Abstract
The mucopolysaccharidoses (MPS) result from attenuation or loss of enzyme activities required for lysosomal degradation of the glycosaminoglycans, hyaluronan, heparan sulfate, chondroitin/dermatan sulfate, and keratan sulfate. This review provides a summary of glycan biomarkers that have been used to characterize animal models of MPS, for diagnosis of patients, and for monitoring therapy based on hematopoietic stem cell transplantation and enzyme replacement therapy. Recent advances have focused on the non-reducing terminus of the glycosaminoglycans that accumulate as biomarkers, using a combination of enzymatic digestion with bacterial enzymes followed by quantitative liquid chromatography/mass spectrometry. These new methods provide a simple, rapid diagnostic strategy that can be applied to samples of urine, blood, cerebrospinal fluid, cultured cells and dried blood spots from newborn infants. Analysis of the non-reducing end glycans provides a method for monitoring enzyme replacement and substrate reduction therapies and serves as a discovery tool for uncovering novel biomarkers and new forms of mucopolysaccharidoses.
Keywords: Carbohydrate biomarkers, Lysosomal storage disorders, Mucopolysaccharidoses, Glycosaminoglycans, Mass spectrometry, Sensi-Pro assay
1. Introduction
Lysosomal storage diseases (LSDs) are a heterogeneous collection of over 50 diseases caused by deficiencies in key components of the lysosomal degradation system [1]. Depending on the nature of the lysosomal deficiency, a wide range of metabolites can accumulate including glycans, lipids and proteins, leading to deleterious effects in multiple tissues and organs. LSDs exhibit a great variation in the age of onset and rate of disease progression due to the degree of enzyme deficiency, genotypic modifiers and poorly defined environmental factors. Thus, both severe and attenuated forms of the disease exist, which do not correlate well with genotype. When symptoms are present, most patients begin what has been called a “diagnostic odyssey” to correctly diagnose the disease and to select appropriate treatment [2]. The absence of early diagnosis, especially in infants, can lead to irreversible developmental, neurological, and physiological changes. Thus, there is a great need for simple, reliable biomarkers for early diagnosis. Such biomarkers could also prove useful for monitoring of disease progression and for optimization of therapy.
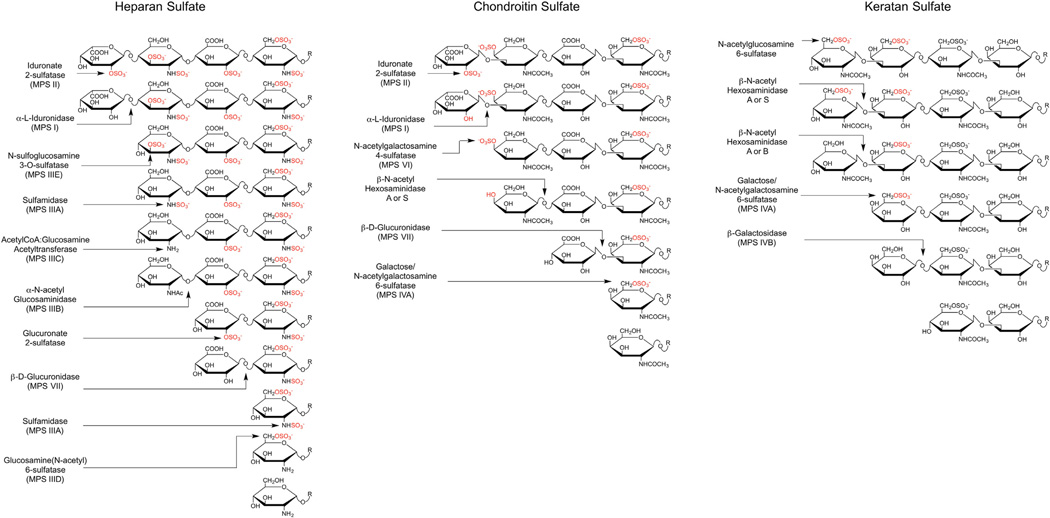
Mucopolysaccharidoses (MPS) refer to a subset of LSDs in which deficiencies occur in one or more enzymes involved in the degradation of glycosaminoglycans (GAGs) [3]. Five types of GAGs exist: heparan sulfate (HS), chondroitin sulfate (CS), dermatan sulfate (DS), hyaluronan (HA), and keratan sulfate (KS). A family of at least 11 enzymes catalyzes the lysosomal degradation of GAGs, including several glycosidases and sulfatases, an acetyltransferase, and an enzyme required for generating the catalytically active form of all known sulfatases (Table 1). Degradation of the chains occurs in a directional manner by removal or processing of the terminal sugar on the non-reducing end (NRE) of the GAG chain (Fig. 1). Due to the sequential nature of the degradative process, mutations in any enzyme in the pathway result in lysosomal storage of undegraded GAGs, the composition of which depends on the specific enzyme deficiency (Table 1). In addition to the lysosomal enzymes, an extracellular endoglycosidase (heparanase) can cleave HS chains at specific sites [4], giving rise to new NREs that are acted on by the catabolic exo-enzymes. The normal action of heparanase coupled with a deficiency in a lysosomal enzyme results in an increase in the number of fragments, i.e., in an accumulation of “ends,” in addition to an increase in total mass of GAGs. Hyaluronidases that can cleave HA and CS into fragments in some tissues have also been described [5].To date, no MPS disorders associated with heparanase deficiency have been reported, presumably because the exolytic enzymes are able to degrade with efficiency even large HS chains.
Table 1.
MPS disorders and GAG storage.
| Disease | Defective enzyme (gene) | Primary storage material |
Symptoms |
|---|---|---|---|
| MPS I (Hurler, Hurler/Scheie or Scheie) | α-L-Iduronidase (IDUA) | Dermatan sulfate Heparan sulfate |
Mental retardation, micrognathia, coarse facial features, macroglossia, retinal degeneration, corneal clouding, cardiomyopathy, hepatosplenomegaly |
| MPS II (Hunter) | Iduronate-2-sulfatase (IDS) | Dermatan sulfate Heparan sulfate |
|
| MPS IIIA (Sanfilippo A) | N-sulfoglucosamine sulfohydrolase (Sulfamidase, SGSH) | Heparan sulfate | Developmental delay, cognitive deterioration, severe hyperactivity, spasticity, motor dysfunction |
| MPS IIIB (Sanfilippo B) | N-acetyl-α-glucosaminidase (NAGLU) | Heparan sulfate | |
| MPS IIIC (Sanfilippo C) | Acetyl-CoA:α-glucosaminide N-acetyltransferase (HGSNAT) | Heparan sulfate | |
| MPS IIID (Sanfilippo D) | Glucosamine (N-acetyl)-6-sulfatase (GNS) | Heparan sulfate | |
| MPS IIIE (Sanfilippo E)a | N-sulfoglucosamine-3-sulfatase (ARSG) | Heparan sulfate | No human patients identified to date |
| MPS IVA (Morquio A) | Galactose/N-acetylgalactosamine-6-sulfatase (GALNS) | Keratan sulfate Chondroitin-6-sulfate Dermatan sulfate |
Skeletal dysplasia, motor dysfunction, joint hyper-extendibility, corneal clouding, but cognitive function unimpaired |
| MPS IVB (Morquio B) | β-Galactosidase (GLB1) | Keratan sulfate | |
| MPS VI (Maroteaux–Lamy) | N-acetylgalactosamine-4-sulfatase (ARSB) | Chondroitin sulfate Dermatan sulfate |
Skeletal dysplasia, motor dysfunction, kyphosis, cardiac defects, corneal clouding, but cognitive function unimpaired |
| MPS VII (Sly) | β-Glucuronidase (GUSB) | Heparan sulfate Chondroitin sulfate Dermatan sulfate |
Hydrops fetalis, hepatomegaly, skeletal dysplasia, corneal clouding, developmental delay |
| MPS IX (Natowicz) | Hyaluronidase (HYAL1) | Hyaluronan | Periarticular soft-tissue masses, mild facial changes, short stature |
| Multiple Sulfatase Deficiency | Multiple sulfatases (SUMF1) | Heparan sulfate Chondroitin sulfate Dermatan sulfate Keratan sulfate |
Coarse facial features, ichthyosis, mental retardation, deafness, hepatosplenomegaly |
Deficiency of N-sulfoglucosamine-3-sulfate sulfatase deficiency and lysosomal storage of heparan sulfate has been reported in arylsulfatase deficient mice [86].
Fig. 1.
Glycosaminoglycan catabolism. The schemes show the different enzymatic activities required for the sequential catabolism of a hypothetical NREs from heparan sulfate, dermatan sulfate and keratan sulfate. It should be noted that the glucuronic acid 2-O-sulfatase in heparan sulfate degradation has been demonstrated in vitro, but has not yet been identified genetically.
Scheme modified from [3] according to findings from Lawrence et al. [18] and Kowalewski et al. [87].
Treatment for MPS currently consists of palliative care and management of secondary symptoms. Attempts to correct or slow the course of the disease by allogeneic stem cell transplantation have met with some success for treatment of MPS I, VI and VII patients [6–8]. In spite of successful restoration of enzyme activity in peripheral tissues, neurological deterioration occurs unabated. Viral vectors and stem cell transplantation strategies are under development with the hope that gene replacement therapy might one day be possible [9,10]. Other approaches include chaperone therapy to partially restore endogenous enzyme activity [10], and substrate reduction therapy to reduce the metabolic load biosynthetically [11]. Enzyme replacement therapy has met with great success for treatment of non-neurological manifestations of MPS I (Aldurazyme™), MPS II (Elaprase™) and MPS VI (Naglazyme™), suggesting that a similar approach for other MPS disorders might prove successful [12,13]. Conventional ERT depends on transport of exogenous recombinant enzyme via mannose-6-phosphate/insulin-like growth factor II (M6P/IGFR) or C-type mannose receptors on cells. Developmental and tissue-specific differences in receptor expression, however, prevent efficient uptake in some tissues and across the blood–brain barrier [14]. To circumvent the blood–brain barrier and treat neurological complications of MPS, intrathecal injection of enzyme is currently being explored [15,16]. The need for biomarkers becomes obvious for assessment of the efficacy of any of these therapeutic options and for monitoring the natural history of the disease [17].
