Abstract
Fibroblast growth factor 21 (FGF21) is a hepatokine that acts as a global starvation signal to modulate fuel partitioning and metabolism, and repress growth1; however the site of action of these diverse effects remains unclear. FGF21 signals through a heteromeric cell surface receptor composed of one of three FGF receptors (FGFR1c, 2c, or 3c) in complex with β-Klotho2-4, a single-pass transmembrane protein that is enriched in metabolic tissues5. Here we show that in addition to its known effects on peripheral metabolism, FGF21 increases systemic glucocorticoid levels, suppresses physical activity, and alters circadian behavior, all features of the adaptive starvation response. These effects are mediated through β-Klotho expression in the suprachiasmatic nucleus (SCN) of the hypothalamus and the dorsal vagal complex (DVC) of the hindbrain. Mice lacking the β-Klotho gene (Klb) in these regions are refractory to these effects, as well as those on metabolism, insulin, and growth. These findings demonstrate a crucial role for the nervous system in mediating the diverse physiologic and pharmacologic actions of FGF21.
Previous studies showed that FGF21 can cross the blood-brain barrier6, is detectable in human cerebrospinal fluid7, and modulates phosphorylation cascades and gene expression in whole hypothalamus8,9. Intracerebroventricular injection of FGF21 into rats increases metabolic rate and insulin sensitivity10, although the site of action is unknown. To determine how and where FGF21 may be acting centrally, we first determined the expression of Klb in the nervous system using anatomically-guided laser capture microdissection (LCM) of specific hypothalamic and brainstem nuclei, and sensory ganglia from parasympathetic and sympathetic divisions of the peripheral nervous system. Quantitative PCR (QPCR) showed that Klb is selectively expressed in the hypothalamus in the SCN (Fig. 1a), the site of a circadian pacemaker that orchestrates entrainment of animals to their external environment and gates the timing of diverse behavioral, physiological, and hormonal rhythms11-14. We found that Klb is also expressed in the hindbrain in key sets of neurons of the DVC, including the area postrema (AP) and the nucleus tractus solitarii (NTS), and in the nodose ganglia, where vagal nerve sensory neurons reside (Fig. 1a). The components of the vagus nerve and its targets in the brainstem comprise a sensory unit that receives and integrates information from the viscera regarding metabolic and physiologic status15. The localization of Klb in the SCN and DVC was confirmed by in situ hybridization (Fig. 1b; Supplementary Fig. 1). In contrast to Klb, Fgfr1c, Fgfr2c, and Fgfr3c are broadly expressed in the nervous system (Fig. 1a). Fgf21 mRNA is undetectable in any of these regions (data not shown).
Figure 1.
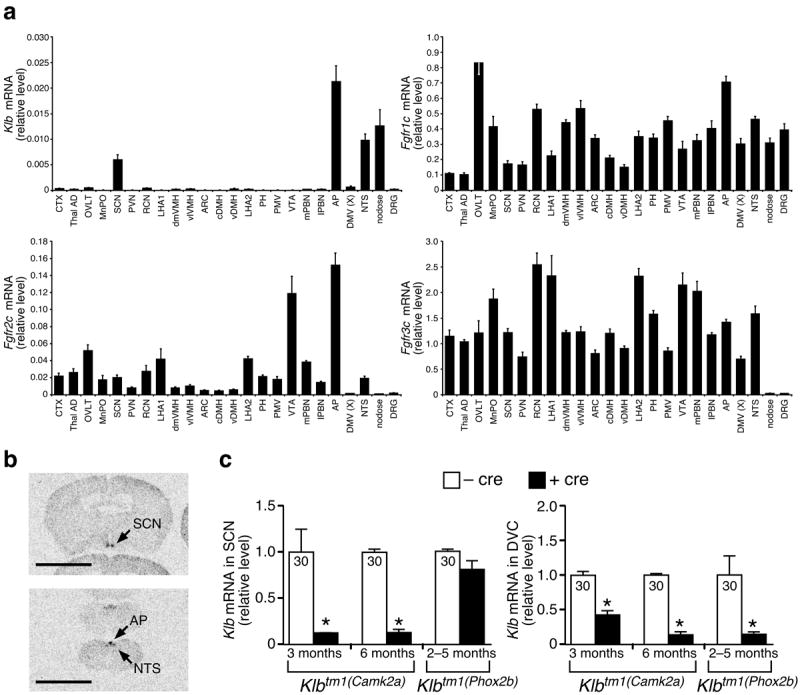
Klb and Fgfr expression in the nervous system. (a) LCM mRNA expression in nonhypothalamic (CTX, Thal AD), hypothalamic (OVLT to PMV), midbrain (VTA to mPBN), hindbrain (AP to NTS), and peripheral nervous system (nodose, DRG) regions of male C57BL/6J mice (n = 4–5) harvested at the start of the light phase (see Supplementary Table 4 for abbreviations). (b) In situ hybridization of Klb from hypothalamus (top) and hindbrain (bottom) coronal sections (see Supplementary Fig. 1 for full panel of coronal sections; bar = 45 mm). (c) Klb expression in blunt-dissected SCN and DVC from Klbtm1(Camk2a) and Klbtm1(Phox2b) mice. Cycle time (Ct) values shown inside bars. Data represent the mean ± SEM. Asterisks indicate significant differences (P < 0.05) relative to –cre controls.
