Abstract
Metabolic Syndrome (MetS) is a phenotype cluster predisposing to type 2 diabetes and cardiovascular diseases. In extended families of Northern European ancestry, we previously identified two significant QTLs 3q27 and 17p12 that were linked with multiple representative traits of MetS. To determine the genetic basis of these linkage signals, QTL-specific genomic and transcriptomic analyses were performed in 1,137 individuals from 85 extended families that contributed to the original linkage. We tested in SOLAR association of MetS phenotypes with QTL-specific haplotype-tagging SNPs as well as transcriptional profiles of peripheral blood mononuclear cells (PBMCs). SNPs significantly associated with phenotypes under the prior hypothesis of linkage mapped to seven genes at 3q27 and seven at 17p12. Prioritization based on biologic relevance, SNP association, and expression analyses identified two genes: insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) at 3q27 and tumor necrosis factor receptor 13B (TNFRSF13B) at 17p12. Prioritized genes could influence cell-cell adhesion and adipocyte differentiation, insulin/glucose responsiveness, cytokine effectiveness and plasma lipids and lipoprotein densities. In summary, our results combine genomic, transcriptomic, and bioinformatic data to identify novel candidate loci for MetS.
INTRODUCTION
Obesity is a common disorder with both environmental and genetic determinants (1). Its frequency has now reached epidemic proportions with the number of affected individuals in the United States approaching 100 million (2). The Metabolic Syndrome (MetS) is a form of obesity characterized by a cluster of phenotypes that include enlargement of the abdominal/visceral fat compartment, insulin resistance and glucose intolerance, dyslipidemia with increased plasma levels of triglycerides and decreased levels of HDL-cholesterol, increased blood pressure, and elevated circulating cytokines and adipokines. It is associated with increased risk for type 2 diabetes and cardiovascular disease (3).
Twin, familial and population studies have confirmed existence of a genetic basis for MetS (4). Several studies have identified a number of candidate and/or animal homology genes that are associated with its phenotypic expression (5, 6). Recent studies have employed genome-wide association scans (GWAS) and genome-wide expression profiling (7, 8, 9,10). However, these discoveries account for only a small percentage of the total variance of the MetS traits, and the full spectrum of contributing loci is likely to be larger.
We previously performed a genome-wide linkage scan using microsatellite markers at 10 centiMorgan (cM) intervals in 2,209 members of 507 nuclear families of predominantly Northern European ancestry (11). Quantitative trait linkage analysis revealed a highly significant locus (QTL) at chr3q27 associated with body composition and insulin/glucose responsiveness. A second locus was linked to plasma leptin levels at chr17p12. Plasma levels of leptin, an adipose tissue product, have been found correlated with total body adiposity. These findings were replicated in several studies of different ethnic groups (12, 13). These QTLs were also found to be epistatically interactive and pleiotropic, influencing a broad spectrum of the MetS phenotypes (11).
The present study was undertaken in 1,137 individuals in 352 nuclear families (85 extended families) of Northern European descent from the cohort that contributed to the QTL linkage. We conducted an in-depth phenotyping procedure by measuring 42 MetS traits that included both clinical outcome and biologic precursor phenotypes. In addition, we obtained dense haplotype-tagging SNP genotypes and transcriptional profiles of peripheral blood mononuclear cells (PBMCs) for a subset of these individuals. Gene prioritization was based upon biologic relevance as well as SNP and PBMC expression associations with the MetS phenotypes.
METHODS AND PROCEDURES
Subjects and phenotypes
The study cohort consists of 1,137 individuals representing 85 extended nuclear families. Analysis of distribution of pairwise relationships showed the majority of the number of pairs were highest of parent-offspring (n=1300) and between siblings (n=1494). Details of recruitment and phenotyping procedures have been described previously (11). Briefly, each family was recruited through an obese proband (BMI ≥ 30) with the minimal requirement of the availability of one obese sibling and one never-obese (BMI ≤ 27) and availability of at least one, preferably both, of the parents. Clinical phenotypes included weight, height, BMI, waist circumference (WC), hip circumference (HC), waist to hip ratio (WHR), fasting glucose (FG), fasting insulin (FI), insulin to glucose ratio (IGR), homeostasis model assessment (HOMA), plasma triglycerides (TG), total cholesterol (TC), LDL-cholesterol (LDL-c) and calculated LDL-cholesterol levels (cal. LDL-c), HDL-cholesterol (HDL-c), systolic and diastolic blood pressure (sBP and dBP) and pulse. Biological phenotypes were determined according to standard published procedures, included measurement of total fat mass in kilogram and percentage (Fatkg and Fatpct) and lean mass in kilogram and percentage (Leankg and Leanpct) by Dual-emission X-ray absorptiometry (DEXA) (14); total abdominal, visceral and subcutaneous fat sizes (TAF, VF and SubQF) by computed tomography (CT/MRI) scans average of three sections at the fourth lumbar spine (15); resting energy expenditure (REE) and respiratory quotient (RQ) were measured in resting subjects using a Deltatrac Indirect Calorimeter (Sensor Medics, VIASYS Healthcare, Conshohocken,PA) after a 10 hr fast; insulin/glucose responsiveness indices: insulin sensitivity (SI), glucose effectiveness (SG), acute insulin response to glucose (AIRG) and disposition index (DI) by Minimal Model (16); lipids/lipoprotein sizing [HDL median diameter (HMED), LDL-cholesterol median diameter (LMEDn), LDL-cholesterol dominant peak diameter (LDLppd) and apoB-containing non-HDL median diameter (BMED) which includes VLDL, ILDL, LPα and LDL] were measured by polyacrylamide gradient gel electrophoresis (17); circulating levels of adiponectin, leptin by a double antibody, equilibrium RIA (Millipore Corporation, Billerica, MA); and TNF-alpha, interleukin-1beta (IL-1β) and interleukin-6 (IL-6)] as described (18). All study procedures for adults and children were approved by the Institutional Review Boards of the Medical College of Wisconsin and Children’s Hospital of Wisconsin, respectively.
SNP Genotyping and Data Cleaning
Genomic DNA was extracted and prepared from whole blood using commercial kits (Puregene, Minneapolis, MN). Genome-wide SNP genotyping was performed using Affymetrix Genome-Wide Human SNP 6.0 arrays and SNP calls were generated by Genotype Console 3.2. Individuals with fewer than 95% of available markers called were excluded. 869,222 autosomal SNPs were prepared by Preswalk and checked for Mendelian consistency with SimWalk2. A SNP was eliminated if: 1) fewer than 95% of the cohort were typed successfully; 2) the SNP was monoallelic; 3) the SNP had more than two alleles; 4) fewer than five copies of the SNP existed in the study cohort. Hardy-Weinberg equilibrium (HWE) was tested for each SNP using SOLAR (19); SNPs with excessive deviation from HWE (p < 10−8) were excluded. Missing SNP data were imputed with MERLIN (20).
Transcriptional profiling
Genome-wide transcriptional profiles of a subset of the SNP genotyping cohort (369 individuals from 55 nuclear families) were obtained as previously described (21) with modifications. Briefly, for each individual 2.5ml blood was collected into a PAXgene® Blood RNA Tube (BD, Franklin Lakes, NJ) following an overnight fast. Total RNA was isolated from each tube using the PAXgene Blood RNA Kit (Qiagen, Valencia, CA) and anti-sense RNA (aRNA) was synthesized using the MessageAmp II-Biotin aRNA kit (Ambion, Austin, TX). A total of 1.5 μg aRNA was hybridized to Illumina HumanWG-6 version 2 or version 3 chips (Illumina, San Diego, CA) and expression detected on the Illumina® BeadArray™ 500GX Reader. Illumina GenomeStudio software (version 2010.3) was used for preliminary data analysis with standard background normalization.
