimulation by chemoattractants (6, 73). PDZ proteins implicated in cell polarity, such as Scrib and PAR3/PAR6, are well conserved structurally and functionally from invertebrates to vertebrates, as are some of their associated protein networks (74). Besides their conservation, the importance of PDZ proteins in the homeostasis of tissues is also highlighted by the dramatic defects observed when their function is disrupted in pathological situations such as cancer. Deregulation of PDZ protein functions, either by overexpression or by down-regulation, is now well established in various cancers of epithelial origin (9–11). Of particular interest are the defects of PDZ functions generated by viral proteins that, as in the case of the E6 oncoprotein produced by HPV-16 or HPV-18, participate in cell transformation (9, 12, 14).
Canonical PDZ binding sites were initially described as short motifs found at the very carboxy-terminal end of receptors such as glutamate receptor and potassium channels (8). Diverse unbiased screening approaches later identified consensus sequences specific for three major classes of PDZ domains with distinct binding specificities (18–21). However, a number of studies have described a more complex landscape for PDZ interactions, with many of them occurring via carboxy-terminal sequences falling outside the canonical consensus sequences, or even via non-carboxy-terminal sequences. In a recent effort to search for PDZ interactions at a genome-wide level, we screened a C. elegans cDNA library with 93 C. elegans PDZ domains by means of Y2H in yeast and revealed a substantial number of PDZ partners devoid of canonical PDZ binding sites at the carboxy-terminal end (23). These alternative modes of interaction underline the versatility of PDZ domain binding specificities (23, 27–29).
Searches for protein interactions usually employ classical techniques such as Y2H, phage display, or purification/mass spectrometry analysis that can be limited by the low amount of partners in protein extracts or by their low representation in non-normalized cDNA libraries. Here, we described a human PDZ domain resource that includes 96% of human PDZ domains organized in an array adapted for extensive Y2H screens. To our knowledge, this is the most exhaustive human PDZ resource available, as one of the most achieved until now included only around 40% of human PDZ domains (96/254) (5). We have demonstrated the value and the robustness of this resource using the E6 oncoprotein and protein kinase C-terminal sequences as models to validate the array. Indeed, through repetitive screens, in addition to already known HPV-16 E6 PDZ partners, we were able to identify SNX27 and CASK as potential new binders of the carboxy-terminal class I sequence of E6. Interactions were further validated by means of a proteomic approach. The contribution of these potential novel interactions in HPV-16-mediated tumorigenesis is worth further investigation. Indeed, SNX27 associates to the retromer recycling complex (62, 63), which has recently emerged as an important protein complex in cell transformation (75, 76). In the case of CASK, a protein containing a single PDZ domain, recent findings demonstrated that this protein kinase has an unexpected mode of regulation (77). It would be interesting to evaluate how E6 binding interferes with CASK scaffolding and enzymatic properties. Moreover, our approach underlines another advantage of our array relative to global Y2H of cDNA libraries, in that the bait directly selects the PDZ domain(s) of interest in a particular protein. This is particularly interesting in the case of multi-PDZ proteins such as MUPP1 (13 PDZ domains) or PATJ (10 PDZ domains). In our screen, E6 could specifically and directly select MUPP1-PDZ7 (44) and PATJ-PDZ5 domains (43). The screen of the human PDZome resource should now be carried out with sequences from human T-lymphotropic virus Type I, influenza virus, and hepatitis B virus that are known to interact with PDZ domains in order to refine the PDZ web associated with the corresponding viral proteins and address the pathological consequences of these interactions (12). As for the selected protein kinases used to screen the PDZome array, we confirmed the already published interaction between PDGFRβ and the NHERF family, and between HER4 and the DLG family (52, 53). Furthermore, our study revealed novel PDZ-containing domain interactors for HER4, BRSK2, PCTK1, and ACVR2B protein kinases that deserve further functional validation. It would be of particular interest to test whether PDZ proteins modulate enzymatic activity, subcellular localization, and signaling functions of these protein kinases (4).