In this review, we summarize various approaches to glycan-based biomarker development for MPS with a discussion of a new approach that has identified unique glycan NRE biomarkers [18]. We refer the reader to other recent reviews that cover other types of biomarkers based on enzyme mass, enzyme activity and pathological consequences of disease [19–22]. Unique glycan structures have long been associated with initiation and progression of diverse diseases, including cancer and inflammation [23]. In cancer, a number of changes in glycans occur that correlate with disease, but only a few changes have demonstrated the specificity to serve as useful biomarkers [24]. In contrast to cancer, in which complex genetic and environmental factors interact to drive a heterogeneous disease, MPS are comparatively homogenous in their root cause. Each enzyme deficiency leads to selective accumulation of glycans that contain a terminal sugar residue that is normally modified or removed by the affected lysosomal enzyme (Fig. 1). Thus, both the GAGs that accumulate and the ends of the chains become unique biomarkers for MPS.
2. Biomarkers based on total GAG accumulation
GAG storage resulting from loss of lysosomal enzyme activity is the primary biochemical event in MPS; thus biomarkers based on GAG storage can report directly the severity of the disease. However, genetic and environmental factors can modulate the severity of GAG accumulation independently of enzyme deficiency, which could explain why patients with identical etiological mutations can present with profoundly different disease severity [25]. Nevertheless, assessment of overall levels of GAG in urine, cerebrospinal fluid or in cell culture provides a simple, convenient biomarker for MPS that has been exploited for diagnosis and for monitoring disease progression and therapy. In this section, we discuss various approaches for assessing GAG accumulation in MPS patients and its use as a biomarker.
2.1. Dye binding methods
MPS patients excrete significant amounts of GAG fragments in the urine. The most common assay involves measurement of GAGs in urine samples using dye-binding assays with dimethylmethylene blue [26,27]. This approach has been used for diagnosis as well as for determining response to therapies in clinical trials for MPS I, II, and VI [28–30]. The method works best with isolated GAGs or urine samples, but can be adapted to tissue samples as well [31]. Drawbacks of the assay include low specificity due to the formation of a non-specific complex of the dye with polyanions, including nucleic acids, and inability to distinguish the kind of GAG excreted without further enzymatic or separation methods. This method exhibits few false-negative results compared to other dye-based assays, but lacks reliability for detecting attenuated forms of MPS [32–34]. The sensitivity of the dye binding methods is also low compared to other methods described below, generally restricting their use to urine samples due to the high concentration of GAGs in MPS patients and general lack of other interfering substances. Using urine as a reporter of the overall GAG storage burden of the body has been criticized because it may reflect storage in the kidney rather than other tissues [22]. Despite these limitations, the method enjoys widespread use presumably because of its simplicity, the availability of commercial kits (Blyscan™) and adaptation to an inexpensive qualitative visual test [35].
2.2. Antibody-based assays
There have been several reports describing the use of anti-GAG antibodies in ELISA format to measure urine and blood GAG levels in MPS patients [36,37]. However, immunological detection of GAGs suffers from lack of definition of the reactive epitope, cross reactivity with other polyanions, or exclusion caused by recognition of a pattern of sulfation and/or epimerization that may not be represented in all GAG chains present in a sample. This latter problem is highly relevant, because of natural variation in GAG structure across individuals, effects due to age, and from variation in sulfation and epimerization of GAGs that accumulate in MPS compared to GAGs present in normal patients [38–42]. Despite these limitations, ELISA based assays have been shown to be able to detect an increase in GAGs in plasma and urine from MPS patients in multiple MPS classes [36,37].
2.3. Ligand-binding assays
In theory, any ligand that binds to GAG can be used to measure the concentration of GAG in a biological sample relative to a standard curve. The high affinity ligand fibroblast growth factor-2 (FGF2; basic FGF) has been used to detect HS on cells, in tissue sections from mice, and in solution [43–45]. High sensitivity is achieved by using fluorescent derivatives of FGF2 or biotinylated FGF2 and enzyme-conjugated streptavidin. This strategy has not yet been applied to MPS samples, but warrants further consideration because several ligands can be used simultaneously (e.g., different FGFs or other cytokines [46–48]), adding potential robustness to the assay.
A related approach for quantification of GAG storage was recently described based on the accumulation of heparin cofactor II-thrombin (HCII-T) complexes in the plasma. In an elegant study, Randall and co-workers identified by proteomic analysis of plasma samples significantly elevated levels of HCII-T complexes in MPS I animal models and patients [49]. These complexes arise from activation of HCII by DS fragments of 6 or more monosaccharides that contain 4-sulfated N-acetylgalactosamine that is either additionally 6-O sulfated or 2-O-sulfated on the adjacent iduronic acid, and subsequent covalent inactivation of thrombin [50,51]. Thus, the presence of HCII-T complexes in blood, which can be readily detected through Western blotting and ELISA, acts as a surrogate marker for DS accumulation. Subsequent studies showed that the HCII-T levels respond to bone marrow transplantation and enzyme replacement therapy. Interestingly, HCII-T levels decline rapidly after enzyme replacement therapy in MPS I, II and VI patients, whereas urine DS levels respond more slowly [52]. In part, this difference may reflect the preferentially detection of larger, more highly sulfated GAGs by dye binding compared to the detection of those GAG chains with the capacity to bind HCII-T. Limitations of the HCII-T biomarker include a significant loss of signal after repetitive freeze–thawing of plasma samples, limitations to detection of disease in MPS classes that have significant DS accumulation, and the dependence of the assay on DS with high affinity for HCII, which might vary naturally between individuals. Nevertheless, the method has been validated and found reliable as a biomarker in a clinical setting [52–54].
2.4. Dermatan:chondroitin sulfate ratio
The ratio of DS to CS (DS/CS) has been found to be a reliable marker of disease for MPS resulting from mutations in enzymes affecting DS turnover (Table 1) [55]. A simple procedure involves electrophoretic separation of GAGs on polyacrylamide gels, followed by staining of the gels with Alcian Blue. The DS/CS ratio correlates with the level of restored enzyme activity after bone marrow transplantation and ERT suggesting that the ratio is a sensitive measure of biochemical response [8,56]. Direct comparison between the HCII-T biomarker and the DS/CS ratio demonstrated that the two biomarkers generally correlate, with notable exceptions at certain time points [52]. The lack of perfect correlation between these assays is not surprising given the unique GAG subset that each assay detects. The DS/CS ratio method uses dye precipitation to prepare the GAG sample, thus the method preferentially measures larger DS and CS fragments, whereas the HCII-T method detects a subset of DS fragments that bind and activate HCII.
2.5. GAG derived oligosaccharides
Early on it was observed that monosaccharides and oligosaccharides derived from GAGs accumulate in plasma and urine from MPS patients through partially characterized degradative pathways that appear to become active when GAGs levels are elevated. Di-, tri-, tetra-, and penta-, and hexasaccharides have been isolated from the urine of MPS I patients. Derivatization using 1-phenyl-3-methyl-5-pyrazolone (PMP) allowed further characterization of their structure by electrospray ionization (ESI)-tandem mass spectrometry (MS/MS) [57], which delineates their structural composition. As predicted, the non-reducing end consisted of iduronic acid. A similar approach demonstrated di- to pentasaccharides derived from HS and DS in the urine of MPS II patients. King and co-workers validated an HS-derived disaccharide (N-sulfoglucosamine– hexuronic acid) that accumulates in the brain, liver and spleen of a mouse model of MPS IIIA [58]. Presumably, the disaccharide arises from degradation of HS fragments containing this disaccharide as the reducing terminal end of the chain. Intracerebral delivery of recombinant human sulfamidase led to a reduction in the amount of the disaccharide biomarker. Therefore, the disaccharide might prove useful for monitoring future therapies for MPS IIIA, which does not currently exist.
A number of years ago, Hopwood and Elliot demonstrated that N-acetylhexosamines were present in human urine and most likely derived from an alternative degradative pathway mediated by β-N-acetylhexosaminidase cleavage of non-reducing end sulfated N-acetylglucosamine from KS and sulfated N-acetylgalactosamine from DS and CS [59–61]. These sulfated monosaccharides would presumably arise in lysosomes and subsequently appear in the urine of sulfatase-deficient patients after transport out of the lysosome or efflux from the cell. Both the amount and type of urinary sulfated monosaccharides depended on the type of MPS and clinical severity of the disease. Although these original discoveries utilized tedious paper chromatography to separate the sulfated monosaccharides, Ramsay and colleagues developed a ratiometric method for quantification of sulfated N-acetylhexosamine-containing mono- and disaccharides based on isomeric product ions generated by ESI-MS/MS of PMP-derivatized samples [62]. Urine from MPS I, II, IIIA, IIIB, IIIC, IIID, IVA, VI, and multiple sulfatase deficient patients had significant increases in di- and/or monosulfated N-acetylhexosamines (GalNAc4,6S [a10], GalNAc6S [a6], GalNAc4S [a4], or GlcNAc6S [A6]) and monosulfated N-acetylhexosamine-uronic acid (UA) disaccharides (GalNAc6S-UA [a6U], GalNAc4S-UA [a4U], or GlcNAc6S-UA [A6U], see legend to Fig. 2 for Disaccharide Structure Code). Urine samples from MPS IVA and VI patients showed decreases in mono and disulfated N-acetylhexosamine residues and sulfated N-acetylhexosamine-UA after bone marrow transplantation, which correlated with clinical improvement. In theory, this assay can be made completely quantitative by inclusion of suitably mass-tagged multiple standards.