Statistical Analysis
Variance-component-based pedigree analysis
The heritability of each of the 42 MetS traits was first established, then a bivariate analysis in SOLAR was used to partition phenotypic correlations between the traits into presumed environmental and additive genetic components, as described previously (22).
Measured genotype analysis
In SOLAR, each SNP genotype was converted to a covariate measure equal to 0, 1 or 2 copies of the minor allele (or, for missing genotypes, a weighted covariate based on imputation). These covariates were included in variance-components mixed models for measured genotype analyses (23) versus null models that incorporated the random effect of kinship and fixed effects such as age, age2, sex and their interactions. Individual scores from a principal components analysis of representative SNPs were also included to correct for possible population stratification (24).
Within chromosomal regions showing the strongest prior evidence of linkage (as logarithm of odds, LOD, score) with MetS traits in our MRC-OB cohort, we selected all available SNPs on the Affymetrics 6.0 array that mapped within a 1-LOD confidence interval of the maximum LOD score in each region. Each SNP covariate was tested independently in a 1 degree of freedom likelihood ratio test. To account for the linkage disequilibrium between SNPs, we calculated the effective number of independent SNPs (Neff) using the method of Moskvina & Schmidt (25). Critical p-values were calculated using Bonferroni/Šidák correction for each linkage region based on its Neff.
Gene expression analysis
Microarray data were available in two batches, one of Version 2 arrays (48,701 probes, 307 samples) and the other of Version 3 (48,803 probes, 230 samples). To guard against possible batch effects and probe differences, each batch was analyzed separately: The number of probe transcripts detectable at p≤0.05 by BeadStudio software was counted, a false discovery rate (FDR) was computed across all probes, and transcripts detectable at 5% FDR were retained. Expression levels were log2 transformed and inverse-quartile normalized. Transformed and normalized expression levels for probes that mapped to the 1-LOD QTL regions were tested for association with each phenotype in models that included the random effect of kinship. Gene-centric p-values were calculated by combining independent p-values from the two microarray batches and multiple probes using Stouffer’s weighted Z-score method (26) implemented in R.
RESULTS
Heritability and genetic inter-correlation
A total of 1,137 individuals, representing extended families that contributed to the linkage on chromosomes 3 and 17 (11), were recruited and measured for the clinical phenotypes. 503 of those also underwent the biologic phenotyping procedures and 369 individuals also provided blood PBMCs for the transcriptional profiling. Their mean (±SD) age was 40.29y±SD=17.68y and ranged from 6 to 90y, 11.7% of the subjects were younger than age 18 and 58.3% of the subjects were female. Table 1 shows means (± SD) of the 42 phenotypes and their levels of heritability. All phenotypes measured were significantly heritable (p < 0.05), justifying further genetic analysis.
Table1. MetS Cohort phenotypic characteristics and heritability levels.
Table 1 shows mean (±SD) and additive heritability of 42 clinical and biologic phenotypes of MetS. For cohort recruitment and phenotyping, see Methods. Clinical phenotypes were measured in 1,137 individuals from 85 extended families that contributed highly to the linkages at 3q27 and 17p12 loci. Biologic phenotypes were measured in 503 individuals.
| Trait | Mean | Heritability ± SE | |
|---|---|---|---|
| Body composition and insulin responsiveness | Weight, kg | 87.57 ± 27.36 | 0.47 ± 0.06 |
| Height, cm | 167.7 ± 12.45 | 0.56 ± 0.05 | |
| BMI, kg/m2 | 30.94 ± 9.31 | 0.44 ± 0.06 | |
| Waist circumference (WC), cm | 95.56 ± 21 | 0.40 ± 0.06 | |
| Hip circumference (HC), cm | 107.94 ± 22.04 | 0.57 ± 0.05 | |
| Waist to Hip ratio (WHR) | 0.89 ± 0.1 | 0.31 ± 0.06 | |
| Total Fat Mass (Fatkg), kg | 32.36 ± 16.47 | 0.50 ± 0.1 | |
| Total Fat Mass (Fatpct),% | 36.61 ± 12.14 | 0.48 ± 0.1 | |
| Total Lean Mass (Leankg), kg | 51.87 ± 13.56 | 0.66 ± 0.09 | |
| Total Lean Mass (Leanpct), % | 65.58 ± 31.75 | 0.44 ± 0.1 | |
| Subcutaneous Fat (SubQF), g | 311.9 ± 176.7 | 0.57 ± 0.1 | |
| Visceral Fat (VF), g | 177.9 ± 111.3 | 0.32 ± 0.07 | |
| Total Abdominal Fat (TAF), g | 489.8 ± 248.8 | 0.51 ± 0.1 | |
| Respiratory Quotient (RQ) | 0.83 ± 0.08 | 0.35 ± 0.09 | |
| Resting Energy Expenditure (REE), kcal/24 hrs | 1740.5 ± 342.21 | 0.31 ± 0.09 | |
| REE/weight, kcal/24hrs/kg | 19.26 ± 3.47 | 0.32 ± 0.11 | |
| REE/Lean mass (REE/LM), kcal/24hrs/kg | 33.48 ± 6.18 | 0.36 ± 0.1 | |
| Fasting Glucose (FG), mmol/l | 4.86 ± 1.64 | 0.38 ± 0.05 | |
| Fasting Insulin (FI),pmol/l | 107.5 ± 105.6 | 0.30 ± 0.05 | |
| Insulin/glucose (IGR) | 0.25 ± 0.62 | 0.25 ± 0.05 | |
| Homeostasis model assessment (HOMA) | 4.31 ± 8.5 | 0.30 ± 0.05 | |
| Insulin Sensitivity (SI), (× 10−4/min/μU/ml) | 3.05 ± 2.83 | 0.15 ± 0.09 | |
| Glucose Effectiveness (SG), min−1 | 0.02 ± 0.01 | 0.13 ± 0.08 | |
| Acute Insulin Response to glucose (AIRG), μU/ml × 10min | 467.02 ± 400.51 | 0.52 ± 0.08 | |
| Disposition Index (DI), AUC (Insulin0-10 min) × SI | 1191.18 ± 1081.76 | 0.26 ± 0.09 | |
|
| |||
|
Lipids/lipoprotein profiles
and cardiovascular performanc |
Triglycerides (TG), mmol/l | 1.33 ± 1.29 | 0.38 ± 0.06 |
| Total Cholesterol (TC), mmol/l | 4.88 ± 1.16 | 0.34 ± 0.05 | |
| LDL-cholesterol (LDL-c), mmol/l | 3.19 ± 0.98 | 0.28 ± 0.05 | |
| Calculated LDL-C (cal. LDL-c), mmol/l | 3.25 ± 1.16 | 0.35 ± 0.05 | |
| HDL-cholesterol (HDL-c), mmol/l | 1.06 ± 0.37 | 0.64 ± 0.06 | |
| HMED, nm | 8.93 ± 0.55 | 0.53 ± 0.07 | |
| LMEDn, nm | 27.03 ± 0.78 | 0.59 ± 0.07 | |
| LDLppd, nm | 27.35 ± 1.2 | 0.65 ± 0.07 | |
| BMED, nm | 27.55 ± 1.17 | 0.63 ± 0.07 | |
| Systolic Blood Pressure (sBP), mmHg | 124.58 ± 18.57 | 0.24 ± 0.05 | |
| Diastolic Blood Pressure (dBP), mmHg | 77.13 ± 11.27 | 0.24 ± 0.06 | |
| Pulse, beats/min | 71.17 ± 14.14 | 0.20 ± 0.05 | |
|
| |||
|
Adipokine
and cytokines |
Adiponectin, ng/ml | 9.51 ± 5.6 | 0.48 ± 0.06 |
| Leptin, ng/ml | 19.28 ± 17.3 | 0.24 ± 0.06 | |