Lastly, we took advantage of our resource to screen the typical class I PDZ binding site of Vangl2, a cell polarity receptor. This allowed the discovery of both known and novel Vangl2 interactors. We decided to first further study the Vangl2–Scrib interaction in cell migration. It was recently shown that Vangl proteins are required for cell migration of breast cancer cells, but no mechanism was assigned to this function (78, 79). Using biochemical and HTRF approaches, we demonstrated the specificity of Scrib PDZ domains toward Vangl2, βPIX, and E6 sequences and provided approximate EC50 values for each interaction. The promigratory function of Vangl2 was recently shown in the context of cancer cell migration (80). In that study, the authors focused on the role of Vangl2 in trafficking and shedding of membrane type-1 matrix metalloproteinase but did not investigate the contribution of the Vangl2 PDZ binding sites (80). As we previously demonstrated that Scrib is implicated in exocytosis in neuronal cells (36), there is a possibility that Scrib might participate in membrane type-1 matrix metalloproteinase shedding and/or trafficking in migrating cells. We provide here compelling evidence that the carboxy-terminal PDZ binding site, but not the Dishevelled binding site, is required for Vangl2 promigratory properties and that Scrib is required for this function.
Among the new Vangl2 interactors found in our screens, we focused on SNX27, as multiple binding assays demonstrated a strong interaction of this PDZ protein with the Vangl2 carboxy-terminal sequence. SNX27 is a versatile protein containing a unique PDZ domain and participates in the membrane localization and intracellular trafficking of various membrane proteins (35, 63). To gain more insight about the potential role of SNX27 in the Vangl2 pathway, we took advantage of the Xenopus embryo, which is a classical model for studying PCP functions. We demonstrated that, like Vangl2, SNX27 contributes to convergence-extension and that it cooperates with Vangl2 in morphogenetic movements. To our knowledge, this is not only the first attempt to study this conserved PDZ protein in Xenopus, but also the first description of its implication in morphogenetic events.
The underlying mechanism responsible for SNX27 function is currently unknown. As an endosomal protein, SNX27 may contribute to the regulation of the trafficking of Vangl2, which is important for its function (81). As SNX27 and Vangl2 colocalize in intracellular structures (Fig. 6), this hypothesis is worth investigating. We noted that SNX27 knockdown caused more dramatic PCP phenotypes than Vangl2 knockdown, suggesting that additional important interactors of SNX27 implicated in morphogenetic movements remain to be found. Previous work has shown that knockout of the mouse SNX27 gene leads to a significant decrease of the expected number of born SNX27−/− pups, suggesting embryonic development defects (82). Apparently, the role played by SNX27 during embryonic development is not absolutely necessary, as a majority of SNX27−/− embryos developed to term and were born, although functional redundancy with other factors cannot be excluded. Nevertheless, they presented with severe defects, with delayed body weight gain, reduced sizes of multiple organs, and lethality before weaning. Given that a functional interaction exists between SNX27 and Vangl2 in Xenopus, it would be interesting to test whether Vangl2 is implicated in SNX27 functions in the mouse. Altogether, our results demonstrated the usefulness of our combined approach in identifying novel PDZ interactions amenable to further functional validation. Of note, we recently demonstrated that GIPC1 (GAIP C terminus Interacting Protein 1), another PDZ protein isolated by our screen (Fig. 4D), regulates Vangl2 membrane localization in neurosensory hair cells (83).
Despite the potential of our resource, we anticipate that the screening of the human PDZome array by Y2H will present some limitations for certain baits. Indeed, PDZ interactions can be regulated by post-translational modifications such as phosphorylation (84), by allosteric conformation and cooperation between PDZ domains present in the same protein (27, 85), or by lipid binding to PDZ domains (28, 29). Obviously, these particular interactions cannot be reconstituted in our Y2H assay. However, we have maximized the flexibility of the PDZome resource through the use of a recombinant strategy that allows easy shuttling of the resource into various acceptor vectors. This versatile format is thus amenable to other potential uses, in particular the expression of each individual PDZ domain fused to GFP or to GST in mammalian cells and bacteria (32). As an example, the PDZome has been used to screen novel PDZ–lipid interactions (86). This array is thus adapted for the screen of PDZ interactors and, beyond, should facilitate the understanding of their functions.