Since Klb localized to the SCN, we evaluated the circadian effect of long-term FGF21 exposure on gene expression and metabolic parameters using Tg(Fgf21) mice, in which an Fgf21 transgene is under the control of the liver-selective apolipoprotein E promoter16. Plasma FGF21 levels varied between 1000 and 2000 ng ml−1 in Tg(Fgf21) mice (Supplementary Fig. 2a). In hypothalamus, the circadian pattern of Klb expression was unchanged in Tg(Fgf21) mice (Supplementary Fig. 2b). There also was no change in the expression of Fgfr1c or the circadian genes, Bmal1, Clock and Per2, in the hypothalamus of Tg(Fgf21) mice (Supplementary Fig. 2b). In liver, Klb was consistently higher in the Tg(Fgf21) compared to wild-type mice (Supplementary Fig. 2c). There were modest changes in the expression of the clock genes Bmal1, Clock and Per2 in liver, which are likely a consequence of changes in SCN output and corticosterone levels (see below). Liver expression of insulin-like growth factor binding protein-1 (Igfbp1), cytochrome P450 (Cyp) 2d9 and hydroxysteroid dehydrogenase (Hsd) 3b5, which are regulated by growth hormone, was reciprocally regulated in Tg(Fgf21) mice throughout the cycle (Supplementary Fig. 2c). Plasma insulin, glucose, triglyceride and cholesterol concentrations were consistently lower in Tg(Fgf21) mice throughout the light cycle (Supplementary Fig. 2a). Taken together, these data indicate that FGF21 does not affect central clock genes but might affect SCN output.
To test directly whether the SCN is a target of FGF21 action, we deleted the Klb gene from the SCN by crossing Klbflox/flox (Klbtm1) mice17 with a calcium/calmodulin-dependent protein kinase IIa (Camk2a)-Cre line in which cre recombinase is expressed widely in the forebrain and hindbrain18. In the resulting Klbtm1(Camk2a) mice, Klb mRNA expression was reduced in both the SCN and DVC to 15% of wild-type levels by 6 months of age (Fig. 1c). Klb expression was unaffected in liver and adipose tissue (Supplementary Fig. 3a). To control for deletion of Klb in the DVC, we crossed Klbtm1 mice with a Phox2b-Cre line that is expressed in the hindbrain19, including neurons in the dorsal motor nucleus of the vagus, NTS, AP, and nodose ganglia. In the resulting Klbtm1(Phox2b) mice, Klb expression was lower in the DVC but not in the SCN, liver or adipose tissues compared to Klbtm1 controls (Fig. 1c; Supplementary Fig. 3b).
To study the effects of FGF21 on the nervous system, Klbtm1(Camk2a) or Klbtm1(Phox2b) mice were bred to the Tg(Fgf21) mice. Plasma FGF21 concentrations were elevated to similar levels in all Tg(Fgf21) mouse lines (Supplementary Table 1). Since the primary function of the SCN is to drive circadian behavior and to entrain these rhythms to light cycles, we measured the effect of FGF21 overexpression on daily wheel-running behavior under various lighting conditions. Representative actograms for wild-type and Tg(Fgf21) mice are shown (Fig. 2a, 2b, and Supplementary Fig. 4a). Tg(Fgf21) mice exhibited significantly lower total wheel-running activity compared to wild-type mice (Fig. 2c, 2d; Supplementary Table 2), resulting in lower amplitude rhythms as assessed by fast Fourier transform (FFT) analysis (Supplementary Table 2). Additionally, the Tg(Fgf21) mice failed to consolidate their running to the dark phase (Fig. 2c, 2d; Supplementary Table 2). Overall, activity remained nocturnal in both wild-type and Tg(Fgf21) mice (Fig. 2c; Supplementary Table 2), demonstrating that entrainment to light was intact, although the phase angle of entrainment was advanced for the Tg(Fgf21) mice. To determine whether the pace of the circadian clock was affected, we measured the free-running period length of the activity rhythm in constant darkness. The circadian period for all mice was identical (23.5 ± 0.1 hours; Supplementary Table 2), demonstrating that the behavioral alterations seen in the Tg(Fgf21) mice are likely due to an inhibition of clock output rather than an effect on the molecular clock per se.
Figure 2.