| TNF-alpha, pg/ml | 4.44 ± 6.28 | 0.32 ± 0.06 | |
| Interleukin-1 beta (IL-1β), pg/ml | 1.33 ± 18.42 | 0.16 ± 0.06 | |
| Interleukin-6 (IL-6), pg/ml | 7.68 ± 53.48 | 0.25 ± 0.07 | |
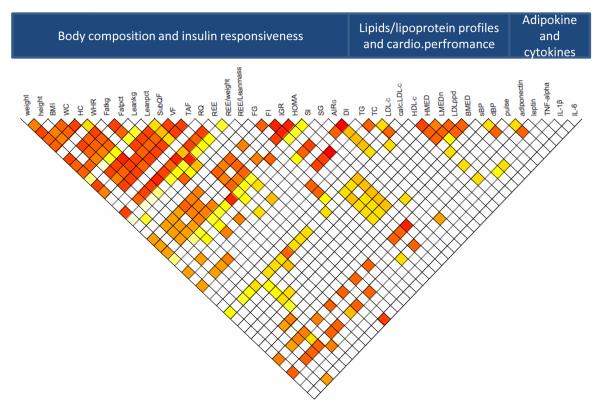
Figure 1 illustrates the traits that significantly inter-correlated genetically. BMI, the trait most commonly used in GWA analyses as the phenotype representing the degree of obesity, was correlated with the other phenotypes of body composition (WC and HC), insulin responsiveness (FI, IGR and HOMA), and lipid panel (TG and HDL-c). BMI was also significantly correlated with both total body fat and lean masses (Fatkg, Fatpct, Leankg and Leanpct. WC, another commonly used phenotype to represent central body or visceral fat distribution, was significantly correlated with clinical phenotypes of insulin responsiveness (IGR and HOMA), plasma lipids (TG and HDL-c) and cardiovascular performance measures (resting pulse and sBP). As with BMI, it was correlated both with visceral and subcutaneous fat masses (VF and SubQF). WHR, another clinical phenotype of body fat distribution, on the other hand was correlated with VF, REE, lipoprotein density profile and circulating levels of TNF-alpha. Of the biologic phenotypes, Fatkg was genetically correlated with REE, phenotypes of insulin responsiveness (SI, SG and AIRG), LDL peak size (LDLppd), and plasma leptin. VF was also genetically correlated with measures of body composition, REE, insulin sensitivity (SI), and plasma of IL-6. AIRG was correlated with Leankg, insulin responsiveness (DI), and leptin. Finally, SI, a direct marker of insulin sensitivity, was genetically correlated with all biologic measures of body composition (Fatkg, Leankg, Fatpct and Leanpct) and clinical parameters of insulin response.
Figure 1. Genetic inter-correlation between MetS phenotypes.
Heat map of pairwise genetic intercorrelation of 42 phenotypes. Colored tones represent only significant (p<0.05) correlations, red the strongest and yellow the weakest. A genetic correlation significantly different from zero suggests that the trait pair is influenced by the same gene or by genes in linkage disequilibrium. For abbreviations and units, see Table 1.
SNP genotype-phenotype association
Tables 2 and 3 show the highest levels of association of SNPs bordered by 1 LOD score drop from the linkage peak at 3q27 QTL (LODBMI=3.3) and 17p12 (LODLeptin=5.0), respectively. The strength of the association is indicated by the negative logarithm of p-values. The genes representative of this association annotated from those SNPs (hg18/NCBI36 human genome assembly) are also shown. The regional threshold for associations was determined by Bonferroni/Šidák correction for the effective number of independent SNPs given their linkage disequilibrium (nSNPs=6305, neffective=3881.47, pα=0.05=1.32×10−5, 3q27; nSNPs=1629, neffective=1412.93, pα=0.05=3.63×10−5, 17p12) (28).
Table 2. MetS SNP association with phenotypes and their gene annotation at 3q27 QTL.
SNPs within 1 LOD reduction from the linkage peak were tested against the 42 phenotypes. For phenotypic abbreviations, see Table 1. SNPs were mapped to genes and the known functions of these genes summarized, based on data from the NCBI build 36 human genome assembly. Annotations were determined at the position and within 250kb up- and downstream of the associated SNP. Data shown in column “−Log10(p)” are levels of the highest association. The QTL-specific significance threshold based on the effective number of SNPs was p0.05=1.32×10−5, −log(p-value)=4.941. Prioritized biologically relevant genes are shaded in gray and SNP associations that exceed statistical QTL-specific significance threshold are bolded. MAF, minor allele frequency.
| Variant | Minor Allele |
MAF | −Log10(p) | Associated phenotype |
Nearest gene | Location | Annotation | |
|---|---|---|---|---|---|---|---|---|
| Body composition and insulin responsiveness | rs6443434 | A | 0.43 | 3.76 | Weight | TBL1XR1 | upstream | signal transduction |
| rs11710431 | A | 0.19 | 4.55 | Height | TBL1XR1 | intergenic | signal transduction | |
| rs6443434 | A | 0.43 | 3.49 | BMI | TBL1XR1 | upstream | signal transduction | |
| rs6772734 | A | 0.02 | 3.82 | WC | LOC131583 | intergenic | unknown function | |
| rs13068101 | T | 0.28 | 3.06 | HC | C3orf59 | intergenic | unknown function | |
| rs12639517 | T | 0.33 | 4.03 | WHR | IGF2BP2 | intergenic | controls IGF translation | |
| rs11708240 | G | 0.14 | 3.54 | Fatkg | NAALADL2 | intergenic | dipeptidase | |
| rs13098487 | G | 0.22 | 3.59 | Fatpct | NLGN1 | intronic | brain synapses | |
| rs569221 | A | 0.19 | 4.47 | Leankg | NLGN1 | intronic | brain synapses | |
| rs301177 | C | 0.23 | 3.39 | Leanpct | TBL1XR1 | intergenic | signal transduction | |
| rs4074869 | C | 0.44 | 3.70 | SubQF | EPHB3 | intergenic | ephrinB receptor | |
| rs7433602 | T | 0.29 | 5.03 | VF | NAALADL2 | intronic | dipeptidase | |
| rs12496403 | A | 0.01 | 3.76 | TAF | TBL1XR1 | intergenic | signal transduction | |
| rs13324849 | T | 0.09 | 4.39 | RQ | NAALADL2 | intergenic | dipeptidase | |
| rs6443434 | A | 0.43 | 3.51 | REE | TBL1XR1 | upstream | signal transduction | |
| rs4074869 | C | 0.44 | 4.47 | REE/weight | EPHB3 | intergenic | ephrinB receptor | |
| rs13321995 | A | 0.40 | 4.89 | REE/Leanmass | C3orf21 | intronic | unknown function | |
| rs6807927 | A | 0.27 | 3.96 | FG | SENP2 | intronic | SUMO protease | |
| rs10933727 | G | 0.32 | 4.43 | FI | LOC100131551 | intergenic | unknown function | |
| rs2513 | A | 0.10 | 3.57 | IGR | AK091265 | downstream | unknown function | |
| rs10933727 | G | 0.32 | 5.93 | HOMA | LOC100131551 | intergenic | unknown function | |
| rs9814412 | T | 0.27 | 3.90 | SI | HRG | 5’UTR | thrombosis | |
| rs1463332 | C | 0.32 | 3.29 | SG | TBL1XR1 | intergenic | signal transduction | |
| rs6785028 | T | 0.26 | 3.87 | AIR | LPP | intronic | cell adhesion | |
| rs937589 | T | 0.02 | 3.70 | DI | C3orf59 | intronic | unknown function | |
|
| ||||||||
|
Lipids/lipoprotein profiles
and cardiovascular performance |
rs10804867 | A | 0.26 | 3.74 | TG | TBL1XR1 | intergenic | signal transduction |
| rs1895070 | A | 0.10 | 4.66 | TC | mRNA FLJ31690 | downstream | unknown function | |