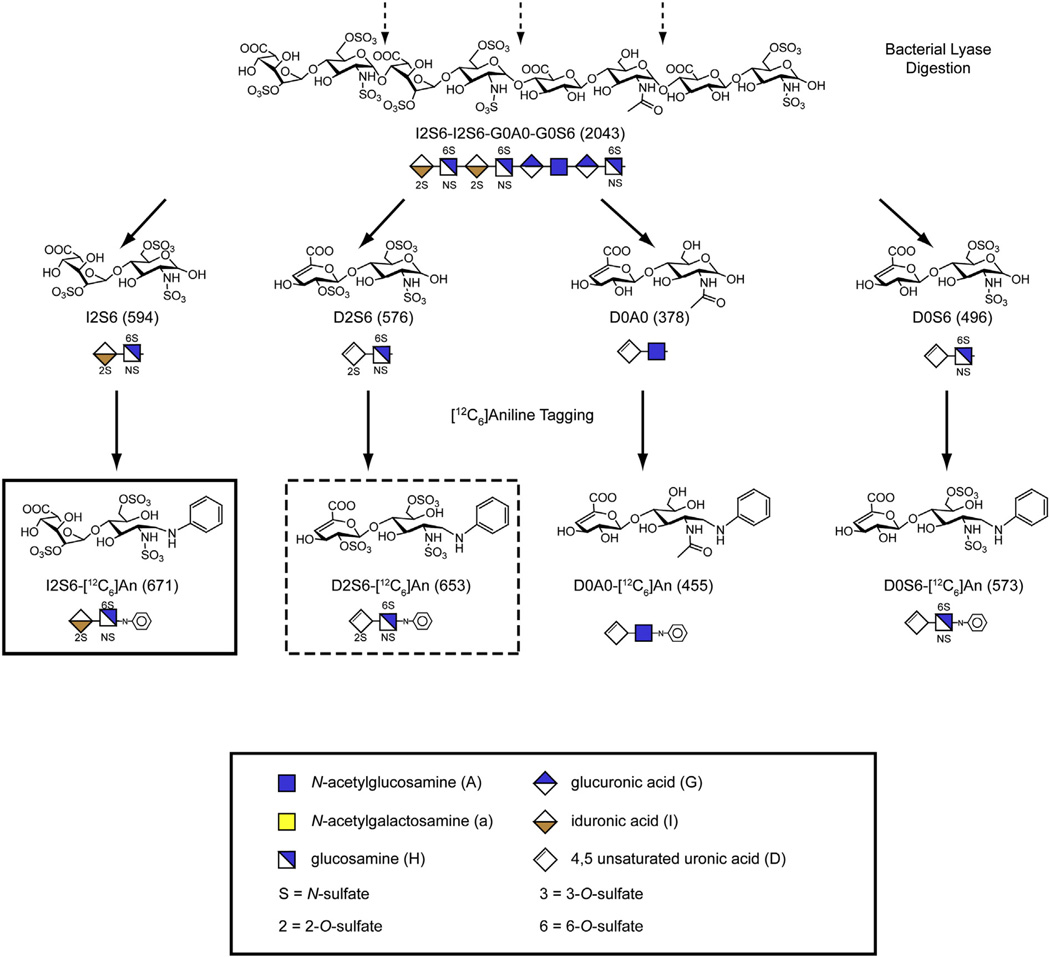
Fig. 2.
Scheme for determining NREs and internal disaccharides. Enzymatic depolymerization of HS with heparan lyases releases internal disaccharides (dashed arrows) that contain an unsaturated uronic acid. The NRE liberated from the left end of the chain as drawn lacks the Δ4,5-double bond and is 18 Da greater in mass than a corresponding internal disaccharide. Reductive amination with [12C6]aniline facilitates separation of the various disaccharides by LC/MS and gives the indicated m/z values for the molecular ions shown in parentheses. The glycan structures are graphically represented by geometric symbols, which are defined in the lower part of the figure [88]. To simplify the representation of constituent disaccharides, we use a structure code (DSC) [89]. In the DSC, a uronic acid is designated as U, G, I or D for an unspecified hexuronic acid, d-glucuronic acid, l-iduronic acid or Δ4,5-unsaturated uronic acid, respectively. The hexosamines are designated in upper case for glucosamine and lower case for galactosamine, and the N substituent is either H, A,S or R for hydrogen, acetate, sulfate or some other substituent, respectively. The presence and location of ester-linked sulfate groups are depicted by the number of the carbon atom on which the sulfate group is located or by 0 if absent. For example, I2S6 refers to a disaccharide composed of 2-sulfoiduronic acid-N-sulfoglucosamine-6-sulfate, whereas D2S6 refers to a similarly structured disaccharide that instead has a Δ4,5-double bond in the uronic acid.
Adapted from [18] with permission.
2.6. Total GAG analysis by mass spectrometry
Mass spectrometry has been used to assess total GAG in blood and urine from MPS patients. Quantitation of total GAG by mass spectrometry typically involves depolymerization of the chains with bacterial lyases (chondroitinase ABC for CS/DS and heparin lyases for HS). These enzymes act by a beta-eliminative mechanism, resulting in a cleavage of the bond between the hexosamine residue and the uronic acid and the production of disaccharides containing a Δ4,5-unsaturated uronic acid (stereochemistry of the uronic acid is lost upon eliminative cleavage) linked to an N-acetylated/N-sulfated hexosamine. KS also can be depolymerized by keratanases, but these enzymes act by hydrolysis, generating disaccharides containing variably sulfated galactose and N-acetylglucosamine residues. Similarly, hyaluronidases hydrolytically cleave HA into disaccharides. These disaccharides can then be separated by liquid chromatography, analyzed by mass spectrometry, and quantitated by comparison to the signal obtained from chemical standards.
de Ruijter and colleagues have determined plasma HS concentration from MPS III patients from the sum of seven lyase-derived disaccharides, and found that plasma HS determined in this way correlates with disease severity and risk of speech loss [63]. The same group analyzed KS, HS and DS levels by LC–MS/MS for clinical diagnosis of MPS I, II, III and VI [64], confirming earlier work by Tomatsu and colleagues [40,65,66]. Monitoring total DS and HS in this way has proven effective for determining the efficacy of ERT in a mouse model of MPS VII [67]. Tomatsu and co-workers identified DS and HS in this way from serum and urine of ERT-treated MPS I patients. The outcome of their analysis showed a marked reduction in DS and HS after ERT [39,40].
With ERT under development for MPS IVA, the identification of biomarkers to evaluate disease progression and response to treatment has become important. To date, most studies have focused on KS, which accumulates in MPS IVA patients and has been identified as an important biomarker. Tomatsu and co-workers have validated that LC–MS/MS can be used to identify levels of KS derived disaccharides in the blood of MPS IVA patients [66]. Their findings showed that blood KS derived disaccharides varied with age and clinical severity, suggesting that this assay is suitable for both early diagnosis and longitudinal assessment of disease severity [68].
Care must be taken using the various depolymerizing enzymes to ensure complete depolymerization of the chains, e.g., by monitoring the production of the unsaturated uronic acids, which absorb light at 232 nm, and comparing the values to samples of standard GAGs treated under identical conditions. Some domains in HS and DS tend to resist digestion, giving rise to tetrasaccharides and hexasaccharides, which are often ignored [69]. Variations in the GAGs that accumulate in patients might complicate these analyses as well, if they had an unusual structure. Nevertheless, the combination of enzyme digestion coupled with LC/MS provides a powerful tool for quantitating GAGs and sets the stage for methods based on the analysis of the NRE of the chains, as explained in the next section.
3. Detection of diagnostic lyase generated non-reducing ends
3.1. Enzymatic modification of the NRE
As discussed above, each type of MPS accumulates GAGs with a char-acteristic non-reducing terminus, whose structure depends on the enzymatic deficiency. Thus, the NREs represent natural biomarkers for each type of mucopolysaccharidosis. One approach to exploit the NRE for diagnosis consists of treating the GAG chains with recombinant sulfatase or exoglycosidase to liberate either sulfate or a monosaccharide from the NRE, respectively. In the original application of this method, Byers et al. showed that enzymatic treatment of urinary GAGs from MPS I,II,IIIA, IIIB, IIIC, IIID, IVA and VI patients resulted in mobility shifts when the samples were analyzed by polyacrylamide gel electrophoresis, providing a definitive diagnosis of different MPS [70]. Digestion of GAGs from urine and brain with recombinant human sulfamidase yielded a definitive diagnosis of sulfamidase deficiency (MPS IIIA) in a spontaneous mouse variant that had the hallmarks of lysosomal storage [71]. In theory, one could also monitor the release of free sulfate or a monosaccharide to assess the structure of the NRE instead of analyzing the electrophoretic mobility of the GAGs. To be broadly applicable, one would need recombinant forms of all of the enzymes involved in GAG degradation.
3.2. Sensi-Pro assay
Recently, we adapted glycan reductive isotope labeling-liquid chromatography/mass spectrometry (GRIL-LC/MS) to analyze the disaccharide composition of GAG chains [72,73]. In this method, the GAG chains are degraded with bacterial lyases and the resulting disaccharides are derivatized with isotopically pure [12C6]aniline by reductive amination (Fig. 2). The aniline tag improves resolution of the disaccharides by high-pressure liquid chromatography on reverse phase resins in the presence of an ion-pairing agent (dibutylamine). The effluent of the column is then analyzed by mass spectrometry, adding a second dimension to the analysis. A third dimension is easily realized by selective daughter ion fragmentation. Adding a known amount of disaccharide standards tagged with [13C6]aniline allows recovery and quantitation of each disaccharide in the biological sample by ratiometric analysis. Thus, GRIL-LC/MS provides a way to determine not only the disaccharide composition of GAG chains, but also the total amount of GAG in a sample.