Supplementary Material
Acknowledgments
We thank Samuel Granjeaud for his contribution to the improvement of mass spectrometry data analysis.
Footnotes
* This work was supported by grants from Association pour la Recherche contre le Cancer (fellowship to E.B. and “Projet ARC” program to MS) and La Ligue Contre le Cancer (Label Ligue (J.P.B.) and Ligue fellowship to A.M.D.), EUCAAD (FP7 program), INCa and IBiSA, ANR-08-MNPS-040-01 (M.M.), and Region Regional Aquitaine. The Marseille Proteomic IBiSA platform (MaP) is supported by Institut Paoli-Calmettes, INCa, Canceropôle PACA.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are::
- ELISA
- enzyme-linked immunosorbent assay
- GFP
- green fluorescent protein
- HA
- hemagglutinin
- HPV
- human papillomavirus
- HRG
- heregulin
- HTRF
- homogeneous time-resolved fluorescence
- MAGUK
- membrane-associated guanylate kinase
- MEF
- mouse embryonic fibroblast
- MO
- morpholino-modified antisense oligonucleotide
- PCP
- planar cell polarity
- PDZ
- PSD95-disc large-zonula occludens
- SNX27
- sorting nexin 27
- STREP-HA
- streptavidine-hemagglutinin
- Y2H
- yeast two-hybrid.
REFERENCES
- 1. Pawson T., Gish G. D., Nash P. (2001) SH2 domains, interaction modules and cellular wiring. Trends Cell Biol. 11, 504–511 [DOI] [PubMed] [Google Scholar]
- 2. Scott J. D., Pawson T. (2009) Cell signaling in space and time: where proteins come together and when they're apart. Science 326, 1220–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawson T. (2007) Dynamic control of signaling by modular adaptor proteins. Curr. Opin. Cell Biol. 19, 112–116 [DOI] [PubMed] [Google Scholar]
- 4. Nourry C., Grant S. G., Borg J. P. (2003) PDZ domain proteins: plug and play! Sci. STKE 2003, RE7. [DOI] [PubMed] [Google Scholar]
- 5. Tonikian R., Zhang Y., Sazinsky S. L., Currell B., Yeh J. H., Reva B., Held H. A., Appleton B. A., Evangelista M., Wu Y., Xin X., Chan A. C., Seshagiri S., Lasky L. A., Sander C., Boone C., Bader G. D., Sidhu S. S. (2008) A specificity map for the PDZ domain family. PLoS Biol. 6, e239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Roh M. H., Margolis B. (2003) Composition and function of PDZ protein complexes during cell polarization. Am. J. Physiol. Renal Physiol. 285, F377–F387 [DOI] [PubMed] [Google Scholar]
- 7. Bilder D. (2001) PDZ proteins and polarity: functions from the fly. Trends Genet. 17, 511–519 [DOI] [PubMed] [Google Scholar]
- 8. Kim E., Niethammer M., Rothschild A., Jan Y. N., Sheng M. (1995) Clustering of Shaker-type K+ channels by interaction with a family of membrane-associated guanylate kinases. Nature 378, 85–88 [DOI] [PubMed] [Google Scholar]
- 9. Javier R. T. (2008) Cell polarity proteins: common targets for tumorigenic human viruses. Oncogene 27, 7031–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pearson H. B., Perez-Mancera P. A., Dow L. E., Ryan A., Tennstedt P., Bogani D., Elsum I., Greenfield A., Tuveson D. A., Simon R., Humbert P. O. (2011) SCRIB expression is deregulated in human prostate cancer, and its deficiency in mice promotes prostate neoplasia. J. Clin. Invest. 121, 4257–4267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhan L., Rosenberg A., Bergami K. C., Yu M., Xuan Z., Jaffe A. B., Allred C., Muthuswamy S. K. (2008) Deregulation of scribble promotes mammary tumorigenesis and reveals a role for cell polarity in carcinoma. Cell 135, 865–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Javier R. T., Rice A. P. (2011) Emerging theme: cellular PDZ proteins as common targets of pathogenic viruses. J. Virol. 