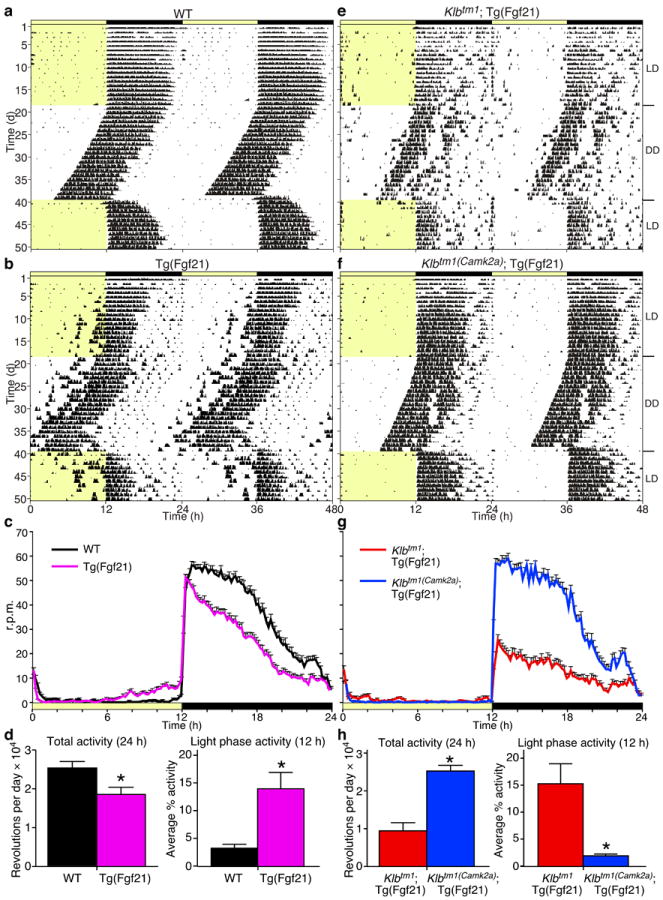
Transgenic overexpression of FGF21 alters wheel-running behavior through the SCN. Representative actograms are shown double-plotted for (a) wild-type (WT), (b) Tg(Fgf21), (e) Klbtm1∷Tg(Fgf21), and (f) Klbtm1(Camk2a)∷Tg(Fgf21) males. Yellow indicates light phase (LD, 12 hours light/12 hours dark; DD, constant darkness). (c, g) Average wave plots summarize wheel-running activity during days 6–15 of LD for indicated genotypes (n = 16–28). Time on x-axis refers to zeitgeber time (ZT) 0 at lights on. (d, h) Daily total and % light phase wheel-running activity for the indicated genotypes are plotted as mean ± SEM (*P < 0.05).
To assess whether the behavioral alterations observed for Tg(Fgf21) mice were mediated by the SCN, we evaluated Klbtm1∷Tg(Fgf21) and Klbtm1(Camk2a)∷Tg(Fgf21) mice on running wheels. Compared to wild-type mice and similar to Tg(Fgf21) mice, Klbtm1∷Tg(Fgf21) animals exhibited lower overall activity, reduced robustness of rhythm, and higher running during the light phase (Fig. 2e, 2h; Supplementary Fig. 4b; Supplementary Table 2). Deletion of β-Klotho in the nervous system normalized these parameters (Fig. 2f, 2g, 2h; Supplementary Fig. 4b; Supplementary Table 2) demonstrating the importance of the SCN in mediating these behaviors in response to chronically elevated FGF21. These data provide further evidence that elevated FGF21 relaxes the inhibitory function of the SCN on activity during the light phase.
To test whether endogenous FGF21 regulates circadian behaviors, we performed wheel-running experiments with wild-type and Fgf21−/− mice fed a ketogenic diet. Although it was not technically possible to administer FGF21 to mice while they are in the wheel-running experiments, the ketogenic diet offers an alternative method by mimicking the starvation response and increasing Fgf21 mRNA in liver and FGF21 concentrations in blood to near fasting levels (ref. 20; Supplementary Fig. 5a). In our experiments, the ketogenic diet increased plasma FGF21 levels to 10–30 ng ml−1 (Supplementary Fig. 5a; Supplementary Table 3). When challenged with the ketogenic diet, wild-type mice reduced overall activity while increasing activity during the light phase (Fig. 3a–c, 3g, 3h; Supplementary Fig. 5b; Supplementary Table 3), similar to that seen in Tg(Fgf21) mice (Fig. 2c). Chow-fed Fgf21−/− mice behaved like their wild-type counterparts (Fig. 3d). While total activity was similarly lower in wild-type and Fgf21−/− mice in response to the ketogenic diet (Fig. 3e–g; Supplementary Table 3), the diet-induced increase in light phase activity was significantly attenuated in Fgf21−/− mice (Fig. 3h; Supplementary Table 3; Supplementary Fig. 5b). Thus, physiologic levels of FGF21 can modulate circadian behavior.
Figure 3.