Analysis of GAGs from MPS patients demonstrated the utility of GRIL-LC/MS for determining total storage and uncovered one or more additional peaks of [12C6]aniline-tagged material that varied in elution position and mass dependent upon the MPS disorder [18]. Mass spectral analysis revealed that the additional peaks were derived from the non-reducing end of GAG chains. Samples from MPS I,II, and VII, diseases that affect the activity of enzymes that act on NRE uronic acids, yielded a characteristic NRE disaccharide of general structure, uronic acid-hexosamine. Unlike the disaccharides liberated from internal segments of the chains, these NRE disaccharides do not contain an unsaturated uronic acid and thus have a unique m/z signature distinguishable from otherwise identical “internal” residues (the m/z value for an NRE disaccharide is 18 amu larger than that of a corresponding internal disaccharide, Figs. 2 and 3). In contrast to these findings, samples from MPS patients or mice with MPS IIIA, IIIB, IIIC, IIID (Sanfilippo) or MPS VI yielded either a monosaccharide (a hexosamine) or trisaccharides (hexosamine–uronate–hexosamine). Thus, the lyases exposed the NRE determinants diagnostic for each MPS. The combination of lyase digestion, GRIL–LC/MS, and inclusion of mass-tagged NRE standards is called the Sensi-Pro assay. An example is shown in Fig. 3A, which illustrates the analysis of two MPS disorders.
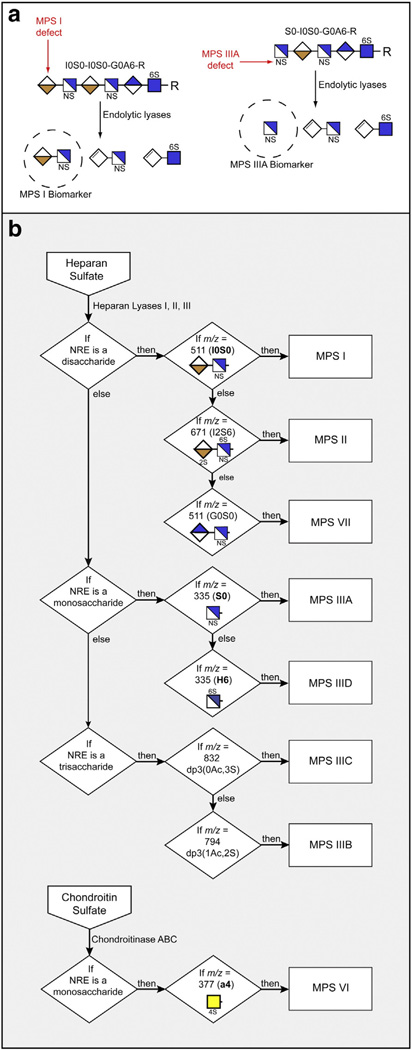
Fig. 3.
Systematic diagnostic screening of GAG samples for various MPS disorders. (a) As an example, the process of uncovering the NRE biomarkers in MPS I and MPS IIIA samples is shown. Dashed circles indicate specific NRE structures for these two disorders. (b) The flow chart illustrates how MPS diagnosis can be carried out. The detection criteria are tied to the exact structure of the biomarkers based on size (monosaccharide, disaccharide or trisaccharide) and structural features such as the number of acetates (Ac) and sulfates (S). For a complete unknown, NRE analysis would be carried out on both HS and CS/DS as indicated. The key to the symbols is shown in Fig. 2.
NRE structures are typically heterogeneous and were only detected in trace amounts in normal samples [74,18]. A likely explanation for this difference derives from the understanding that the abundance of ends results from the combination of interrupted degradation caused by the missing lysosomal enzyme and in the case of HS heparanase activity, which can cleave the intact HS chains into multiple fragments. Unique CS/DS NREs accumulate to high levels in MPS I, II and VI, but CS/DS may only undergo limited internal cleavage reactions [75].
In order to make Sensi-Pro a credible means of MPS diagnosis, we investigated the NRE profile of MPS I, II, IIIA, IIIB, IIIC, IIID, VI and VII using multiple samples. We rationalized all possible candidate structures, assuming that the enzymes liberate a terminal disaccharide if the chain ends in a uronic acid, or a monosaccharide (hexosamine), trisaccharide (hexosamine–uronate–hexosamine) or both a monosaccharide and trisaccharide if the chain ends in a hexosamine. It was then possible to select single unique NREs as biomarkers for each MPS disorder and combine them into a decision tree on the basis of NRE size (mono-, di-and trisaccharides), degree of sulfation, retention time, and co-migration with NRE standards during liquid chromatography. The specific NREs indicated in the scheme outlined in Fig. 3B are sufficient to simultaneously diagnose any of the eight MPS disorders listed in the decision tree. These MPS biomarkers were tested in blinded studies to prove their reliability. Using this approach we have diagnosed successfully the MPS subtype in many different types of samples, including tissue, cells, urine, plasma and blood spots (see below) derived from MPS patients or animal models.
3.3. Morquio syndrome
Diagnosis of Morquio syndrome (MPS IVA and IVB) present unique challenges. Morquio patients accumulate KS, and like GAGs that accumulate in other MPS, the KS that accumulates should contain a unique NRE (N-acetylglucosamine-6-sulfate in MPS IVA and galactose in MPS IVB). Unfortunately, the bacterial enzymes available for depolymerization of KS and liberation of the NREs are endolytic hydrolases and produce NREs that are indistinguishable from disaccharides liberated from the internal parts of the chains. Thus, analysis of KS accumulation has been limited to quantitation of the total amount of GAG using dimethylmethylene blue binding, by ELISA using anti-KS monoclonal antibody (5D4) or by mass spectrometry of products generated by digestion with keratanase in blood or urine samples [39,68,76]. A comparison of ELISA and mass spectrometry showed greater sensitivity afforded by mass spectrometry [37,77]. Urine KS level varies with age and clinical severity and accumulates in other MPS disorders as a secondary consequence of other GAG accumulation [59,76,78]. Although the blood KS levels in MPS IVA patients (0.4–26 µg/ml) were higher than those in age-matched controls (0.67–4.6 µg/ml), the fold-difference between patients with attenuated disease and normal controls makes diagnosis and therapeutic monitoring challenging [40]. As mentioned above, MPS IVA patients also accumulate sulfated hexosamines in urine, presumably reflecting the alternative degradative route of KS by β-N-acetylhexosaminidase (Fig. 1) [60–62].
MPS IVA results from a deficiency in N-acetylgalactosamine 6-sulfatase (GALNS). The enzyme acts on both galactose-6-sulfate, which is found in KS, and on N-acetylgalactosamine-6-sulfate, which is found in CS (Fig. 1). Thus, the absence of enzyme activity results in accumulation of both KS and CS. This well-known fact should render MPS IVA amenable to analysis by Sensi-Pro; the relevant biomarker would be the release of N-acetylgalactosamine-6-sulfate from CS in samples using chondroitinase ABC. Detection of MPS IVB, which results from a deficiency in β-galactosidase (GLB1) is more challenging, but should be amenable to methods that target terminal galactose residues in KS or by parallel analysis of glycolipids that also contain a β-linked galactose moiety.
4. Newborn screening
Early clinical intervention is important to avoid many of the debilitating and life threatening symptoms of MPS. Thus, much interest exists in early detection, either by amniocentesis or in neonates. Early detection might be afforded by analysis of amniotic fluid because it contains fetal urine and cells commensurate with fetal age. Ramsay et al. analyzed amniotic fluid samples from patients for oligosaccharide biomarkers using PMP-derivatization and mass spectrometry. Although glycan biomarkers were not observed in samples from MPSI,II, IIIC, amniotic fluid from MPS VII, IVA and VI and multiple sulfatase deficiency demonstrated accumulation of storage material [79]. Using a similar methodology, Meikle et al showed in a retrospective study that glycan biomarkers accumulate in dried blood smears from babies with MPS IVA and IIIA, but not for MPS II [80]. HCII-T may be a reliable marker for MPS I in blood spots prepared from mice, suggesting that it might be a useful biomarker for newborn screening [81]. Ruijter and colleagues utilized LC–MS/MS method to successfully identify elevated levels of HS and DS in newborn blood spots from suspected MPS I, II and III patients and easily distinguished affected individuals from controls and heterozygous carriers. To our knowledge, more comprehensive studies of glycan biomarkers for MPS in blood spots have not been pursued.
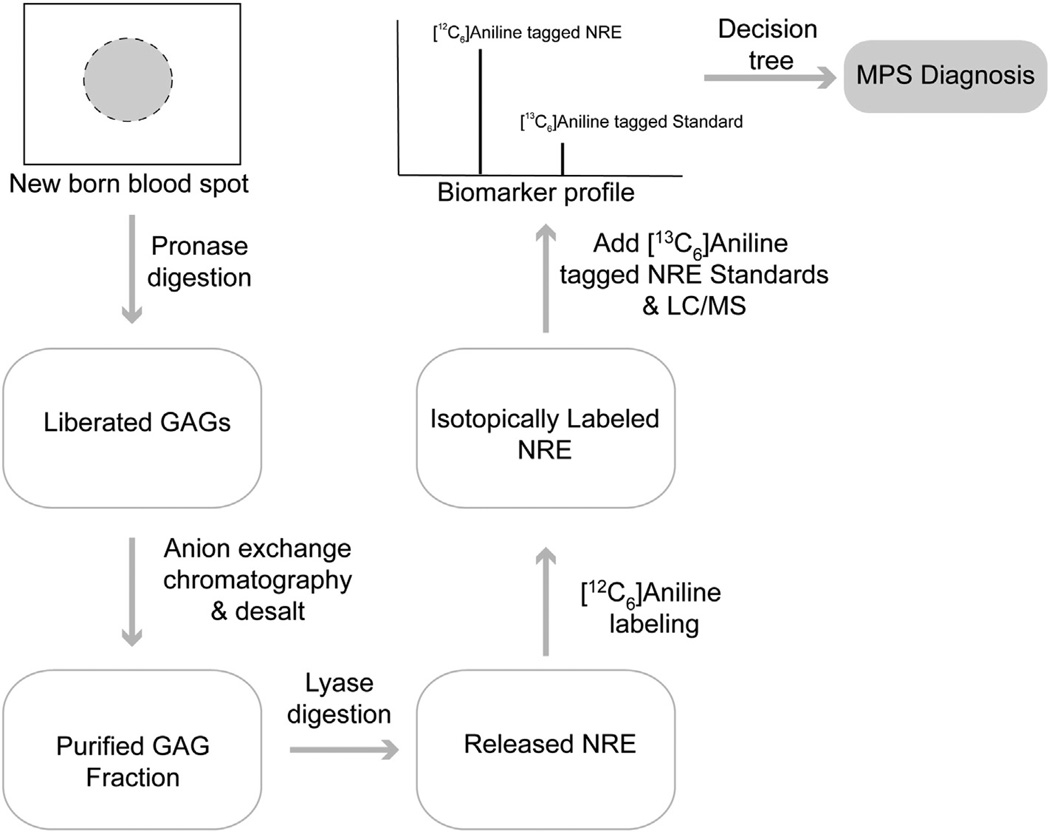
To test if Sensi-Pro might be reliable for newborn screening, we obtained from the California Department of Health a blinded panel of newborn bloodspots from both normal individuals and MPS patients. The blood spots were processed and evaluated as previously described for other samples (Fig. 4). The results were expressed as a biomarker profile for each sample comparing detected NREs with standards or known NRE signatures. No significant MPS NRE structures were detected in samples from normal individuals, whereas large amounts of MPS NRE structures were detected in samples from MPS individuals (Table 2). In all cases, NRE analysis correctly determined the MPS condition, easily discriminating between normal and different individuals affected with MPS I, II, IIIA and IIIB. Despite being purified from sections of small bloodspots (between one quarter and a half of the available blood spot), the biomarker signals were high, making the correlation to a particular MPS disorder unambiguous. These initial studies clearly warrant additional development to establish the accuracy and reliability of NRE analysis in blood spots for early diagnosis. If the technique proves reliable, definitive diagnosis can take place within a very short period of time, allowing early therapeutic intervention.