85, 11544–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Spanos W. C., Hoover A., Harris G. F., Wu S., Strand G. L., Anderson M. E., Klingelhutz A. J., Hendriks W., Bossler A. D., Lee J. H. (2008) The PDZ binding motif of human papillomavirus type 16 E6 induces PTPN13 loss, which allows anchorage-independent growth and synergizes with ras for invasive growth. J. Virol. 82, 2493–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thomas M., Narayan N., Pim D., Tomaic V., Massimi P., Nagasaka K., Kranjec C., Gammoh N., Banks L. (2008) Human papillomaviruses, cervical cancer and cell polarity. Oncogene 27, 7018–7030 [DOI] [PubMed] [Google Scholar]
- 15. Doyle D. A., Lee A., Lewis J., Kim E., Sheng M., MacKinnon R. (1996) Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ. Cell 85, 1067–1076 [DOI] [PubMed] [Google Scholar]
- 16. Morais Cabral J. H., Petosa C., Sutcliffe M. J., Raza S., Byron O., Poy F., Marfatia S. M., Chishti A. H., Liddington R. C. (1996) Crystal structure of a PDZ domain. Nature 382, 649–652 [DOI] [PubMed] [Google Scholar]
- 17. Harris B. Z., Lim W. A. (2001) Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 18. Songyang Z., Fanning A. S., Fu C., Xu J., Marfatia S. M., Chishti A. H., Crompton A., Chan A. C., Anderson J. M., Cantley L. C. (1997) Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science 275, 73–77 [DOI] [PubMed] [Google Scholar]
- 19. Giallourakis C., Cao Z., Green T., Wachtel H., Xie X., Lopez-Illasaca M., Daly M., Rioux J., Xavier R. (2006) A molecular-properties-based approach to understanding PDZ domain proteins and PDZ ligands. Genome Res. 16, 1056–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stiffler M. A., Chen J. R., Grantcharova V. P., Lei Y., Fuchs D., Allen J. E., Zaslavskaia L. A., MacBeath G. (2007) PDZ domain binding selectivity is optimized across the mouse proteome. Science 317, 364–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. te Velthuis A. J., Sakalis P. A., Fowler D. A., Bagowski C. P. (2011) Genome-wide analysis of PDZ domain binding reveals inherent functional overlap within the PDZ interaction network. PLoS One 6, e16047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bezprozvanny I., Maximov A. (2001) Classification of PDZ domains. FEBS Lett. 509, 457–462 [DOI] [PubMed] [Google Scholar]
- 23. Lenfant N., Polanowska J., Bamps S., Omi S., Borg J. P., Reboul J. (2010) A genome-wide study of PDZ-domain interactions in C. elegans reveals a high frequency of non-canonical binding. BMC Genomics 11, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Penkert R. R., DiVittorio H. M., Prehoda K. E. (2004) Internal recognition through PDZ domain plasticity in the Par-6-Pals1 complex. Nat. Struct. Mol. Biol. 11, 1122–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang Y., Appleton B. A., Wiesmann C., Lau T., Costa M., Hannoush R. N., Sidhu S. S. (2009) Inhibition of Wnt signaling by Dishevelled PDZ peptides. Nat. Chem. Biol. 5, 217–219 [DOI] [PubMed] [Google Scholar]
- 26. Zhang Y., Appleton B. A., Wu P., Wiesmann C., Sidhu S. S. (2007) Structural and functional analysis of the ligand specificity of the HtrA2/Omi PDZ domain. Protein Sci. 16, 1738–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van den Berk L. C., Landi E., Walma T., Vuister G. W., Dente L., Hendriks W. J. (2007) An allosteric intramolecular PDZ-PDZ interaction modulates PTP-BL PDZ2 binding specificity. Biochemistry 46, 13629–13637 [DOI] [PubMed] [Google Scholar]