| rs1895070 | A | 0.10 | 4.25 | LDL-c | mRNA FLJ31690 | downstream | unknown function | |
| rs1895070 | A | 0.10 | 4.57 | cal.LDL-c | mRNA FLJ31690 | downstream | unknown function | |
| rs4452373 | G | 0.00 | 3.90 | HDL-c | mRNA HQ448050 | intergenic | unknown function | |
| rs9716 | C | 0.24 | 3.55 | HMED | ST6GAL1 | downstream | glycosyltransferase | |
| rs6793161 | T | 0.01 | 3.62 | LMEDn | mRNA AF338197 | intergenic | unknown function | |
| rs2700839 | C | 0.12 | 3.70 | LDLppd | ATP11B | intergenic | ATPase | |
| rs10937009 | G | 0.00 | 5.14 | BMED | NDUFB5 | intergenic | repiratory chain | |
| rs260589 | G | 0.33 | 3.27 | sBP | TPRG1 | intergenic | tumor protein regulated | |
| rs6444517 | G | 0.10 | 4.64 | dBP | OSTN | intergenic | osteoblastic differentiation | |
| rs7631640 | C | 0.09 | 4.51 | Pulse | MCF2L2 | downstream | guanine exchange factor | |
|
| ||||||||
|
Adipokine
and cytokines |
rs17427594 | C | 0.10 | 3.60 | Adiponectin | CLDN1 | intergenic | cell junction |
| rs6772734 | A | 0.02 | 3.45 | Leptin | FAM43A | intergenic | unknown function | |
| rs13070418 | T | 0.36 | 5.06 | TNFalfa | SOX2 | intergenic | transcription factor | |
| rs9877500 | C | 0.22 | 4.11 | IL-1β | IQCG | intronic | unknown function | |
| rs6807480 | A | 0.36 | 3.60 | IL-6 | FGF12 | intronic | fibroblast growth factor | |
Table 3. MetS SNP association with phenotypes and their gene annotation at 17p12 QTL.
SNPs within 1 LOD reduction from the linkage peak were tested against the 42 phenotypes. For phenotypic abbreviations, see Table 1. SNPs were mapped to genes and the known functions of these genes summarized, based on data from the NCBI build 36 human genome assembly. Annotations were determined at the position and within 250kb up- and downstream of the associated SNP. Data shown in column “−Log10(p)” are levels of the highest association. The QTL-specific significance threshold based on the effective number of SNPs was ρα=0.05=3.63×10−5, −log(p-value)=4.361. Prioritized biologically relevant genes are shaded in gray and SNP associations that exceed statistical QTL-specific significance threshold are bolded. MAF, minor allele frequency.
| Variant | Minor Allele |
MAF | −Log10(p) | Associated phenotype |
Nearest gene | Location | Annotation | |
|---|---|---|---|---|---|---|---|---|
| Body composition and insulin responsiveness | rs9889434 | T | 0.46 | 2.56 | Weight | MYOCD | intergenic | Cardiogenesis |
| rs12943502 | C | 0.49 | 3.15 | Height | NM_001001684 | intergenic | unknown function | |
| rs2052003 | T | 0.33 | 2.77 | BMI | MYOCD | intronic | Cardiogenesis | |
| rs2052003 | T | 0.33 | 2.99 | WC | MYOCD | intronic | Cardiogenesis | |
| rs4792371 | G | 0.46 | 3.35 | HC | HS3ST3A1 | intronic | heparan sulfate | |
| rs7219760 | T | 0.09 | 2.96 | WHR | NM_001001684 | intergenic | unknown function | |
| rs9893022 | A | 0.03 | 2.75 | Fatkg | HS3ST3A1 | intergenic | heparan sulfate | |
| rs2190699 | G | 0.34 | 2.95 | Fatpct | RICH2 | intronic | GTPase activator | |
| rs3944086 | C | 0.11 | 2.80 | Leankg | HS3ST3A1 | intergenic | heparan sulfate | |
| rs9907078 | C | 0.33 | 3.02 | Leanpct | CDRT15 | upstream | unknown function | |
| rs12949155 | G | 0.43 | 3.47 | SubQF | NM_001001684 | intergenic | unknown function | |
| rs4343329 | C | 0.26 | 6.54 | VF | TNFRSF13B | intronic | pro-inflammation | |
| rs7225344 | C | 0.26 | 5.68 | VF | TNFRSF13B | intronic | pro-inflammation | |
| rs4985700 | C | 0.40 | 5.13 | VF | TNFRSF13B | intronic | pro-inflammation | |
| rs7226097 | C | 0.41 | 4.22 | VF | TNFRSF13B | intronic | pro-inflammation | |
| rs11651352 | A | 0.23 | 4.17 | VF | TNFRSF13B | downstream | pro-inflammation | |
| rs4343329 | C | 0.26 | 5.23 | TAF | TNFRSF13B | intronic | pro-inflammation | |
| rs8081054 | A | 0.33 | 4.74 | TAF | NM_001001684 | intergenic | unknown function | |
| rs7225344 | C | 0.26 | 4.51 | TAF | TNFRSF13B | intronic | pro-inflammation | |
| rs8079010 | A | 0.34 | 3.98 | RQ | TRPV2 | intronic | stress response | |
| rs4985700 | C | 0.40 | 4.01 | REE | TNFRSF13B | intronic | pro-inflammation | |
| rs9303119 | G | 0.45 | 2.81 | REE/weight | NM_001001684 | intergenic | unknown function | |
| rs7222088 | G | 0.43 | 3.53 | REE/Leanmass | AK123100 | intronic | unknown function | |
| rs2108683 | A | 0.11 | 3.48 | FG | COX10 | intronic | Respiratory chain | |
| rs3744342 | G | 0.48 | 3.04 | FI | HS3ST3A1 | intergenic | heparan sulfate | |
| rs3744342 | G | 0.48 | 2.91 | IGR | HS3ST3A1 | intergenic | heparan sulfate | |
| rs6502302 | A | 0.01 | 2.87 | HOMA | HS3ST3A1 | intergenic | heparan sulfate | |
| rs9908356 | G | 0.21 | 3.19 | SI | PMP22 | intergenic | Peripheral nervous | |
| rs2106795 | T | 0.14 | 3.79 | SG | HS3ST3A1 | intergenic | heparan sulfate | |
| rs2996033 | T | 0.25 | 2.95 | AIR | HS3ST3A1 | intergenic | heparan sulfate | |
| rs12453295 | A | 0.01 | 3.02 | DI | MYOCD | intronic | Cardiogenesis | |
|
| ||||||||
|
Lipids/lipoprotein and
cardiovascular performance |
rs4792303 | A | 0.05 | 2.80 | TG | RICH2 | intronic | GTPase activator |
| rs173160 | C | 0.34 | 3.22 | TC | MYOCD | intergenic | Cardiogenesis | |
| rs11870163 | T | 0.42 | 3.11 | LDL-c | HS3ST3B1 | intronic | heparan sulfate | |
| rs173160 | C | 0.34 | 2.72 | cal. LDL-c | MYOCD | intergenic | Cardiogenesis | |
| rs8077611 | A | 0.01 | 3.54 | HDL-c | NM_001001684 | intergenic | unknown function | |
| rs230922 | T | 0.30 | 5.21 | HMED | PMP22 | intergenic | Peripheral nervous | |
| rs7214429 | G | 0.45 | 3.64 | LMEDn | MPRIP | intronic | Myosin phosphatase | |
| rs7214429 | G | 0.45 | 3.20 | LDLppd | MPRIP | intronic | Myosin phosphatase | |
| rs7214429 | G | 0.45 | 3.58 | bMED | MPRIP | intronic | Myosin phosphatase | |
| rs7214091 | C | 0.40 | 4.05 | sBP | TNFRSF13B | intergenic | heparan sulfate | |
| rs1233861 | A | 0.23 | 3.36 | dBP | MYOCD | intergenic | Cardiogenesis | |
| rs12944789 | C | 0.36 | 4.36 | Pulse | NM_001001684 | intergenic | unknown function | |
|
| ||||||||
|
Adipokine
and cytokines |
rs8070118 | G | 0.41 | 2.22 | Adiponectin | HS3ST3A1 | intergenic | heparan sulfate |
| rs2906985 | G | 0.06 | 3.75 | Leptin | CDRT4 | intronic | unknown function | |
| rs9915627 | A | 0.21 | 4.13 | TNF-alpha | HS3ST3A1 | intergenic | heparan sulfate | |
| rs9904641 | T | 0.27 | 3.48 | IL-1β | CDRT4 | downstream | unknown function | |