Fig. 4.
Newborn screening of various MPS disorders. A portion of a newborn bloodspot sample was immersed in a solution of Pronase and incubated overnight to solubilize GAGs. The GAGs were then purified by anion exchange chromatography using DEAE-Sepharose, washing away contaminants with 0.2 M NaCl. The GAGs were eluted with1 M NaCl. After desalting, the GAG chains were enzymatically depolymerized with heparan lyases and the released component residues including both NRE and internal unsaturated disaccharides were derivatized with [12C6]aniline. The derivatized samples were mixed with standards NRE biomarkers labeled with [13C6]aniline to distinguish them from the NRE residues present in the biological samples. The mixed samples were next analyzed by LC/MS and based on the biomarker profile a tentative diagnosis was determined using the logic mapped out in the decision tree shown in Fig. 3.
Table 2.
Newborn screening for various MPS disorders among a blind panel of patient samples.
| Anonomyzer code | NRE profile (m/z) | Associated NRE structures | Sensi-Pro Dx | Patient history |
|
|---|---|---|---|---|---|
| Age at first symptoms | Age at clinical Dx | ||||
| g000400885 | None | Normal | |||
| g000401205 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | At birth | <1 year |
| g000401688 | None | Normal | |||
| g000402008 | 335, 832 | S0, S0U2S0 | MPS IIIA – Sanfilippo A | Tested b/c of sibs | 2 years, 5 months |
| g000402587 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | Unknown | 6 months |
| g000402971 | 794 | A0U2S0 | MPS IIIB – Sanfilippo B | 3 years, 11 months | 3 years, 11 months |
| g000403390 | 335, 832 | S0, S0U2S0 | MPS IIIA – Sanfilippo A | 2 years | 3 years |
| g000403486 | None | Normal | |||
| g000403870 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | 4 months | 4 year, 5 months |
| g000404289 | 335, 832 | S0, S0U2S0 | MPS IIIA – Sanfilippo A | 2 years | 5 years, 3 months |
| g000404673 | 511, 591 | I0S0, I0S6 | MPS I – Hurler | 4 months | 3 years |
| g000405092 | 335, 832 | S0, S0U2S0 | MPS IIIA – Sanfilippo A | 2 years | 3 years, 10 months |
| g000405476 | None | Normal | |||
| g000405860 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | 1 year | 1 year, 6 months |
| g000406279 | 511, 591 | I0S0, I0S6 | MPS I – Hurler | 1 month | 4 years |
| g000406663 | 511, 591 | I0S0, I0S6 | MPS I – Hurler | 4 years, 6 months | 13 years, 6 months |
| g000407082 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | 5 years | 5 years |
| g000442004 | 511, 591 | I0S0, I0S6 | MPS I – Hurler | 1 year, 6 months | 9 years, 6 months |
| g000442583 | 591, 671 | I2S0, I2S6 | MPS II – Hunter | 1 year | 2 years, 5 months |
| g000443386 | None | Normal | |||
| g000443770 | 511, 591 | I0S0, I0S6 | MPS I – Hurler | 2 months | 8 months |
5. Other uses for NRE analysis
NRE analysis potentially has many other uses, for example in determining the efficacy of ERT and substrate reduction therapy (SRT). Lawrence et al. showed that treating cells from MPS IIIA patients with recombinant sulfamidase resulted in a precipitous drop of the cognate biomarkers to levels near those of normal control cells [18]. To test directly whether substrate reduction might be feasible for treating MPS disease, we developed a genetic model for SRT by crossing MPS IIIA mice with animals partially deficient in HS biosynthesis due to heterozygosity in Ext1 and Ext2, genes that encode the copolymerase required for HS chain assembly [75]. Reduction of HS by 30–50% using this genetic strategy ameliorated the amount of disease-specific biomarker and pathology in multiple tissues, including the brain. Genetic SRT also improved the efficacy of ERT in cell culture and in mice based on biomarker reduction. High doses of genistein, a non-specific soy isoflavone that modulates cell signaling and viability, appear to reduce GAG biosynthesis [82]. Continuous treatment of MPS IIIB mice over a 9-month period significantly reduced the NRE biomarker. Analysis of MPS I dogs that received intrathecal enzyme replacement demonstrated significantly reduced NRE biomarker in the brain and cerebrospinal fluid in all treated animals [83].
NRE analysis also provides a way to assess secondary storage. For example, significant accumulation of CS/DS occurs in cells derived from MPS III patients [84]. Treating cells with sulfamidase reversed both HS accumulation as well as CS/DS accumulation, suggesting that the HS that accumulated in the lysosome might block one or more enzymes involved in CS/DS turnover. Enzyme studies demonstrated that stored HS can inhibit iduronate 2-sulfatase and thus could explain the secondary storage effect. Screening of these samples for CS/DS NRE structures in the future could verify this idea. This strategy might be applied to other LSDs or even diseases not known to affect lysosomal function, possibly yielding new biomarkers for other disorders.
Finally, NRE analysis has proven useful as a discovery tool. Over 17 sulfatases are known to exist in the human genome, but the biological significance of over half of these enzymes remains obscure [85]. Recently, we analyzed mutant mice containing a deletion of arylsulfatase G (Arsg−/−), which had been previously suggested to result in ceroid lipofucsinosis in dogs [86]. The application of GRIL–LC/MS demonstrated that Arsg−/− mice accumulate large amounts of HS and NRE analysis demonstrated the release of monosaccharide and trisaccharides resembling a Sanfilippo syndrome [87]. Subsequent analysis showed that the NRE consisted of 3-sulfo-N-sulfoglucosamine, demonstrating that ARSG is the long sought after glucosamine-3-O-sulfatase and thus defining a new potential form of Sanfilippo syndrome (MPS IIIE) [87]. The characterization of a novel NRE in Arsg−/− mice provides the impetus for analyzing MPS patients lacking molecular diagnosis. This approach could also yield insights into the function of other uncharacterized arylsulfatases in the genome.
6. Summary
Over the years, much attention has been focused on glycan biomarkers for MPS. Anaysis of total GAG in cells, tissues, or biological fluids provides a direct assessment of GAG storage. However, quantitation of total GAG for molecular diagnosis is limited without further analysis of the type of GAG that accumulates and analysis of the NRE. Other strategies based on unusual glycans that accumulate are useful, but restricted to the certain subtypes of MPS. In contrast, techniques that focus on the NRE provide accurate diagnosis and only depend on having a small set of bacterial lyases, that are commercially available, and synthetic standards. Sensi-Pro has the advantage of allowing simultaneous analysis of multiple NRE biomarkers in patient samples in a single analysis. It also has enormous potential for identification of MPS in neonates, to improve current treatment through monitoring of the NRE biomarker, and can aid in the development of new therapies for MPS. Further development and validation of NRE biomarkers as surrogate markers are clearly warranted and could accelerate the development and FDA approval of new therapies.
Acknowledgments
This work was supported by grants GM077471 and GM093131 from the National Institutes of Health (to J.D.E.) and grants from the National MPS Society to J.D.E. and B.E.C.
Footnotes
Conflict of interest
Jillian R. Brown and Brett E. Crawford were employees of Zacharon Pharmaceuticals, Inc. at the time that the paper was written and Roger Lawrence and Jeffrey D. Esko were paid consultants to the company.