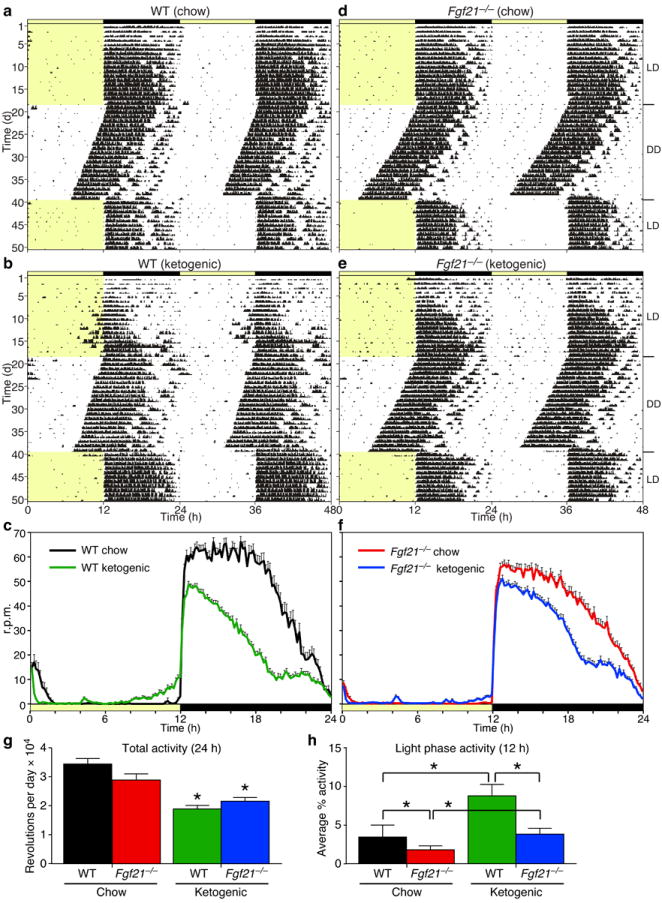
Endogenous FGF21 alters wheel-running behavior. Representative actograms are shown double-plotted for wild-type (WT) (a, b) and Fgf21−/− (d, e) males on standard chow (a, d) or ketogenic (b, e) diets. Yellow indicates light phase (LD, 12 hours light/12 hours dark; DD, constant darkness). (c, f) Average wave plots summarize wheel-running activity during days 6–15 of LD for indicated genotypes (n = 6–24). Time on x-axis refers to ZT0 at lights on. (g, h) Daily total and % light phase wheel-running activity for the indicated genotypes are plotted as mean ± SEM. Asterisks indicate significant differences (P < 0.05) compared to chow diets in (g) and as indicated in (h).
We next evaluated the consequence of eliminating β-Klotho from the nervous system on metabolic parameters. Neither the Klbtm1(Camk2a) or Klbtm1(Phox2b) mice had changes in body weight or other metabolic parameters under ad libitum chow-fed conditions (Fig 4a; Supplementary Table 1). As expected16,21, Klbtm1∷Tg(Fgf21) mice had significantly lower body weight, tibia length and plasma insulin and cholesterol concentrations than control Klbtm1 mice but no change in percent lean mass or fat mass (Fig. 4a; Supplementary Fig. 6a; Supplementary Table 1). Notably, the changes in body weight, tibia length and plasma insulin and cholesterol concentrations observed in Klbtm1∷Tg(Fgf21) mice were reversed in Klbtm1(Camk2a)∷Tg(Fgf21) mice (Fig. 4a; Supplementary Fig. 6a; Supplementary Table 1). These changes were not reversed in Klbtm1(Phox2b)∷Tg(Fgf21) mice with the exception of cholesterol concentrations (Fig. 4a; Supplementary Table 1). These data indicate that FGF21 action on the SCN mediates the effects of FGF21 on body weight and insulin levels.
Figure 4.
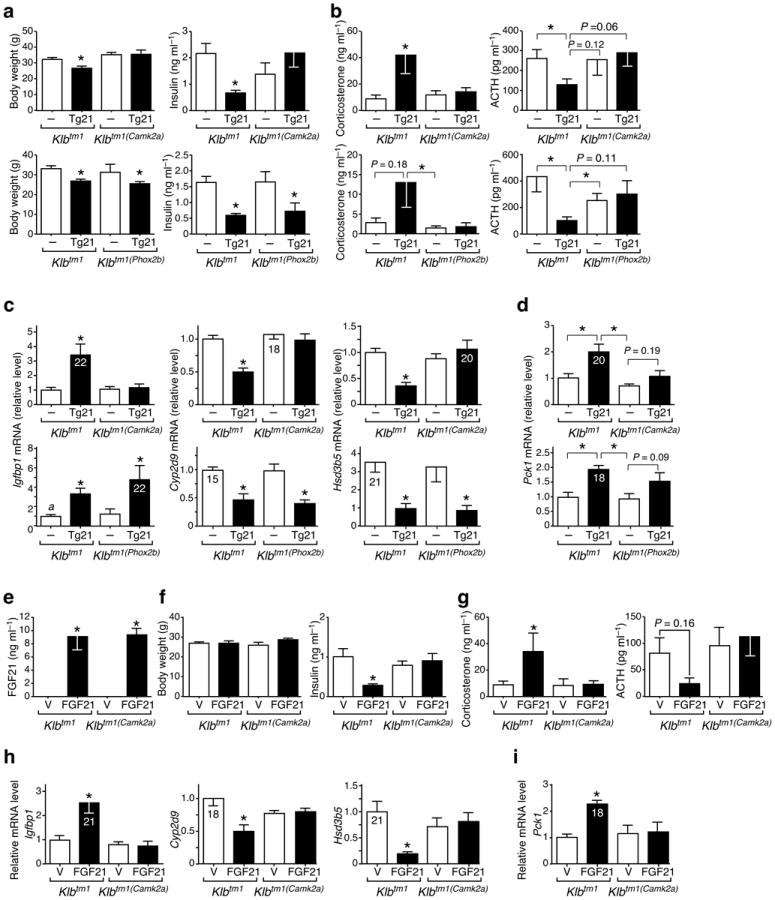
Klb expression in the SCN is required for FGF21 suppression of growth and insulin, and stimulation of glucocorticoid activity. (a – d) Male mice of indicated genotypes without (–) or with (Tg21) the Tg(Fgf21). (e – i) Male mice of indicated genotypes implanted with osmotic mini-pumps delivering vehicle (V) or human FGF21. Animals were sacrificed at ZT8 (a, c), ZT3 (e – i), or at ZT0 (b, d), and body weight, plasma hormone levels, and liver gene expression were determined. Data represent mean ± SEM, n = 5–9 for (a – d) and n = 6 for (e – i). Ct values shown inside bars. Asterisks indicate significant differences (P < 0.05) compared to (–) or (V) controls, or as indicated.