| rs1736202 | C | 0.41 | 3.60 | IL-6 | COPS3 | intronic | signal transduction | |
QTL3q27
Body composition and insulin responsiveness
Table 2 shows four biologically relevant genes identified in this region to be associated with body composition and insulin/glucose responsiveness. Transducing (beta)-like 1 X-linked receptor 1 (TBL1XR1) (rs6443434 and rs11710431), a signal transducer influencing multiple biological functions including cytoskeletal assembly, was associated with both clinical (weight and BMI) and biologic (Leanpct, REE, TAF and SG) traits. Insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2) (rs12639517), known to regulate IGF2 mRNA translation, was associated with the identifier phenotype WHR. Neuroligin 1 (NLGN1) (rs569221), a brain synapses protein, was associated with Leankg. Lipoma-preferred partner (LPP) (rs6785028), a cell adhesion and growth signal communicator, was associated with AIRG. Within this region, there were two associations that exceeded the regional significance threshold. SNP rs7433602 mapped to N-acetylated alpha-linked acidic dipeptidase-like 2 (NAALADL2), a dipeptidase that lacks a functional domain, was associated with VF. SNP rs10933727 was associated with HOMA and was mapped to a hypothetical gene LOC100131551.
Lipids/lipoprotein and cardiovascular performance
Of the lipids/lipoprotein phenotypes, there are two biologically relevant genes disclosed by SNP association. ATP11B (rs2700839) in association with LDLppd is an ATPase functioning in ion transport in cell membrane. Another gene, Rho family guanine nucleotide exchange factor (MCF2L2), referred by rs7631640 was associated with pulse. It was identified as a gene associated with type 2 diabetes (27). Only one association in this category was above the regional significance threshold, that is, rs10937009 with BMED. This SNP is near the gene NADH dehydrogenase (ubiquinone) 1 beta (NDUFB5), which transfers electrons from NADH to the respiratory chain.
Adipokine/cytokines
In the adipokine/cytokines phenotype category, we identified a biologically relevant association between rs6807480 and circulating IL-6 that is annotated to gene FGF12, a fibroblast growth factor. Another association above the threshold was between rs13070418 and TNF-alpha. This SNP was annotated to the gene SOX2, a transcription factor functioning in embryonic development.
QTL17p12
Body composition and insulin responsiveness
There are seven biologically relevant genes annotated to SNP-phenotype associations (Table 3). Three of those were identified by multiple associations, including TNFRSF13B (eight associations), HS3ST3A1 (eight associations) and MYOCD (four associations). Of those, TNFRSF13B, a pro-inflammtory macrophage receptor, was associated with VF and TAF at levels above regional significance threshold. HS3ST3A1 [heparan sulfate (glucosamine) 3-O-sulfotransferase 3A1], a heperan sulfate biosynthesis enzyme, was associated with HC, DEXA measures of body fatness and leanness, and several of the other biological phenotypes which index insulin responsiveness (SG and AIRG). MYOCD (myocardin), a cardiogenesis regulator, was associated with clinical phenotypes of body weight, BMI and WC. There were four associations annotated to potentially important biological regulator genes relevant to MetS. RICH2, a Rho-GTPase activator, was associated with Fatpct. TRPV2 (transient receptor potential cation channel), a heat responsive channel, was associated with RQ. COX10 (cytochrome c oxidase component 10), an essential component of the mitochondrial respiratory chain, was associated with FG. PMP22 (peripheral myelin protein 22) associated with SI, is known to function in the peripheral nervous system.
Lipids/lipoprotein and cardiovascular performance
Five of the body composition associated genes were also associated with phenotypes in this category. RICH2 was associated with TG. MYOCD was associated with TC. HS3ST3B1, a sister to HS3ST3A1, was associated with LDL-c. PMP22 was associated with HMED (above the threshold). TNFRSF13B was associated with sBP. Finally MPRIP, which is a myosin phosphatase Rho interacting protein, was associated with three of the lipoprotein sizing measures (LMEDn, LDLppd and BMED).
Adipokine/cytokines
Associations with adiponectin and TNF-alpha were mapped to HS3ST3A1, which was also associated with body composition and insulin responsiveness. COPS3 (COP9 constitutive photomorphogenic homolog subunit 3), a kinase regulating signal transduction, was associated with IL-6.
PBMC mRNA expression
Of the biologically relevant genes identified by SNP association at 3q27 and 17p12, Tables 4 and 5 show the association (as in p-values) of transcript levels in relation to each of the 42 phenotypes. At 3q27, expression of three biologically relevant genes, LPP, IGF2BP2 and TBL1XR1, were strongly associated with (p<0.01) MetS phenotypes. ATP11B, MCF2L2, and NLGN1 had nominal associations with MetS phenotypes (p<0.05). LPP was found associated with both biologic and clinical phenotypes (REE/weight, IL-1β, FI, TG, and HDL-c). IGF2BP2 was associated with body weight, BMI, WC, HC, Fatkg, Fatpct, Leankg and Leanpct, RQ, REE, SI, DI, LDL-c, HMED, LDLppd and leptin. TBL1XR1 was associated with Leankg and resting pulse. ATP11B was associated with WHR, MCF2L2 with SI, LDLppd and BMED, and NLGN1 with FG. Of the seven genes identified by SNP-phenotype association as biological relevant, the transcript levels of only one gene, FGF12, did not pass any significant level. At 17p12, of the eight biologically relevant genes, seven had transcription levels significantly associated with related MetS phenotypes: TNFRSF13B, COX10, RICH2, TRPV2, MPRIP, COPS3 and PMP22. TNFRSF13B was associated with WHR, RQ, LDLppd and BMED. COX10 was associated with weight, BMI, SG and TG. PMP22 was associated with TG and HMED, and RICH2 with Leankg, REE, AIRG and leptin. TRPV2 was associated with fat mass, lean mass and REE/leanmass, and MPRIP with TG, HMED and pulse. HS3ST3B1 was not represented in our transcriptomic profile. However, because of its extensive SNP association with multiple biologic phenotypes, and because of its known relevant function, we prioritized it as a significant candidate.