- 28. Chen Y., Sheng R., Kallberg M., Silkov A., Tun M. P., Bhardwaj N., Kurilova S., Hall R. A., Honig B., Lu H., Cho W. (2012) Genome-wide functional annotation of dual-specificity protein- and lipid-binding modules that regulate protein interactions. Mol. Cell, 46, 226–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J. V., Vandekerckhove J., David G., Gettemans J. (2002) PIP(2)-PDZ domain binding controls the association of syntenin with the plasma membrane. Mol. Cell 9, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 30. Schultz J., Copley R. R., Doerks T., Ponting C. P., Bork P. (2000) SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hillier L., Green P. (1991) OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1, 124–128 [DOI] [PubMed] [Google Scholar]
- 32. Hartley J. L., Temple G. F., Brasch M. A. (2000) DNA cloning using in vitro site-specific recombination. Genome Res. 10, 1788–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Walhout A. J., Temple G. F., Brasch M. A., Hartley J. L., Lorson M. A., van den Heuvel S., Vidal M. (2000) GATEWAY recombinational cloning: application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 328, 575–592 [DOI] [PubMed] [Google Scholar]
- 34. Belotti E., Puvirajesinghe T. M., Audebert S., Baudelet E., Camoin L., Pierres M., Lasvaux L., Ferracci G., Montcouquiol M., Borg J. P. (2012) Molecular characterisation of endogenous Vangl2/Vangl1 heteromeric protein complexes. PLoS One 7, e46213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Joubert L., Hanson B., Barthet G., Sebben M., Claeysen S., Hong W., Marin P., Dumuis A., Bockaert J. (2004) New sorting nexin (SNX27) and NHERF specifically interact with the 5-HT4a receptor splice variant: roles in receptor targeting. J. Cell Sci. 117, 5367–5379 [DOI] [PubMed] [Google Scholar]
- 36. Audebert S., Navarro C., Nourry C., Chasserot-Golaz S., Lecine P., Bellaiche Y., Dupont J. L., Premont R. T., Sempere C., Strub J. M., Van Dorsselaer A., Vitale N., Borg J. P. (2004) Mammalian Scribble forms a tight complex with the betaPIX exchange factor. Curr. Biol. 14, 987–995 [DOI] [PubMed] [Google Scholar]
- 37. Nola S., Sebbagh M., Marchetto S., Osmani N., Nourry C., Audebert S., Navarro C., Rachel R., Montcouquiol M., Sans N., Etienne-Manneville S., Borg J. P., Santoni M. J. (2008) Scrib regulates PAK activity during the cell migration process. Hum. Mol. Genet. 17, 3552–3565 [DOI] [PubMed] [Google Scholar]
- 38. Marchal L., Luxardi G., Thome V., Kodjabachian L. (2009) BMP inhibition initiates neural induction via FGF signaling and Zic genes. Proc. Natl. Acad. Sci. U.S.A. 106, 17437–17442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mitchell B., Stubbs J. L., Huisman F., Taborek P., Yu C., Kintner C. (2009) The PCP pathway instructs the planar orientation of ciliated cells in the Xenopus larval skin. Curr. Biol. 19, 924–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Heasman J., Kofron M., Wylie C. (2000) Beta-catenin signaling activity dissected in the early Xenopus embryo: a novel antisense approach. Dev. Biol. 222, 124–134 [DOI] [PubMed] [Google Scholar]
- 41. Nakagawa S., Huibregtse J. M. (2000) Human scribble (Vartul) is targeted for ubiquitin-mediated degradation by the high-risk papillomavirus E6 proteins and the E6AP ubiquitin-protein ligase. Mol. Cell. Biol. 20, 8244–8253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thomas M., Massimi P., Navarro C., Borg J. P., Banks L. (2005) The hScrib/Dlg apico-basal control complex is differentially targeted by HPV-16 and HPV-18 E6 proteins. Oncogene 24, 6222–6230 [DOI] [PubMed] [Google Scholar]