Since glucocorticoids play a crucial role in the starvation response and are regulated by both the SCN and hindbrain 22,23, we examined glucocorticoid homeostasis in Tg(Fgf21) mice. Consistent with lowered SCN clock output, arginine vasopressin (Avp) expression was lower and corticotropin releasing hormone (Crh) expression was higher in hypothalamus of Tg(Fgf21) mice compared to wild-type mice (Supplementary Fig. 2b). These changes were accompanied by higher concentrations of plasma corticosterone and lower concentrations of adrenocorticotropic hormone (ACTH) (Supplementary Fig. 2a). In Klbtm1∷Tg(Fgf21) mice, plasma corticosterone was ~4-fold higher compared to control Klbtm1 mice, and this effect was lost in Klbtm1(Camk2a)∷Tg(Fgf21) mice (Fig. 4b). Plasma ACTH concentrations were regulated in a reciprocal manner (Fig. 4b), indicating that feedback regulation at the level of the pituitary is intact in the Klbtm1∷Tg(Fgf21) mice. A similar pattern of corticosterone and ACTH regulation was seen in experiments performed with Klbtm1(Phox2b) and Klbtm1(Phox2b)∷Tg(Fgf21) mice, although the plasma corticosterone in Klbtm1∷Tg(Fgf21) mice did not reach statistically higher levels relative to Klbtm1 control mice in these experiments (Fig. 4b). Together, these data indicate that FGF21 acts on both the SCN and hindbrain regions of the nervous system to increase corticosterone levels.
We next assessed the consequences of eliminating β-Klotho from the SCN on FGF21-regulated gene expression in liver and adipose tissue. In liver, Klbtm1∷Tg(Fgf21) mice had higher Igfbp1 mRNA levels, and lower Cyp2d9 and Hsd3b5 mRNA levels compared to Klbtm1 control mice (Fig. 4c). The effect of the Fgf21 transgene on the expression of these genes was lost in Klbtm1(Camk2a)∷Tg(Fgf21) mice but not in Klbtm1(Phox2b)∷Tg(Fgf21) mice (Fig. 4c), which is consistent with FGF21 acting on the SCN to regulate growth. Hepatic expression of the gluconeogenic gene, phosphoenolpyruvate carboxykinase (Pck1), which is induced by glucocorticoids, was higher in Klbtm1∷Tg(Fgf21) mice compared to those without Tg(Fgf21) (Fig. 4d). Consistent with the FGF21-dependent CNS effects on plasma corticosterone, Pck1 expression was unchanged in Klbtm1(Camk2a)∷Tg(Fgf21) mice and trended lower in Klbtm1(Phox2b)∷Tg(Fgf21) mice (Fig. 4d). In adipose tissue, adipose triglyceride lipase (Atgl), stearoyl-Coenzyme A desaturase 1 (Scd1), and hydroxysteroid dehydrogenase 11b1 (Hsd11b1) were higher in Klbtm1∷Tg(Fgf21) mice compared to Klbtm1 control mice (Supplementary Fig. 6b). The FGF21-dependent expression of these genes was unaffected in both the Klbtm1(Camk2a)∷Tg(Fgf21) and Klbtm1(Phox2b)∷Tg(Fgf21) mice (Supplementary Fig. 6b), consistent with the notion that FGF21 acts directly on white adipose tissue to regulate their expression17.
Finally, given the pharmacologic levels of circulating FGF21 in the Tg(Fgf21) mice, we performed an experiment in which physiologic levels of FGF21 were delivered by mini-pump for 7 days to Klbtm1 and Klbtm1(Camk2a) mice. Under these conditions, steady-state plasma FGF21 concentrations were ~9 ng ml−1 (Fig. 4e), which is comparable to peak levels obtained during starvation (ref. 19; Supplementary Fig. 6c) and lower than those obtained by ketogenic diet feeding (ref. 20; Supplementary Fig. 5a). There was no effect of FGF21 administration on body weight (Fig. 4f). In agreement with the Tg(Fgf21) studies, FGF21 reduced basal plasma insulin concentrations in Klbtm1 but not Klbtm1(Camk2a) mice (Fig. 4f). Importantly, FGF21 significantly increased plasma corticosterone levels in Klbtm1 but not Klbtm1(Camk2a) mice (Fig. 4g). In gene expression studies, FGF21 increased Igfbp1 and Pck1 expression and decreased Cyp2d6 and Hsd3b5 expression in livers of Klbtm1 but not Klbtm1(Camk2a) mice (Fig. 4h, 4i). These data recapitulate those seen in the Tg(Fgf21) studies and demonstrate that physiologic concentrations of FGF21 act on the nervous system to regulate metabolism.