Table 4. Association of lymphocyte expression levels of prioritized genes with MetS phenotypes at 3q27 QTL.
For gene prioritization strategy, see text. SNP associations with phenotypes are depicted in Table 2. The values shown represent the level of significance (p-value) of association between prioritized gene transcript levels and MetS phenotypes.
| Phenotype | ATP11B | FGF12 | IGF2BP2 | LPP | MCF2L2 | NLGN1 | TBL1XR1 | |
|---|---|---|---|---|---|---|---|---|
| Body composition and insulin responsiveness | weight | 0.51 | 0.35 | 0.00 | 0.62 | 0.96 | 0.18 | 1.00 |
| height | 0.17 | 0.14 | 0.12 | 0.97 | 1.00 | 0.31 | 0.00 | |
| BMI | 1.00 | 0.89 | 0.00 | 0.74 | 0.36 | 0.21 | 0.34 | |
| WC | 0.44 | 0.63 | 0.00 | 0.69 | 0.73 | 0.68 | 0.34 | |
| HC | 0.01 | 0.57 | 0.01 | 0.50 | 0.63 | 0.80 | 0.28 | |
| WHR | 0.02 | 0.39 | 0.12 | 0.37 | 0.51 | 0.92 | 0.69 | |
| Fatkg | 0.61 | 0.73 | 0.02 | 0.34 | 0.84 | 0.16 | 0.09 | |
| Fatpct | 0.93 | 0.96 | 0.27 | 0.48 | 0.18 | 0.18 | 0.65 | |
| Leankg | 0.14 | 0.70 | 0.00 | 0.86 | 0.33 | 0.10 | 0.01 | |
| Leanpct | 0.93 | 1.00 | 0.26 | 0.43 | 0.25 | 0.16 | 0.55 | |
| SubQF | 0.82 | 0.28 | 0.06 | 0.74 | 0.84 | NA | 0.96 | |
| VF | 0.81 | 0.28 | 0.21 | 0.91 | 0.83 | NA | 0.89 | |
| TAF | 0.86 | 0.24 | 0.11 | 0.80 | 0.94 | NA | 0.86 | |
| RQ | 0.18 | 0.90 | 0.01 | 0.22 | 0.06 | NA | 0.84 | |
| REE | 0.81 | 0.63 | 0.00 | 0.20 | 0.45 | NA | 0.52 | |
| REE/weight | 0.24 | 0.99 | 0.09 | 0.03 | 0.12 | NA | 0.29 | |
| REE/leanmass | 0.43 | 0.86 | 0.38 | 0.08 | 0.93 | NA | 0.31 | |
| FG | 0.57 | 0.77 | 0.21 | 0.71 | 0.74 | 0.04 | 0.83 | |
| FI | 0.73 | 0.12 | 0.68 | 0.05 | 0.81 | 0.85 | 0.13 | |
| IGR | 0.55 | 0.20 | 0.68 | 0.05 | 0.99 | 0.58 | 0.13 | |
| HOMA | 0.79 | 0.15 | 0.40 | 0.06 | 0.67 | 0.41 | 0.16 | |
| SI | 0.07 | 0.13 | 0.04 | 0.67 | 0.04 | NA | 0.64 | |
| SG | 0.83 | 0.51 | 0.17 | 0.75 | 0.37 | NA | 0.62 | |
| AIR | 0.19 | 0.15 | 0.45 | 0.94 | 0.98 | NA | 0.31 | |
| DI | 0.18 | 0.26 | 0.02 | 0.49 | 0.83 | 0.57 | 0.73 | |
|
| ||||||||
|
Lipids/lipoprotein profiles and
cardiovascular performance |
TG | 0.46 | 0.35 | 0.90 | 0.01 | 0.30 | 0.44 | 0.62 |
| TC | 0.64 | 0.91 | 0.12 | 0.94 | 0.77 | 0.45 | 0.84 | |
| LDL-c | 0.45 | 0.68 | 0.02 | 0.60 | 0.35 | 0.86 | 0.17 | |
| cal. LDL-c | 0.50 | 0.58 | 0.76 | 0.67 | 0.62 | 0.75 | 0.95 | |
| HDL-c | 0.20 | 0.35 | 0.08 | 0.02 | 0.64 | 0.15 | 0.67 | |
| HMED | 0.81 | 0.97 | 0.02 | 0.49 | 0.60 | 0.08 | 0.56 | |
| LMEDn | 0.81 | 0.93 | 0.10 | 0.67 | 0.44 | 0.46 | 0.76 | |
| LDLppd | 0.56 | 0.58 | 0.03 | 0.89 | 0.01 | 0.31 | 0.35 | |
| BMED | 0.88 | 0.50 | 0.19 | 0.90 | 0.04 | 0.86 | 0.65 | |
| sBP | 0.40 | 0.51 | 0.07 | 0.79 | 0.45 | 0.45 | 0.83 | |
| dBP | 0.79 | 0.25 | 0.71 | 0.62 | 0.37 | 0.49 | 0.67 | |
| Pulse | 0.67 | 0.70 | 0.56 | 0.36 | 0.23 | 0.30 | 0.00 | |
|
| ||||||||
|
Adipokine
and cytokines |
Adiponectin | 0.22 | 0.30 | 0.05 | 0.37 | 0.92 | 0.57 | 0.84 |
| Lepin | 0.76 | 0.61 | 0.05 | 0.73 | 0.10 | 0.77 | 0.40 | |
| TNF-alpha | 0.90 | 0.81 | 0.94 | 0.32 | 0.95 | 0.45 | 0.63 | |
| IL-1β | 0.58 | 0.59 | 0.59 | 0.04 | 1.00 | 0.57 | 0.81 | |
| IL-6 | 0.34 | 0.87 | 0.57 | 0.82 | 0.42 | 0.84 | 0.79 | |
Table 5. Association of lymphocyte expression levels of prioritized genes with MetS phenotypes at 17p12 QTL.
For gene prioritization strategy, see text. SNP associations with phenotypes are depicted in Table 3. The values shown represent the level of significance (p-value) of association between prioritized gene transcript levels and MetS phenotypes.