- 43. Storrs C. H., Silverstein S. J. (2007) PATJ, a tight junction-associated PDZ protein, is a novel degradation target of high-risk human papillomavirus E6 and the alternatively spliced isoform 18 E6. J. Virol. 81, 4080–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee S. S., Glaunsinger B., Mantovani F., Banks L., Javier R. T. (2000) Multi-PDZ domain protein MUPP1 is a cellular target for both adenovirus E4-ORF1 and high-risk papillomavirus type 18 E6 oncoproteins. J. Virol. 74, 9680–9693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Venkatesan K., Rual J. F., Vazquez A., Stelzl U., Lemmens I., Hirozane-Kishikawa T., Hao T., Zenkner M., Xin X., Goh K. I., Yildirim M. A., Simonis N., Heinzmann K., Gebreab F., Sahalie J. M., Cevik S., Simon C., de Smet A. S., Dann E., Smolyar A., Vinayagam A., Yu H., Szeto D., Borick H., Dricot A., Klitgord N., Murray R. R., Lin C., Lalowski M., Timm J., Rau K., Boone C., Braun P., Cusick M. E., Roth F. P., Hill D. E., Tavernier J., Wanker E. E., Barabasi A. L., Vidal M. (2009) An empirical framework for binary interactome mapping. Nat. Methods 6, 83–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Favre-Bonvin A., Reynaud C., Kretz-Remy C., Jalinot P. (2005) Human papillomavirus type 18 E6 protein binds the cellular PDZ protein TIP-2/GIPC, which is involved in transforming growth factor beta signaling and triggers its degradation by the proteasome. J. Virol. 79, 4229–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jeong K. W., Kim H. Z., Kim S., Kim Y. S., Choe J. (2007) Human papillomavirus type 16 E6 protein interacts with cystic fibrosis transmembrane regulator-associated ligand and promotes E6-associated protein-mediated ubiquitination and proteasomal degradation. Oncogene 26, 487–499 [DOI] [PubMed] [Google Scholar]
- 49. Caruana G. (2002) Genetic studies define MAGUK proteins as regulators of epithelial cell polarity. Int. J. Dev. Biol. 46, 511–518 [PubMed] [Google Scholar]
- 50. Blake D. J., Hawkes R., Benson M. A., Beesley P. W. (1999) Different dystrophin-like complexes are expressed in neurons and glia. J. Cell Biol. 147, 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sugihara-Mizuno Y., Adachi M., Kobayashi Y., Hamazaki Y., Nishimura M., Imai T., Furuse M., Tsukita S. (2007) Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells 12, 473–486 [DOI] [PubMed] [Google Scholar]
- 52. Garcia R. A., Vasudevan K., Buonanno A. (2000) The neuregulin receptor ErbB-4 interacts with PDZ-containing proteins at neuronal synapses. Proc. Natl. Acad. Sci. U.S.A. 97, 3596–3601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Maudsley S., Zamah A. M., Rahman N., Blitzer J. T., Luttrell L. M., Lefkowitz R. J., Hall R. A. (2000) Platelet-derived growth factor receptor association with Na(+)/H(+) exchanger regulatory factor potentiates receptor activity. Mol. Cell. Biol. 20, 8352–8363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Park M., Moon R. T. (2002) The planar cell-polarity gene stbm regulates cell behaviour and cell fate in vertebrate embryos. Nat. Cell Biol. 4, 20–25 [DOI] [PubMed] [Google Scholar]
- 55. Wolff T., Rubin G. M. (1998) Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development 125, 1149–1159 [DOI] [PubMed] [Google Scholar]
- 56. Montcouquiol M., Rachel R. A., Lanford P. J., Copeland N. G., Jenkins N. A., Kelley M. W. (2003) Identification of Vangl2 and Scrb1 as planar polarity genes in mammals. Nature 423, 173–177 [DOI] [PubMed] [Google Scholar]
- 57. Torban E., Kor C., Gros P. (2004) Van Gogh-like2 (Strabismus) and its role in planar cell polarity and convergent extension in vertebrates. Trends Genet. 20, 570–577 [DOI] [PubMed] [Google Scholar]
- 58. Torban E., Wang H. J., Groulx N., Gros P. (2004) Independent mutations in mouse Vangl2 that cause neural tube defects in looptail mice impair interaction with members of the Dishevelled family. J. Biol. Chem. 279, 52703–52713 [DOI] [PubMed] [Google Scholar]