References
- 1.Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. Lysosomal Disorders. Metabolic and Molecular Basis of Inherited Disease. San Francisco: MacGraw-Hill; 2001. pp. 3371–3896. [Google Scholar]
- 2.Bouwman MG, Teunissen QG, FA Wijburg, Linthorst GE. ‘Doctor Google’ ending the diagnostic odyssey in lysosomal storage disorders: parents using internet search engines as an efficient diagnostic strategy in rare diseases. Arch. Dis. Child. 2010;95:642–644. doi: 10.1136/adc.2009.171827. [DOI] [PubMed] [Google Scholar]
- 3.Neufeld EF, Muenzer J. The Mucopolysaccharidoses. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editors. Metabolic and Molecular Basis of Inherited Disease. San Francisco: MacGraw-Hill; 2001. pp. 3421–3452. [Google Scholar]
- 4.Peterson S, Liu J. Deciphering mode of action of heparanase using structurally defined oligosaccharides. J. Biol. Chem. 2012;287:34836–34843. doi: 10.1074/jbc.M112.390161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaneiwa T, Mizumoto S, Sugahara K, Yamada S. Identification of human hyaluronidase-4 as a novel chondroitin sulfate hydrolase that preferentially cleaves the galactosaminidic linkage in the trisulfated tetrasaccharide sequence. Glycobiology. 2010;20:300–309. doi: 10.1093/glycob/cwp174. [DOI] [PubMed] [Google Scholar]
- 6.Giugliani R, Federhen A, Rojas MV, Vieira T, Artigalas O, Pinto LL, Azevedo AC, Acosta A, Bonfim C, Lourenco CM, CA Kim, Horovitz D, Bonfim D, Norato D, Marinho D, Palhares D, Santos ES, Ribeiro E, Valadares E, Guarany F, de Lucca GR, Pimentel H, de Souza IN, Correa JN, Fraga JC, Goes JE, Cabral JM, Simionato J, Llerena J, Jr., Jardim L, Giuliani L, da Silva LC, Santos ML, MA Moreira, Kerstenetzky M, Ribeiro M, Ruas N, Barrios P, Aranda P, R Honjo, Boy R, Costa R, Souza C, Alcantara FF, Avilla SG, Fagondes S, Martins AM. Mucopolysaccharidosis I, II, and VI: Brief review and guidelines for treatment. Genet. Mol. Biol. 2010;33:589–604. doi: 10.1590/S1415-47572010005000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lau AA, Hannouche H, Rozaklis T, Hassiotis S, Hopwood JJ, Hemsley KM. Allogeneic stem cell transplantation does not improve neurological deficits in mucopolysaccharidosis type IIIA mice. Exp. Neurol. 2010;225:445–454. doi: 10.1016/j.expneurol.2010.07.024. [DOI] [PubMed] [Google Scholar]
- 8.Church H, Tylee K, Cooper A, Thornley M, Mercer J, Wraith E, Carr T, O’Meara A, Wynn RF. Biochemical monitoring after haemopoietic stem cell transplant for Hurler syndrome (MPSIH): implications for functional outcome after transplant in metabolic disease. Bone Marrow Transplant. 2007;39:207–210. doi: 10.1038/sj.bmt.1705569. [DOI] [PubMed] [Google Scholar]
- 9.Pan D. Cell- and gene-based therapeutic approaches for neurological deficits in mucopolysaccharidoses. Curr. Pharm Biotechnol. 2011;12:884–896. doi: 10.2174/138920111795542679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Ruijter J, Valstar MJ, FA Wijburg. Mucopolysaccharidosis Type III (Sanfilippo syndrome): emerging treatment strategies. Curr. Pharm Biotechnol. 2011;12:923–930. doi: 10.2174/138920111795542651. [DOI] [PubMed] [Google Scholar]
- 11.Jakobkiewicz-Banecka J, Piotrowska E, Gabig-Ciminska M, Borysiewicz E, Slominska-Wojewodzka M, Narajczyk M, Wegrzyn A, Wegrzyn G. Substrate reduction therapies for mucopolysaccharidoses. Curr. Pharm. Biotechnol. 2011;12:1860–1865. doi: 10.2174/138920111798376932. [DOI] [PubMed] [Google Scholar]
- 12.Valayannopoulos V, Wijburg FA. Therapy for the mucopolysaccharidoses. Rheumatology (Oxford) 2011;50:49–59. doi: 10.1093/rheumatology/ker396. [DOI] [PubMed] [Google Scholar]
- 13.Muenzer J. Overview of the mucopolysaccharidoses. Rheumatology (Oxford) 2011;50:4–12. doi: 10.1093/rheumatology/ker394. [DOI] [PubMed] [Google Scholar]
- 14.Neufeld EF. Enzyme replacement therapy - a brief history. In: Mehta A, Beck M, Sunder-Plassmann G, editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford: Oxford PharmaGenesis; 2006. [PubMed] [Google Scholar]
- 15.Dickson PI. Novel treatments and future perspectives: outcomes of intrathecal drug delivery. Int. J. Clin. Pharmacol. Ther. 2009;47:124–127. [PubMed] [Google Scholar]
- 16.Dickson P, McEntee M, Vogler C, Le S, Levy B, Peinovich M, Hanson S, Passage M, Kakkis E. Intrathecal enzyme replacement therapy: successful treatment of brain disease via the cerebrospinal fluid. Mol. Genet Metab. 2007;91:61–68. doi: 10.1016/j.ymgme.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wraith EJ, Hopwood JJ, Fuller M, Meikle PJ, Brooks DA. Laronidase treatment of mucopolysaccharidosis I. BioDrugs. 2005;19:1–7. doi: 10.2165/00063030-200519010-00001. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence R, Brown JR, Al-Mafraji K, Lamanna WC, Beitel JR, Boons GJ, Esko JD, Crawford BE. Disease-specific non-reducing end carbohydrate biomarkers for mucopolysaccharidoses. Nat. Chem. Biol. 2012;8:197–204. doi: 10.1038/nchembio.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemsley KM, Hopwood JJ. Lessons learnt from animal models: pathophysiology of neuropathic lysosomal storage disorders. J. Inherit. Metab. Dis. 2010;33:363–371. doi: 10.1007/s10545-010-9078-6. [DOI] [PubMed] [Google Scholar]
- 20.Aerts JM, Kallemeijn WW, Wegdam W, Joao Ferraz M, van Breemen MJ, Dekker N, Kramer G, Poorthuis BJ, Groener JE, Cox-Brinkman J, Rombach SM, Hollak CE, Linthorst GE, Witte MD, Gold H, van der Marel GA, Overkleeft HS, Boot RG. Biomarkers in the diagnosis of lysosomal storage disorders: proteins, lipids, and inhibodies. J. Inherit. Metab. Dis. 2011;34:605–619. doi: 10.1007/s10545-011-9308-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lehman TJ, Miller N, Norquist B, Underhill L, Keutzer J. Diagnosis of the mucopolysaccharidoses. Rheumatology (Oxford) 2011;50:41–48. doi: 10.1093/rheumatology/ker390. [DOI] [PubMed] [Google Scholar]
- 22.Clarke LA, Winchester B, Giugliani R, Tylki-Szymanska A, Amartino H. Biomarkers for the mucopolysaccharidoses: Discovery and clinical utility. Mol. Genet. Metab. 2012;106:395–402. doi: 10.1016/j.ymgme.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Adamczyk B, Tharmalingam T, Rudd PM. Glycans as cancer biomarkers. Biochim. Biophys. Acta. 2012;1820:1347–1353. doi: 10.1016/j.bbagen.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 24.Fuster MM, Esko JD. The sweet and sour of cancer: glycans as novel therapeutic targets. Nat. Rev. Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 25.Valstar MJ, Neijs S, Bruggenwirth HT, Olmer R, Ruijter GJ, Wevers RA, van Diggelen OP, Poorthuis BJ, Halley DJ, Wijburg FA. Mucopolysaccharidosis type IIIA: clinical spectrum and genotype-phenotype correlations. Ann. Neurol. 2010;68:876–887. doi: 10.1002/ana.22092. [DOI] [PubMed] [Google Scholar]
- 26.Whitley CB, Ridnour MD, Draper KA, Dutton CM, Neglia JP. Diagnostic test for mucopolysaccharidosis I. Direct method for quantifying excessive urinary glycosaminoglycan excretion. Clin. Chem. 1989;35:374–379. [PubMed] [Google Scholar]
- 27.de Jong JG, Wevers RA, Laarakkers C, Poorthuis BJ. Dimethylmethylene blue-based spectrophotometry of glycosaminoglycans in untreated urine: a rapid screening procedure for mucopolysaccharidoses. Clin. Chem. 1989;35:1472–1477. [PubMed] [Google Scholar]
- 28.Clarke LA, Wraith JE, Beck M, Kolodny EH, Pastores GM, Muenzer J, Rapoport DM, Berger KI, Sidman M, Kakkis ED, Cox GF. Long-term efficacy and safety of laronidase in the treatment of mucopolysaccharidosis I. Pediatrics. 2009;123:229–240. doi: 10.1542/peds.2007-3847. [DOI] [PubMed] [Google Scholar]
- 29.Muenzer J, Wraith JE, Beck M, Giugliani R, Harmatz P, Eng CM, Vellodi A, Martin R, Ramaswami U, Gucsavas-Calikoglu M, Vijayaraghavan S, Wendt S, Puga AC, Ulbrich B, Shinawi M, Cleary M, Piper D, Conway AM, Kimura A. A phase II/III clinical study of enzyme replacement therapy with idursulfase in mucopolysaccharidosis II (Hunter syndrome) Genet. Med. 2006;8:465–473. doi: 10.1097/01.gim.0000232477.37660.fb. [DOI] [PubMed] [Google Scholar]
- 30.Harmatz P, Giugliani R, Schwartz I, Guffon N, Teles EL, Miranda MC, Wraith JE, Beck M, Arash L, Scarpa M, Yu ZF, Wittes J, Berger KI, Newman MS, Lowe AM, Kakkis E, Swiedler SJ. Enzyme replacement therapy for mucopolysaccharidosis VI: a phase 3, randomized, double-blind, placebo-controlled, multinational study of recombinant human N-acetylgalactosamine 4-sulfatase (recombinant human arylsulfatase B or rhASB) and follow-on, open-label extension study. J. Pediatr. 2006;148:533–539. doi: 10.1016/j.jpeds.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 31.Davies NP, Roubin RH, Whitelock JM. Characterization and purification of glycosaminoglycans from crude biological samples. J. Agric. Food Chem. 2008;56:343–348. doi: 10.1021/jf072624v. [DOI] [PubMed] [Google Scholar]