| Phenotype | COPS3 | COX10 | PMP22 | RICH2 | TNFRSF13B | TRPV2 | HS3ST3B1 | |
|---|---|---|---|---|---|---|---|---|
| Body composition and insulin responsiveness | weight | 0.30 | 0.04 | 0.58 | 0.26 | 0.43 | 0.92 | 0.58 |
| height | 0.85 | 0.18 | 0.73 | 0.57 | 0.86 | 0.67 | 0.21 | |
| BMI | 0.19 | 0.01 | 0.71 | 0.41 | 0.24 | 0.95 | 0.26 | |
| WC | 0.67 | 0.23 | 0.46 | 0.45 | 0.64 | 0.94 | 0.76 | |
| HC | 0.59 | 0.09 | 0.14 | 0.35 | 0.24 | 1.00 | 0.41 | |
| WHR | 0.66 | 0.13 | 0.54 | 0.95 | 0.03 | 0.37 | 0.26 | |
| Fatkg | 0.99 | 0.22 | 0.41 | 0.14 | 0.65 | 0.03 | 0.46 | |
| Fatpct | 0.93 | 0.16 | 0.62 | 0.19 | 0.36 | 0.03 | 0.19 | |
| Leankg | 1.00 | 0.23 | 0.61 | 0.03 | 0.58 | 0.81 | 0.49 | |
| Leanpct | 0.89 | 0.17 | 0.53 | 0.17 | 0.46 | 0.03 | 0.19 | |
| SubQF | 0.55 | 0.23 | 0.67 | 0.39 | 0.23 | 0.04 | NA | |
| VF | 0.88 | 0.40 | 0.59 | 0.44 | 0.58 | 0.80 | NA | |
| TAF | 0.75 | 0.24 | 0.65 | 0.49 | 0.24 | 0.10 | NA | |
| RQ | 0.83 | 0.20 | 0.84 | 0.16 | 0.01 | 0.39 | NA | |
| REE | 0.45 | 0.94 | 0.59 | 0.02 | 0.94 | 0.75 | NA | |
| REE/weight | 0.85 | 0.97 | 0.99 | 0.00 | 0.83 | 0.99 | NA | |
| REE/leanmass | 0.07 | 1.00 | 0.93 | 0.16 | 0.94 | 0.00 | NA | |
| FG | 0.33 | 0.17 | 1.00 | 0.64 | 0.11 | 0.39 | 0.66 | |
| FI | 0.68 | 0.07 | 0.07 | 0.64 | 0.14 | 0.53 | 0.81 | |
| IGR | 0.26 | 0.20 | 0.15 | 0.34 | 0.20 | 0.58 | 0.95 | |
| HOMA | 0.90 | 0.06 | 0.15 | 0.81 | 0.23 | 0.48 | 0.73 | |
| SI | 0.41 | 0.81 | 0.84 | 0.05 | 0.12 | 0.16 | NA | |
| SG | 0.10 | 0.05 | 0.88 | 0.62 | 0.13 | 0.49 | NA | |
| AIR | 0.28 | 0.38 | 0.65 | 0.03 | 0.60 | 0.42 | NA | |
| DI | 1.00 | 0.59 | 0.54 | 0.07 | 0.34 | 0.35 | 0.34 | |
|
| ||||||||
|
Lipids/lipoprotein profiles and
cardiovascular performance |
TG | 0.70 | 0.04 | 0.01 | 0.79 | 0.91 | 0.19 | 0.47 |
| TC | 0.91 | 0.63 | 0.68 | 0.69 | 0.21 | 0.40 | 0.90 | |
| LDL-c | 0.83 | 0.28 | 0.32 | 0.27 | 0.75 | 0.45 | 0.99 | |
| cal. LDL-c | 0.99 | 0.91 | 0.26 | 0.45 | 0.26 | 0.40 | 0.84 | |
| HDL-c | 0.31 | 0.46 | 0.42 | 0.19 | 0.26 | 0.29 | 0.97 | |
| HMED | 0.26 | 0.22 | 0.04 | 0.78 | 0.03 | 0.19 | 0.21 | |
| LMEDn | 0.71 | 0.55 | 0.16 | 0.82 | 0.06 | 0.63 | 0.28 | |
| LDLppd | 0.74 | 0.81 | 0.03 | 0.08 | 0.00 | 0.15 | 0.90 | |
| BMED | 0.96 | 0.95 | 0.46 | 0.27 | 0.00 | 0.34 | 0.43 | |
| sBP | 0.96 | 0.47 | 0.54 | 0.41 | 0.20 | 0.30 | 0.89 | |
| dBP | 0.54 | 0.89 | 0.09 | 0.95 | 0.33 | 0.22 | 0.43 | |
| pulse | 0.50 | 0.94 | 0.36 | 0.81 | 0.34 | 0.07 | 0.33 | |
|
| ||||||||
|
Adipokine
and cytokines |
adiponectin | 0.95 | 0.76 | 0.77 | 0.47 | 0.29 | 0.45 | 0.34 |
| leptin | 0.55 | 0.69 | 0.45 | 0.05 | 0.08 | 0.99 | 0.14 | |
| TNF-alpha | 0.02 | 0.17 | 0.70 | 0.60 | 0.81 | 0.46 | 0.21 | |
| IL-1β | 0.39 | 0.53 | 0.21 | 0.48 | 0.40 | 0.53 | 0.40 | |
| IL-6 | 0.66 | 0.76 | 0.31 | 0.05 | 0.55 | 0.34 | 0.36 | |
Summary of candidate gene prioritization
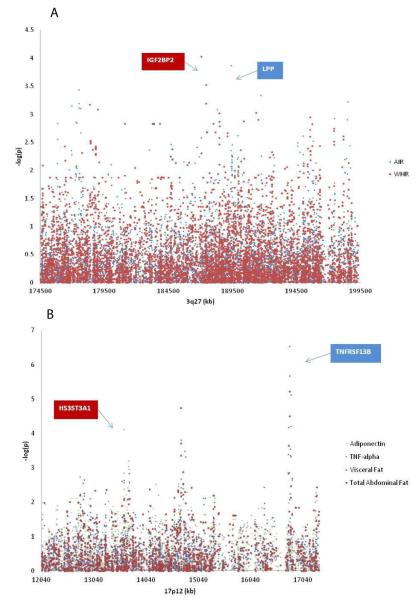
Table 6 summarizes the prioritized genes identified by SNP association in the two QTL regions, their identifier phenotype and the genetic correlates of this identifier. The steps of this prioritizing strategy included the level of significance of the SNP associations, function of identified gene in relation to the MetS and level of PBMC gene expression in association with the MetS phenotypes. Figure 2A and 2B show the final four prioritized genes located within the QTL peaks at 3q27 and 17p12, together with their identifier phenotype(s).
Table 6. Summary illustrating the prioritized genes at 3q27 and 17p12 QTLs.
For prioritized genes see Table 2 and 3. For phenotype abbreviations, see Table 1. Identifier phenotype is the one that was highly associated with a SNP that mapped to that gene. Correlated phenotypes are those that were found to be genetically correlated significantly with the identifier phenotype.