In summary, we show that FGF21 acts directly through its co-receptor β-Klotho in the brain to lower insulin, inhibit growth, and alter light/dark cycle activity, all of which occur during starvation24,25. Coincident with this role, we show that FGF21 acts on the nervous system, both at the level of the hypothalamus and the hindbrain, to increase systemic corticosterone levels, another prominent feature of starvation. This latter finding may explain the liver nonautonomous inductive effect of FGF21 on hepatic gluconeogenesis26. In an accompanying paper (Owen et al.), we show that FGF21 also acts as a starvation signal to inhibit female fertility. These effects on circadian behavior, glucocorticoids, metabolism, and reproduction are consistent with our results showing that FGF21 suppresses the expression of the neuropeptide vasopressin in the SCN (Supplementary Fig. 2b; Owen et al.). The finding that diverse effects of FGF21 are mediated by suppressing the output of the SCN (and by acting on the DVC) reveals a previously unrecognized regulatory circuit between the liver and brain. Although not typically regarded as being influenced by diet, the SCN expresses receptors for other metabolic factors, including leptin and ghrelin27,28. Thus, the SCN appears to play a broad role in gating adaptive responses to nutritional status. Finally, the observation that, in addition to starvation, FGF21 is elevated in conditions related to the metabolic syndrome29 raises the possibility that FGF21 working through the nervous system may play a part in mediating some of the established effects of the metabolic syndrome on circadian and metabolic parameters.
Online Methods
Animals
All procedures and use of animals were approved by the Institutional Animal Care and Use Committee of UT Southwestern Medical Center Dallas. Unless otherwise stated in specific dietary studies, all animals were maintained on 2916 Global Diet (Harlan Teklad). Ketogenic diet was from BioServ F3666. Male mice were used in all studies. C57BL/6J mice used for LCM and in situ hybridization were obtained from Jackson or from the UT Southwestern mouse breeding core, which sources its colony breeders from Jackson. The C57BL/6J Tg(Fgf21), C57BL/6J Fgf21−/−, C57BL/6J;129/Sv Klbtm1, and C57BL/6J Camk2a-cre lines have been previously described16-18,26. The C57BL/6J Phox2b-cre line 4 expresses cre recombinase in the hindbrain to a greater extent than line 3, in addition to some hypothalamic (excluding the SCN) and cortical sites19. Animals were sacrificed at times indicated by rapid decapitation to avoid stress responses. Trunk blood was collected into K+EDTA tubes, and plasma was separated and stored at −80 °C until assayed. Tissues were collected, snap-frozen in liquid nitrogen, and stored at −80 °C until assayed. For mini-pump experiments, Alzet micro-osmotic pumps model 1007D were filled with vehicle (25 mM HEPES, 150 mM NaCl, 50% glycerol) or recombinant human FGF21 (to deliver 1.1 μg in 0.5 μl per hour) and implanted subcutaneously under isofluorane inhalation anaesthesia. Animals were sacrificed by decapitation at ZT3, 7-days after implantation.
Neuroanatomical Mapping
Laser Capture Microdissection and Peripheral Ganglia
At the start of the light cycle, C57BL/6J males were deeply anesthetized with chloral hydrate and decapitated. Left nodose ganglion and mid-thoracic dorsal root ganglia were rapidly collected and frozen in liquid nitrogen. Brains were dissected, slow-frozen on dry ice, and stored at −80 °C until sectioning. Brains were cryosectioned at 30 μm and thaw-mounted onto silanecoated PEN membrane glass slides (Molecular Devices, Sunnyvale, CA) and stored at −80 °C. Slides were lightly fixed in 75% ethanol immediately prior to thionin staining. Slides were then dehydrated in a graded ethanol series followed by 5 minutes in xylenes. The Arcturus Veritas Microdissection System (Molecular Devices) was used to isolate each nucleus based on defined neuroanatomical boundaries30 as described previously31. Nuclei collected are listed in Supplementary Table 4.
Peripheral ganglia RNA was isolated using Ambion’s Ribopure kit, while LCM nuclei RNA was extracted using the PicoPure RNA Isolation Kit (Molecular Devices) with on-column DNAseI treatment (Qiagen, Valencia, CA), and stored at −80 °C. RNA quality was assessed using the Experion Automated Electrophoresis system (Bio-Rad, Hercules, CA).
cDNA was synthesized using the High Capacity cDNA Kit (Applied Biosystems), followed by cDNA pre-amplification of specific gene targets using TaqMan Preamplification Buffer (Applied Biosystems), according to the manufacturer’s directions. Due to sufficiently high expression, it was not necessary to pre-amplify 18S rRNA, which was used as the normalizer gene. All gene expression levels were measured with an Applied Biosystems 7900HT Sequence Detection System using the efficiency-corrected ΔCt method as previously described31,32. The anatomic specificity of the LCM dissections was confirmed by QPCR for the expression of known marker genes in each nucleus.
In situ hybridization
C57BL/6J males were deeply anesthetized with chloral hydrate and transcardially perfused with saline, followed by formalin. Brains were dissected and post-fixed in formalin for several hours prior to overnight incubation in 20% sucrose. Brains were cut at 25 μm, 1:4 series using a sliding microtome, and sections stored in cryoprotectant at −20 °C until use. Free-floating in situ hybridization was carried out as described28 using a 33P-labelled anti-sense probe for Klb cDNA (spanning base pairs 169–692). Sections were mounted onto SuperFrost plus slides, rostral to caudal and exposed to film for approximately 4 days. Developed film was scanned using an Epson ImageScanner III.