- 32.Chih-Kuang C, Shuan-Pei L, Shyue-Jye L, Tuen-Jen W. MPS screening methods, the Berry spot and acid turbidity tests, cause a high incidence of false-negative results in Sanfilippo and Morquio syndromes. J. Clin. Lab. Anal. 2002;16:253–258. doi: 10.1002/jcla.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.de Jong JG, Heijs WM, Wevers RA. Mucopolysaccharidoses screening: dimethylmethylene blue versus alcian blue. Ann. Clin. Biochem. 1994;31:267–271. doi: 10.1177/000456329403100309. [DOI] [PubMed] [Google Scholar]
- 34.Mabe P, Valiente A, Soto V, Cornejo V, Raimann E. Evaluation of reliability for urine mucopolysaccharidosis screening by dimethylmethylene blue and Berry spot tests. Clin. Chim. Acta. 2004;345:135–140. doi: 10.1016/j.cccn.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 35.Lage S, Prieto JA, Andrade F, Sojo A, Sanjurjo P, Aldamiz-Echevarria LJ. Reliability of a visual test for the rapid detection of mucopolysaccharidoses: GAG-test((R)) J. Clin. Lab. Anal. 2011;25:179–184. doi: 10.1002/jcla.20453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomatsu S, Gutierrez MA, Ishimaru T, Pena OM, Montano AM, Maeda H, Velez-Castrillon S, Nishioka T, Fachel AA, Cooper A, Thornley M, Wraith E, Barrera LA, Laybauer LS, Giugliani R, Schwartz IV, Frenking GS, Beck M, Kircher SG, Paschke E, Yamaguchi S, Ullrich K, Isogai K, Suzuki Y, Orii T, Noguchi A. Heparan sulfate levels in mucopolysaccharidoses and mucolipidoses. J. Inherit. Metab. Dis. 2005;28:743–757. doi: 10.1007/s10545-005-0069-y. [DOI] [PubMed] [Google Scholar]
- 37.Hintze JP, Tomatsu S, Fujii T, Montano AM, Yamaguchi S, Suzuki Y, Fukushi M, Ishimaru T, Orii T. Comparison of liquid chromatography-tandem mass spectrometry and sandwich ELISA for determination of keratan sulfate in plasma and urine. Biomark. Insights. 2011;6:69–78. doi: 10.4137/BMI.S7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrosius M, Kleesiek K, Gotting C. Quantitative determination and comparison of the glycosaminoglycan Delta-disaccharide composition in 22 different human cell lines. Cell Biol. Int. 2009;33:848–852. doi: 10.1016/j.cellbi.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 39.Oguma T, Tomatsu S, Montano AM, Okazaki O. Analytical method for the determination of disaccharides derived from keratan, heparan, and dermatan sulfates in human serum and plasma by high-performance liquid chromatography/turbo ion spray ionization tandem mass spectrometry. Anal. Biochem. 2007;368:79–86. doi: 10.1016/j.ab.2007.05.016. [DOI] [PubMed] [Google Scholar]
- 40.Tomatsu S, Montano AM, Oguma T, Dung VC, Oikawa H, Gutierrez ML, Yamaguchi S, Suzuki Y, Fukushi M, Barrera LA, Kida K, Kubota M, Orii T. Validation of disaccharide compositions derived from dermatan sulfate and heparan sulfate in mucopolysaccharidoses and mucolipidoses II and III by tandem mass spectrometry. Mol. Genet. Metab. 2010;99:124–131. doi: 10.1016/j.ymgme.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Wei W, Ninonuevo MR, Sharma A, Danan-Leon LM, Leary JA. A comprehensive compositional analysis of heparin/heparan sulfate-derived disaccharides from human serum. Anal. Chem. 2011;83:3703–3708. doi: 10.1021/ac2001077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holley RJ, Deligny A, Wei W, Watson HA, Ninonuevo MR, Dagalv A, Leary JA, Bigger BW, Kjellen L, Merry CL. Mucopolysaccharidosis type I: Unique structure of accumulated heparan sulfate and increased N-sulfotransferase activity in mice lacking alpha-L-iduronidase. J. Biol. Chem. 2011;286:37515–37524. doi: 10.1074/jbc.M111.287474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maccarana M, Casu B, Lindahl U. Minimal sequence in heparin/heparan sulfate required for binding of basic fibroblast growth factor. J. Biol. Chem. 1993;268:23898–23905. [PubMed] [Google Scholar]
- 44.Bai XM, Esko JD. An animal cell mutant defective in heparan sulfate hexuronic acid 2-O-sulfation. J. Biol. Chem. 1996;271:17711–17717. doi: 10.1074/jbc.271.30.17711. [DOI] [PubMed] [Google Scholar]
- 45.Crawford BE, Garner OB, Bishop JR, Zhang DY, Bush KT, Nigam SK, Esko JD. Loss of the heparan sulfate sulfotransferase, Ndst1, in mammary epithelial cells selectively blocks lobuloalveolar development in mice. PLoS One. 2010;5:e10691. doi: 10.1371/journal.pone.0010691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lortat-Jacob H, Kleinman HK, Grimaud JA. High-affinity binding of interferon-gamma to a basement membrane complex (matrigel) J. Clin. Invest. 1991;87:878–883. doi: 10.1172/JCI115093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salmivirta M, Safaiyan F, Prydz K, Andresen MS, ARyan M, Kolset SO. Differentiation-associated modulation of heparan sulfate structure and function in CaCo-2 colon carcinoma cells. Glycobiology. 1998;8:1029–1036. doi: 10.1093/glycob/8.10.1029. [DOI] [PubMed] [Google Scholar]
- 48.Maccarana M, Lindahl U. Mode of interaction between platelet factor 4 and heparin. Glycobiology. 1993;3:271–277. doi: 10.1093/glycob/3.3.271. [DOI] [PubMed] [Google Scholar]
- 49.Randall DR, Sinclair GB, Colobong KE, Hetty E, Clarke LA. Heparin cofactor II-thrombin complex in MPS I: a biomarker of MPS disease. Mol. Genet. Metab. 2006;88:235–243. doi: 10.1016/j.ymgme.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 50.Halldorsdottir AM, Zhang L, Tollefsen DM. N-Acetylgalactosamine 4,6-O-sulfate residues mediate binding and activation of heparin cofactor II by porcine mucosal dermatan sulfate. Glycobiology. 2006;16:693–701. doi: 10.1093/glycob/cwj117. [DOI] [PubMed] [Google Scholar]
- 51.Tollefsen DM. Heparin cofactor II modulates the response to vascular injury. Arterioscler. Thromb. Vasc. Biol. 2007;27:454–460. doi: 10.1161/01.ATV.0000256471.22437.88. [DOI] [PubMed] [Google Scholar]
- 52.Langford-Smith KJ, Mercer J, Petty J, Tylee K, Church H, Roberts J, Moss G, Jones S, Wynn R, Wraith JE, Bigger BW. Heparin cofactor II-thrombin complex and dermatan sulphate:chondroitin sulphate ratio are biomarkers of short- and long-term treatment effects in mucopolysaccharide diseases. J. Inherit. Metab. Dis. 2011;34:499–508. doi: 10.1007/s10545-010-9254-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Randall DR, Colobong KE, Hemmelgarn H, Sinclair GB, Hetty E, Thomas A, Bodamer OA, Volkmar B, Fernhoff PM, Casey R, Chan AK, Mitchell G, Stockler S, Melancon S, Rupar T, Clarke LA. Heparin cofactor II-thrombin complex: a biomarker of MPS disease. Mol. Genet. Metab. 2008;94:456–461. doi: 10.1016/j.ymgme.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 54.Clarke LA, Hemmelgarn H, Colobong K, Thomas A, Stockler S, Casey R, Chan A, Fernoff P, Mitchell J. Longitudinal observations of serum heparin cofactor II-thrombin complex in treated Mucopolysaccharidosis I and II patients. J. Inherit. Metab. Dis. 2012;35:355–362. doi: 10.1007/s10545-011-9369-6. [DOI] [PubMed] [Google Scholar]
- 55.Whiteman P. Prenatal diagnosis of mucopolysaccharidoses. Lancet. 1973;1:1249. doi: 10.1016/s0140-6736(73)90560-6. [DOI] [PubMed] [Google Scholar]
- 56.Wynn RF, Wraith JE, Mercer J, O’Meara A, Tylee K, Thornley M, Church HJ, Bigger BW. Improved metabolic correction in patients with lysosomal storage disease treated with hematopoietic stem cell transplant compared with enzyme replacement therapy. J. Pediatr. 2009;154:609–611. doi: 10.1016/j.jpeds.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 57.Fuller M, Meikle PJ, Hopwood JJ. Glycosaminoglycan degradation fragments in mucopolysaccharidosis I. Glycobiology. 2004;14:443–450. doi: 10.1093/glycob/cwh049. [DOI] [PubMed] [Google Scholar]
- 58.King B, Savas P, Fuller M, Hopwood J, Hemsley K. Validation of a heparan sulfate-derived disaccharide as a marker of accumulation in murine mucopolysaccharidosis type IIIA. Mol. Genet. Metab. 2006;87:107–112. doi: 10.1016/j.ymgme.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 59.Hopwood JJ, Elliott H. N-acetylglucosamine 6-sulfate residues in keratan sulfate and heparan sulfate are desulfated by the same enzyme. Biochem. Int. 1983;6:141–148. [PubMed] [Google Scholar]
- 60.Hopwood JJ, Elliott H. Isolation and characterization of N-acetylglucosamine 6-sulfate from the urine of a patient with Sanfilippo type D syndrome and its occurrence in normal urine. Biochem. Int. 1983;6:831–836. [PubMed] [Google Scholar]
- 61.Hopwood JJ, Elliott H. Urinary excretion of sulphated N-acetylhexosamines in patients with various mucopolysaccharidoses. Biochem. J. 1985;229:579–586. doi: 10.1042/bj2290579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ramsay SL, Meikle PJ, Hopwood JJ. Determination of monosaccharides and disaccharides in mucopolysaccharidoses patients by electrospray ionisation mass spectrometry. Mol. Genet. Metab. 2003;78:193–204. doi: 10.1016/s1096-7192(03)00018-0. [DOI] [PubMed] [Google Scholar]