- 59. Wallingford J. B., Mitchell B. (2011) Strange as it may seem: the many links between Wnt signaling, planar cell polarity, and cilia. Genes Dev. 25, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kallay L. M., McNickle A., Brennwald P. J., Hubbard A. L., Braiterman L. T. (2006) Scribble associates with two polarity proteins, Lgl2 and Vangl2, via distinct molecular domains. J. Cell. Biochem. 99, 647–664 [DOI] [PubMed] [Google Scholar]
- 61. Babayeva S., Zilber Y., Torban E. (2011) Planar cell polarity pathway regulates actin rearrangement, cell shape, motility, and nephrin distribution in podocytes. Am. J. Physiol. Renal Physiol. 300, F549–F560 [DOI] [PubMed] [Google Scholar]
- 62. Lauffer B. E., Melero C., Temkin P., Lei C., Hong W., Kortemme T., von Zastrow M. (2010) SNX27 mediates PDZ-directed sorting from endosomes to the plasma membrane. J. Cell Biol. 190, 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Temkin P., Lauffer B., Jager S., Cimermancic P., Krogan N. J., von Zastrow M. (2011) SNX27 mediates retromer tubule entry and endosome-to-plasma membrane trafficking of signalling receptors. Nat. Cell Biol. 13, 715–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Valdembri D., Caswell P. T., Anderson K. I., Schwarz J. P., Konig I., Astanina E., Caccavari F., Norman J. C., Humphries M. J., Bussolino F., Serini G. (2009) Neuropilin-1/GIPC1 signaling regulates alpha5beta1 integrin traffic and function in endothelial cells. PLoS Biol. 7, e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Angers S., Thorpe C. J., Biechele T. L., Goldenberg S. J., Zheng N., MacCoss M. J., Moon R. T. (2006) The KLHL12-Cullin-3 ubiquitin ligase negatively regulates the Wnt-beta-catenin pathway by targeting Dishevelled for degradation. Nat. Cell Biol. 8, 348–357 [DOI] [PubMed] [Google Scholar]
- 66. Daulat A. M., Maurice P., Froment C., Guillaume J. L., Broussard C., Monsarrat B., Delagrange P., Jockers R. (2007) Purification and identification of G protein-coupled receptor protein complexes under native conditions. Mol. Cell. Proteomics 6, 835–844 [DOI] [PubMed] [Google Scholar]
- 67. Bilder D., Perrimon N. (2000) Localization of apical epithelial determinants by the basolateral PDZ protein Scribble. Nature 403, 676–680 [DOI] [PubMed] [Google Scholar]
- 68. Dow L. E., Kauffman J. S., Caddy J., Zarbalis K., Peterson A. S., Jane S. M., Russell S. M., Humbert P. O. (2007) The tumour-suppressor Scribble dictates cell polarity during directed epithelial migration: regulation of Rho GTPase recruitment to the leading edge. Oncogene 26, 2272–2282 [DOI] [PubMed] [Google Scholar]
- 69. Montcouquiol M., Sans N., Huss D., Kach J., Dickman J. D., Forge A., Rachel R. A., Copeland N. G., Jenkins N. A., Bogani D., Murdoch J., Warchol M. E., Wenthold R. J., Kelley M. W. (2006) Asymmetric localization of Vangl2 and Fz3 indicate novel mechanisms for planar cell polarity in mammals. J. Neurosci. 26, 5265–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wallingford J. B., Niswander L. A., Shaw G. M., Finnell R. H. (2013) The continuing challenge of understanding, preventing, and treating neural tube defects. Science 339, 1222002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wallingford J. B., Harland R. M. (2002) Neural tube closure requires Dishevelled-dependent convergent extension of the midline. Development 129, 5815–5825 [DOI] [PubMed] [Google Scholar]
- 72. Ohno S. (2001) Intercellular junctions and cellular polarity: the PAR-aPKC complex, a conserved core cassette playing fundamental roles in cell polarity. Curr. Opin. Cell Biol. 13, 641–648 [DOI] [PubMed] [Google Scholar]