Quantitative Real Time PCR
Total RNA from liver, white, and brown adipose was isolated using RNA Stat60 (Teltest), and cDNA was synthesized using the High Capacity cDNA Kit (Applied Biosystems). Gene expression profiles of peripheral tissues were performed using the ΔΔCt assay as described32.
Blood Analyses
Glucose, ketones, cholesterol, and triglycerides were measured by colorimetric assays (Wako, Pointe Scientific); corticosterone and ACTH by radioimmunoassay (MP Biomedical); insulin and mouse or human FGF21 by ELISA (Crystal Chem, Biovendor), according to manufacturers’ instructions17,26,33.
Wheel-Running
Wheel-running experiments were performed as described34. Singly-housed animals were entrained to a 12 hour light, 12 hour dark cycle (LD12:12) for 18 days, followed by 21 days of constant darkness (DD), before return to LD12:12 for 15 days. Data were analyzed using ClockLab35. Total daily activity, % light, and % dark counts were taken from days 6–15 of LD recording. Period and amplitude were calculated from days 29 to 38 during DD. Phase was determined using activity onsets on days 22 to 28 with extrapolation to the first day of DD (day 19).
Statistics
Data are presented as mean ± SEM. GraphPad Prism was used for ANOVA with Bonferroni’s post-hoc test or two-tailed Student’s t-test. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Y. Zhang, K. Vale, and E. Borowicz for help with animal studies; X. Wang and Y. Wan for tibia measurements; D. Lauzon and C. Lee for in situ hybridization; M. Izumo for Camk2a-cre mice; M. Scott for Phox2b-cre mice; C. Cummins for advice on glucocorticoid and ACTH assays; and all members of the Takahashi, Elmquist, and Mango/Kliewer laboratories for critical discussions. This work was supported by US National Institutes of Health grants DK067158, P20RR20691, and 1RL1GM084436-01 (to S.A.K. and D.J.M.), U19DK62434 (to J.K.E. and D.J.M.), P01DK088761, RL1DK081185 (to J.K.E.), and GM007062 (to A.L.B.); the Robert A. Welch Foundation (grant I-1558 to S.A.K., grant I-1275 to D.J.M.); and the Howard Hughes Medical Institute (to J.S.T. and D.J.M.).
Footnotes
Author Contributions
A.L.B. conceived the study and with M.H.M.dG. designed, performed, and analyzed all experiments. B.M.O. designed, performed and analyzed mini-pump experiments. S.L. and L.G. performed anatomical profiling experiments. H.L.L. generated study animals and performed experiments. X.D. created the floxed Klb mouse line. D.J.M., S.A.K., J.S.T., and J.K.E. provided conceptual advice and supervised the project. A.L.B., D.J.M., and S.A.K. wrote the paper.
The authors declare no competing financial interests.
References
- 1.Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: from feast to famine. Genes Dev. 2012;26:312–324. doi: 10.1101/gad.184788.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kharitonenkov A, et al. FGF-21/FGF-21 receptor interaction and activation is determined by betaKlotho. Journal of cellular physiology. 2008;215:1–7. doi: 10.1002/jcp.21357. [DOI] [PubMed] [Google Scholar]
- 3.Ogawa Y, et al. BetaKlotho is required for metabolic activity of fibroblast growth factor 21. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:7432–7437. doi: 10.1073/pnas.0701600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki M, et al. betaKlotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol. 2008;22:1006–1014. doi: 10.1210/me.2007-0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fon Tacer K, et al. Research resource: Comprehensive expression atlas of the fibroblast growth factor system in adult mouse. Mol Endocrinol. 2010;24:2050–2064. doi: 10.1210/me.2010-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsuchou H, Pan W, Kastin AJ. The fasting polypeptide FGF21 can enter brain from blood. Peptides. 2007;28:2382–2386. doi: 10.1016/j.peptides.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan BK, et al. Fibroblast growth factor 21 (FGF21) in human cerebrospinal fluid: relationship with plasma FGF21 and body adiposity. Diabetes. 2011;60:2758–2762. doi: 10.2337/db11-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang C, et al. Differential Specificity of Endocrine FGF19 and FGF21 to FGFR1 and FGFR4 in Complex with KLB. PloS one. 2012;7:e33870. doi: 10.1371/journal.pone.0033870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coskun T, et al. Fibroblast growth factor 21 corrects obesity in mice. Endocrinology. 2008;149:6018–6027. doi: 10.1210/en.2008-0816. [DOI] [PubMed] [Google Scholar]
- 10.Sarruf DA, et al. Fibroblast growth factor 21 action in the brain increases energy expenditure and insulin sensitivity in obese rats. Diabetes. 2010;59:1817–1824. doi: 10.2337/db09-1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. Annual review of physiology. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- 12.Huang W, Ramsey KM, Marcheva B, Bass J. Circadian rhythms, sleep, and metabolism. The Journal of clinical investigation. 2011;121:2133–2141. doi: 10.1172/JCI46043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buijs RM, Kalsbeek A. Hypothalamic integration of central and peripheral clocks. Nature reviews. Neuroscience. 2001;2:521–526. doi: 10.1038/35081582. [DOI] [PubMed] [Google Scholar]