- 63.de Ruijter J, Ijlst L, Kulik W, van Lenthe H, Wagemans T, van Vlies N, Wijburg FA. Heparan sulfate derived disaccharides in plasma and total urinary excretion of glycosaminoglycans correlate with disease severity in Sanfilippo disease. J. Inherit. Metab. Dis. 2013;36:271–279. doi: 10.1007/s10545-012-9535-5. [DOI] [PubMed] [Google Scholar]
- 64.de Ruijter J, de Ru MH, Wagemans T, Ijlst L, Lund AM, Orchard PJ, Schaefer GB, Wijburg FA, van Vlies N. Heparan sulfate and dermatan sulfate derived disaccharides are sensitive markers for newborn screening for mucopolysaccharidoses types I, II and III. Mol. Genet. Metab. 2012;107:705–710. doi: 10.1016/j.ymgme.2012.09.024. [DOI] [PubMed] [Google Scholar]
- 65.Tomatsu S, Montano AM, Oguma T, Dung VC, Oikawa H, de Carvalho TG, Gutierrez ML, Yamaguchi S, Suzuki Y, Fukushi M, Sakura N, Barrera L, Kida K, Kubota M, Orii T. Dermatan sulfate and heparan sulfate as a biomarker for mucopolysaccharidosis I. J. Inherit. Metab. Dis. 2010;33:141–150. doi: 10.1007/s10545-009-9036-3. [DOI] [PubMed] [Google Scholar]
- 66.Tomatsu S, Montano AM, Oguma T, Dung VC, Oikawa H, de Carvalho TG, Gutierrez ML, Yamaguchi S, Suzuki Y, Fukushi M, Kida K, Kubota M, Barrera L, Orii T. Validation of keratan sulfate level in mucopolysaccharidosis type IVA by liquid chromatog-raphy–tandem mass spectrometry. J. Inherit. Metab. Dis. 2010 doi: 10.1007/s10545-009-9013-x. http://dx.doi.org/10.1007/s10545-009-9013-x. [DOI] [PubMed] [Google Scholar]
- 67.Montano AM, Oikawa H, Tomatsu S, Nishioka T, Vogler C, Gutierrez MA, Oguma T, Tan Y, Grubb JH, Dung VC, Ohashi A, Miyamoto K, Orii T, Yoneda Y, Sly WS. Acidic amino acid tag enhances response to enzyme replacement in mucopolysaccharidosis type VII mice. Mol. Genet. Metab. 2008;94:178–189. doi: 10.1016/j.ymgme.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 68.Hendriksz CJ, Harmatz P, Beck M, Jones S, Wood T, Lachman R, Gravance CG, Orii T, Tomatsu S. Review of clinical presentation and diagnosis of mucopolysaccharidosis IVA. Mol. Genet. Metab. 2013 doi: 10.1016/j.ymgme.2013.04.002. http://dx.doi.org/10.1016/j.ymgme.2013.04.002 (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Linhardt RJ, Galliher PM, Cooney CL. Polysaccharide lyases. Appl. Biochem. Biotechnol. 1986;12:135–176. doi: 10.1007/BF02798420. [DOI] [PubMed] [Google Scholar]
- 70.Byers S, Rozaklis T, Brumfield LK, Ranieri E, Hopwood JJ. Glycosaminoglycan accumulation and excretion in the mucopolysaccharidoses: characterization and basis of a diagnostic test for MPS. Mol. Genet. Metab. 1998;65:282–290. doi: 10.1006/mgme.1998.2761. [DOI] [PubMed] [Google Scholar]
- 71.Bhaumik M, Muller VJ, Rozaklis T, Johnson L, Dobrenis K, Bhattacharyya R, Wurzelmann S, Finamore P, Hopwood JJ, Walkley SU, Stanley P. A mouse model for mucopolysaccharidosis type III A (Sanfilippo syndrome) Glycobiology. 1999;9:1389–1396. doi: 10.1093/glycob/9.12.1389. [DOI] [PubMed] [Google Scholar]
- 72.Lawrence R, Olson SK, Steele RE, Wang L, Warrior R, Cummings RD, Esko JD. Evolutionary differences in glycosaminoglycan fine structure detected by quantitative glycan reductive isotope labeling. J. Biol. Chem. 2008;283:33674–33684. doi: 10.1074/jbc.M804288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Xia B, Feasley CL, Sachdev GP, Smith DF, Cummings RD. Glycan reductive isotope labeling for quantitative glycomics. Anal. Biochem. 2009;387:162–170. doi: 10.1016/j.ab.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Staples GO, Shi X, Zaia J. Extended N-sulfated domains reside at the nonreducing end of heparan sulfate chains. J. Biol. Chem. 2010;285:18336–18343. doi: 10.1074/jbc.M110.101592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lamanna WC, Lawrence R, Sarrazin S, Lameda-Diaz C, Gordts PL, Moremen KW, Esko JD. A genetic model of substrate reduction therapy for mucopolysaccharidosis. J. Biol. Chem. 2012;287:36283–36290. doi: 10.1074/jbc.M112.403360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tomatsu S, Okamura K, Taketani T, Orii KO, Nishioka T, Gutierrez MA, Velez-Castrillon S, Fachel AA, Grubb JH, Cooper A, Thornley M, Wraith E, Barrera LA, Giugliani R, Schwartz IV, Frenking GS, Beck M, Kircher SG, Paschke E, Yamaguchi S, Ullrich K, Isogai K, Suzuki Y, Orii T, Kondo N, Creer M, Noguchi A. Development and testing of new screening method for keratan sulfate in mucopolysaccharidosis IVA. Pediatr. Res. 2004;55:592–597. doi: 10.1203/01.PDR.0000113767.60140.E9. [DOI] [PubMed] [Google Scholar]
- 77.Martell LA, Cunico RL, Ohh J, Fulkerson W, Furneaux R, Foehr ED. Validation of an LC-MS/MS assay for detecting relevant disaccharides from keratan sulfate as a biomarker for Morquio A syndrome. Bioanalysis. 2011;3:1855–1866. doi: 10.4155/bio.11.172. [DOI] [PubMed] [Google Scholar]
- 78.Tomatsu S, Okamura K, Maeda H, Taketani T, Castrillon SV, Gutierrez MA, Nishioka T, Fachel AA, Orii KO, Grubb JH, Cooper A, Thornley M, Wraith E, Barrera LA, Laybauer LS, Giugliani R, Schwartz IV, Frenking GS, Beck M, Kircher SG, Paschke E, Yamaguchi S, Ullrich K, Haskins M, Isogai K, Suzuki Y, Orii T, Kondo N, Creer M, Okuyama T, Tanaka A, Noguchi A. Keratan sulphate levels in mucopolysaccharidoses and mucolipidoses. J. Inherit. Metab. Dis. 2005;28:187–202. doi: 10.1007/s10545-005-5673-3. [DOI] [PubMed] [Google Scholar]
- 79.Ramsay SL, Maire I, Bindloss C, Fuller M, Whitfield PD, Piraud M, Hopwood JJ, Meikle PJ. Determination of oligosaccharides and glycolipids in amniotic fluid by electrospray ionisation tandem mass spectrometry: in utero indicators of lysosomal storage diseases. Mol. Genet. Metab. 2004;83:231–238. doi: 10.1016/j.ymgme.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 80.Meikle PJ, Ranieri E, Simonsen H, Rozaklis T, Ramsay SL, Whitfield PD, Fuller M, Christensen E, Skovby F, Hopwood JJ. Newborn screening for lysosomal storage disorders: clinical evaluation of a two-tier strategy. Pediatrics. 2004;114:909–916. doi: 10.1542/peds.2004-0583. [DOI] [PubMed] [Google Scholar]
- 81.Langford-Smith K, Arasaradnam M, Wraith JE, Wynn R, Bigger BW. Evaluation of heparin cofactor II-thrombin complex as a biomarker on blood spots from mucopolysaccharidosis I, IIIA and IIIB mice. Mol. Genet. Metab. 2010;99:269–274. doi: 10.1016/j.ymgme.2009.10.175. [DOI] [PubMed] [Google Scholar]
- 82.Malinowska M, Wilkinson FL, Langford-Smith KJ, Langford-Smith A, Brown JR, Crawford BE, Vanier MT, Grynkiewicz G, Wynn RF, Wraith JE, Wegrzyn G, Bigger BW. Genistein improves neuropathology and corrects behaviour in a mouse model of neurodegenerative metabolic disease. PLoS One. 2010;5:e14192. doi: 10.1371/journal.pone.0014192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dickson PI, Ellinwood NM, Brown JR, Witt RG, Le SQ, Passage MB, Vera MU, Crawford BE. Specific antibody titer alters the effectiveness of intrathecal enzyme replacement therapy in canine mucopolysaccharidosis I. Mol. Genet. Metab. 2012;106:68–72. doi: 10.1016/j.ymgme.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lamanna WC, Lawrence R, Sarrazin S, Esko JD. Secondary storage of dermatan sulfate in Sanfilippo disease. J. Biol. Chem. 2011;286:6955–6962. doi: 10.1074/jbc.M110.192062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Diez-Roux G, Ballabio A. Sulfatases and human disease. Annu. Rev. Genomics Hum. Genet. 2005;6:355–379. doi: 10.1146/annurev.genom.6.080604.162334. [DOI] [PubMed] [Google Scholar]
- 86.Abitbol M, Thibaud JL, Olby NJ, Hitte C, Puech JP, Maurer M, Pilot-Storck F, Hedan B, Dreano S, Brahimi S, Delattre D, Andre C, Gray F, Delisle F, Caillaud C, Bernex F, Panthier JJ, Aubin-Houzelstein G, Blot S, Tiret L. A canine Arylsulfatase G (ARSG) mutation leading to a sulfatase deficiency is associated with neuronal ceroid lipofuscinosis. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14775–14780. doi: 10.1073/pnas.0914206107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kowalewski B, Lamanna WC, Lawrence R, Damme M, Stroobants S, Padva M, Kalus I, Frese MA, Lubke T, Lullmann-Rauch R, D’Hooge R, Esko JD, Dierks T. Arylsulfatase G inactivation causes loss of heparan sulfate 3-O-sulfatase activity and mucopolysaccharidosis in mice. Proc. Natl. Acad. Sci. U. S. A. 2012;109:10310–10315. doi: 10.1073/pnas.1202071109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Marth JD, Bertozzi CR, Hart GW, Etzler ME. Symbol nomenclature for glycan representation. Proteomics. 2009;9:5398–5399. doi: 10.1002/pmic.200900708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat. Methods. 2008;5:291–292. doi: 10.1038/nmeth0408-291. [DOI] [PubMed] [Google Scholar]