- 73. Etienne-Manneville S. (2004) Cdc42—the centre of polarity. J. Cell Sci. 117, 1291–1300 [DOI] [PubMed] [Google Scholar]
- 74. McCaffrey L. M., Macara I. G. (2011) Epithelial organization, cell polarity and tumorigenesis. Trends Cell Biol. 21, 727–735 [DOI] [PubMed] [Google Scholar]
- 75. Kingston D., Chang H., Ensser A., Lee H. R., Lee J., Lee S. H., Jung J. U., Cho N. H. (2011) Inhibition of retromer activity by herpesvirus saimiri tip leads to CD4 downregulation and efficient T cell transformation. J. Virol. 85, 10627–10638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Scott K. L., Kabbarah O., Liang M. C., Ivanova E., Anagnostou V., Wu J., Dhakal S., Wu M., Chen S., Feinberg T., Huang J., Saci A., Widlund H. R., Fisher D. E., Xiao Y., Rimm D. L., Protopopov A., Wong K. K., Chin L. (2009) GOLPH3 modulates mTOR signalling and rapamycin sensitivity in cancer. Nature 459, 1085–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Mukherjee K., Sharma M., Urlaub H., Bourenkov G. P., Jahn R., Sudhof T. C., Wahl M. C. (2008) CASK functions as a Mg2+-independent neurexin kinase. Cell 133, 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Anastas J. N., Biechele T. L., Robitaille M., Muster J., Allison K. H., Angers S., Moon R. T. (2012) A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 31, 3696–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Luga V., Zhang L., Viloria-Petit A. M., Ogunjimi A. A., Inanlou M. R., Chiu E., Buchanan M., Hosein A. N., Basik M., Wrana J. L. (2012) Exosomes mediate stromal mobilization of autocrine Wnt-PCP signaling in breast cancer cell migration. Cell 151, 1542–1556 [DOI] [PubMed] [Google Scholar]
- 80. Williams B. B., Cantrell V. A., Mundell N. A., Bennett A. C., Quick R. E., Jessen J. R. (2012) VANGL2 regulates membrane trafficking of MMP14 to control cell polarity and migration. J. Cell Sci. 125 (Pt9), 2141–2147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Wansleeben C., Feitsma H., Montcouquiol M., Kroon C., Cuppen E., Meijlink F. (2010) Planar cell polarity defects and defective Vangl2 trafficking in mutants for the COPII gene Sec24b. Development 137, 1067–1073 [DOI] [PubMed] [Google Scholar]
- 82. Cai L., Loo L. S., Atlashkin V., Hanson B. J., Hong W. (2011) Deficiency of sorting nexin 27 (SNX27) leads to growth retardation and elevated levels of N-methyl-D-aspartate receptor 2C (NR2C). Mol. Cell. Biol. 31, 1734–1747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Giese A. P., Ezan J., Wang L., Lasvaux L., Lembo F., Mazzocco C., Richard E., Reboul J., Borg J. P., Kelley M. W., Sans N., Brigande J., Montcouquiol M. (2012) Gipc1 has a dual role in Vangl2 trafficking and hair bundle integrity in the inner ear. Development 139, 3775–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Pangon L., Van Kralingen C., Abas M., Daly R. J., Musgrove E. A., Kohonen-Corish M. R. (2012) The PDZ-binding motif of MCC is phosphorylated at position -1 and controls lamellipodia formation in colon epithelial cells. Biochim. Biophys. Acta 1823, 1058–1067 [DOI] [PubMed] [Google Scholar]
- 85. Grootjans J. J., Reekmans G., Ceulemans H., David G. (2000) Syntenin-syndecan binding requires syndecan-synteny and the co-operation of both PDZ domains of syntenin. J. Biol. Chem. 275, 19933–19941 [DOI] [PubMed] [Google Scholar]
- 86. Ivarsson Y., Wawrzyniak A. M., Kashyap R., Polanowska J., Betzi S., Lembo F., Vermeiren E., Chiheb D., Lenfant N., Morelli X., Borg J. P., Reboul J., Zimmermann P. (2013) Prevalence, specificity and determinants of lipid-interacting PDZ domains from an in-cell screen and in vitro binding experiments. PLoS One 8, e54581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.