- 14.Welsh DK, Takahashi JS, Kay SA. Suprachiasmatic nucleus: cell autonomy and network properties. Annual review of physiology. 2010;72:551–577. doi: 10.1146/annurev-physiol-021909-135919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowey AD, Spyer M. Central Regulation of Autonomic Functions. Oxford University Press; USA: 1990. [Google Scholar]
- 16.Inagaki T, et al. Endocrine regulation of the fasting response by PPARalpha-mediated induction of fibroblast growth factor 21. Cell metabolism. 2007;5:415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 17.Ding X, et al. betaKlotho Is Required for Fibroblast Growth Factor 21 Effects on Growth and Metabolism. Cell metabolism. 2012;16:387–393. doi: 10.1016/j.cmet.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Casanova E, et al. A CamKIIalpha iCre BAC allows brain-specific gene inactivation. Genesis. 2001;31:37–42. doi: 10.1002/gene.1078. [DOI] [PubMed] [Google Scholar]
- 19.Scott MM, Williams KW, Rossi J, Lee CE, Elmquist JK. Leptin receptor expression in hindbrain Glp-1 neurons regulates food intake and energy balance in mice. The Journal of clinical investigation. 2011;121:2413–2421. doi: 10.1172/JCI43703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Badman MK, Koester A, Flier JS, Kharitonenkov A, Maratos-Flier E. Fibroblast growth factor 21-deficient mice demonstrate impaired adaptation to ketosis. Endocrinology. 2009;150:4931–4940. doi: 10.1210/en.2009-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inagaki T, et al. Inhibition of growth hormone signaling by the fasting-induced hormone FGF21. Cell metabolism. 2008;8:77–83. doi: 10.1016/j.cmet.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buijs RM, Kalsbeek A, van der Woude TP, van Heerikhuize JJ, Shinn S. Suprachiasmatic nucleus lesion increases corticosterone secretion. The American journal of physiology. 1993;264:R1186–1192. doi: 10.1152/ajpregu.1993.264.6.R1186. [DOI] [PubMed] [Google Scholar]
- 23.Ritter S, Dinh TT, Zhang Y. Localization of hindbrain glucoreceptive sites controlling food intake and blood glucose. Brain research. 2000;856:37–47. doi: 10.1016/s0006-8993(99)02327-6. [DOI] [PubMed] [Google Scholar]
- 24.Cahill GF., Jr Fuel metabolism in starvation. Annual review of nutrition. 2006;26:1–22. doi: 10.1146/annurev.nutr.26.061505.111258. [DOI] [PubMed] [Google Scholar]
- 25.Wang T, Hung CC, Randall DJ. The comparative physiology of food deprivation: from feast to famine. Annual review of physiology. 2006;68:223–251. doi: 10.1146/annurev.physiol.68.040104.105739. [DOI] [PubMed] [Google Scholar]
- 26.Potthoff MJ, et al. FGF21 induces PGC-1alpha and regulates carbohydrate and fatty acid metabolism during the adaptive starvation response. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:10853–10858. doi: 10.1073/pnas.0904187106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakansson ML, Brown H, Ghilardi N, Skoda RC, Meister B. Leptin receptor immunoreactivity in chemically defined target neurons of the hypothalamus. The Journal of neuroscience : the official journal of the Society for Neuroscience. 1998;18:559–572. doi: 10.1523/JNEUROSCI.18-01-00559.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zigman JM, Jones JE, Lee CE, Saper CB, Elmquist JK. Expression of ghrelin receptor mRNA in the rat and the mouse brain. The Journal of comparative neurology. 2006;494:528–548. doi: 10.1002/cne.20823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woo YC, Xu A, Wang Y, Lam KSL. Fibroblast Growth Factor 21 as an emerging metabolic regulator: clinical perspectives. Clinical endocrinology. 2013;78:489–496. doi: 10.1111/cen.12095. [DOI] [PubMed] [Google Scholar]
- 30.Paxinos G, Franklin KBJ. The Mouse Brain in Stereotaxic Coordinates. Academic Press; San Diego, CA: 2003. [Google Scholar]
- 31.Lee S, et al. Laser-capture microdissection and transcriptional profiling of the dorsomedial nucleus of the hypothalamus. The Journal of comparative neurology. 2012 doi: 10.1002/cne.23116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bookout AL, Cummins CL, Mangelsdorf DJ, Pesola JM, Kramer MF. High-throughput real-time quantitative reverse transcription PCR. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Unit 15. Chapter 15. 2006. p. 18. [DOI] [PubMed] [Google Scholar]
- 33.Cummins CL, et al. Liver X receptors regulate adrenal cholesterol balance. The Journal of clinical investigation. 2006;116:1902–1912. doi: 10.1172/JCI28400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siepka SM, Takahashi JS. Methods to record circadian rhythm wheel running activity in mice. Methods in enzymology. 2005;393:230–239. doi: 10.1016/S0076-6879(05)93008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimomura K, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome research. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