| Annotated associated genes |
Original QTL region |
Identifier phenotype |
Correlated biological phenotypes |
Biological function |
|---|---|---|---|---|
| LPPab | 3q27 | AIR | Fatkg, DI, leptin | signal transduction to celll adhesion sites; cell to cell communication |
| IGF2BP2ab | 3q27 | WHR | VF, TAF, REE, HMED, LMEDn, LDLppd, BMED,TNF-alpha |
neonatal development, body fat regulation and lipid metabolism |
| TBL1XR1a | 3q27 | BMI, weight, Leanpct, TAF, REE,SG, TG |
WHR, Fatkg, Fatpct, Leankg, Leanpct, SubQF, VF, REE/weight, REE/Leanmass, SI, AIR, DI, HMED, leptin, IL-6 |
member of WD40 protein family known to regulate signal transduction, RNA processing, vesicular trafficking, cytoskeletal assembly and cytotypic differentiation. |
| NLGN1a | 3q27 | Leankg | Fatkg, SubQF, VF, TAF, REE, REE/weight, REE/leanmass, SI, AIR, HMED |
member of a family of neuronal cell surface proteins, known to act as splice site-specific ligands for beta-neurexins that could be involved in the formation and remodeling of central nervous system |
| ATP11Ba | 3q27 | LDLppd | WHR, Fatkg, HMED,BMED,TNF- alpha |
member of the ATPase family that participate the uphill transport of ions across cell membranes, including H+,Na+, K+ and Ca++ |
| MCF2L2a | 3q27 | Pulse | Leankg, SubQF, TAF, REE, SI, adiponectin, leptiin |
member of the guanine exchange factor family. Together with ADIPQ, SOX2 polymorphism influence diabetic complications. |
| FGF12 | 3q27 | IL-6 | VF | Member of the fibroblast growth factor family possess broad mitogenic and cell survival activities,involved in several biological functions. |
| TNFRSF13B ab |
17p12 | VF, TAF, REE, sBP |
weight, BMI, WC, HC, WHR,Fatkg,Fatpct,Leankg,Leanpc t,SubQF,REE/weight, FI, IGR,, HOMA, SI,SG,DI, HDL-c, dBP,pulse, leptin, IL-6 |
A lymphocyte-specific member of the tumor necrosis factor (TNF) receptor superfamily, which plays a crutial role in humoral immunity. |
| HS3ST3A1ab | 17p12 | HC, Fatkg, Leankg, FI, IGR, HOMA, SG, AIR, Adiponectin, TNF-alpha |
weight, height,, BMI, WC, WHR,SubQF,VF, TAF,RQ, REE, REE/weight, REE/leanmass, FG, SI, DI,TG,TC, LDL-c, cal. LDL-c, HDL-c, HMED, LMEDn, LDLppd, BMED, sBP,pulse, leptin, IL-1β, IL- 6 |
A biosynthesis enzyme of heparan sulfate find structures, whose substrates are important in the cellular uptake and biological effects of the lipoproteins that carry out multiple biologic activities. |
| MYOCDa | 17p12 | Weight, BMI, WC, DI, TC, cal. LDL-c, dBP |
height, hips,WHR, Fatkg, Fatpct, Leankg, Leanpct, SubQF, VF,TAF,REE, REE/weight, REE/leanmass, FI, IGR, HOMA, SI,SG, AIR,TG, LDL-c, HDL-c, HMED, sBP, pulse, leptin, IL-6 |
A nuclear protein, a transcriptional coactivator that modulates the function of cardiac and smooth-muschle cells. Important in cardiogenesis and smooth muscle cell differentiation. |
| RICH2a | 17p12 | Fatpct, TG | weight, BMI, WC,HC,WHR, Fatkg,Leanpct, SubQF, VF, TAF, REE, REE/weight,REE/leanmass,FI, IGR, HOMA, SI,AIR,TC,HDL-c, leptin |
GTPase activator protein, which activate CDC42 and Rac-1, known to interfere with platelet growth factor BB-induced membrane ruffling. |
| TRPV2a | 17p12 | RQ | Pulse | A channel protein in response to environmental stresses. |
| COX10a | 17p12 | FG | FI, HOMA, AIR, leptin | The terminal component of the mitochodrial respiratory chain, which catalyzes the electron transfer from reduced cytochrome c to oxygen. Mutations were linked to cytochrome c function deficiency. |
| PMP22a | 17p12 | SI, HMED | weight, BMI, WC, HC, WHR, Fatkg, Fatpct,Leankg, Leanpct,SubQF, VF, TAF, REE/weight, FI, IGR, HOMA, TG, HDL-c, LMEDn, LDLppd, BMED,pulse |
the major component of the myelin structure in the peripheral nervous system that might function in growth regulation, and in myelinization in the peripheral nervous system |
| MPRIPa | 17p12 | LMEDn, LDLppd, BMED |
WHR,Fatkg, FI, IGR, HOMA, TG, HDL-c, HMED,TNF-alpha |
Target myosin phosphatases interacting with RhoA and ROCK1 to regulate actin cytoskeleton. |
| COPS3 | 17p12 | IL-6 | BMI, VF | The protein encoded by this gene possesses kinase activity that phosphorylates regulators involved in signal transduction and protects proteins from ubiquitin-dependent degradation. |
Candidate genes prioritized from SNP association analysis whose transcript levels are associated with MetS phenotypes
Biologic candidate gens with the highest prioritization
Figure 2. Plots of SNP associations with MetS phenotypes within QTLs at 3q27 (panel A) and 17p12 (panel B).
Dots depict levels of association of identifier phenotypes with all SNPs in the QTL region. Vertical axis represents minus logarithm of the p-values and horizontal represents the chromosomal position (kb). In panel A, LPP and IGF2BP2 are the highly prioritized genes in the 3q27 QTL region. In panel B, TNFRSF13B and HS3ST3A1 are the highly prioritized genes in the 17p12 QTL region.
Discussion
The present study focused on identification of genes situated in previously-determined QTLs at chr3q27 and chr17p12. Both targeted SNP genotype association and mRNA expression analyses were performed. The results provided for the first time clarification of the differential informativeness of the clinical phenotypes vs. concomitantly measured biologic traits. Although BMI and waist circumferences, the commonly used phenotypes in the GWAS meta-analyses, were genetically intercorrelated with other clinical phenotypes of MetS, they were not reflecting their biologically targeted traits. BMI was intercorrelated with both total body fat and lean mass. Waist circumferences were also intercorrelated with subcutaneous and visceral fat sizes. Within each of the QTLs a number of genes were found to be associated with the multiple phenotypes of body composition, insulin responsiveness, lipoprotein density profiles and cytokines. Our results revealed two novel genes (IGF2BP2 and TNFRSF13B), whose function could account for the biologic pathways influencing MetS phenotypes. IGF2BP2 is also known to mediate glucose/insulin response effectiveness. TNFRSF13B is a mediator of cytokine activities like TNF-alpha and its influence on the pro-inflammatory response. Our results thus unraveled the genetic origin of the epistatic and pleiotropic effects exerted by the identified QTLs with their potential influences on MetS. It also emphasizes the rationale behind the extensive efforts and the cost encountered in measuring the biologic phenotypes in order to identify those genes participating in the evolution of MetS phenotypic clusters.
To prioritize genes mapped to the associated SNPs, we stressed their biologic function, statistical level and association of selected genes’ expression levels with related MetS phenotypes. In this respect, our highest level of priority at 3q27 was IGF2BP2 whose sequence polymorphism was associated with WHR, the only clinical phenotype that was genetically intercorrelated with visceral fat size (VF). WHR was also correlated with other biologic markers including REE, all lipoprotein sizes profile and cytokine TNF-alpha. The transcript levels of IGF2BP2 were also significantly associated with biologic measures of body composition phenotypes (Fatkg, Leankg, RQ and REE), insulin/glucose responsiveness (SI and DI), lipoprotein sizes profile (HMED and LDLppd) and plasma leptin. The protein product of IGF2BP2 binds the mRNA of insulin-like growth factor 2 (IGF2) and therefore regulates its translation. IGF2 is a hormone that affects growth and development in neonatal stages (28). In adults, IGF2 is known to influence regulation of lipids/lipoprotein metabolism and body weight (29). Lean type 2 diabetic men and women whose weight gain within the next five years can be predicted by circulating levels of IGF2 (30). In a number of genome-wide association studies, IGF2BP2 has also been found to be associated with the onset of type 2 diabetes (31, 32).
At 17p12 QTL, our first priority gene is TNFRSF13B, a member of a well-characterized receptor family for tumor necrosis factors. Polymorphisms in the proximity of TNFRSF13B were associated with four MetS phenotypes. High levels of associations were observed for multiple variants near or within TNFRSF13B with VF and TAF, two of the most informative biological phenotypes of body fat distribution. Plasma leptin levels, the identifier phenotype of this linkage region has been recognized to be a surrogate of adverse adiposity between VF and SubQF (33). In our cohort leptin levels were genetically correlated with SubQF and TAF. We also observed association of the transcript levels of TNFRSF13B with WHR, RQ, LDLppd and BMED. TNFRSF13B, sometimes referred to as TACI (transmembrane activator) or CAML (calcium-modulating cyclophilin ligand interactor), is a receptor shared by two members of the tumor necrosis factor superfamily, APRIL (TNFSF13) and BAFF (the B-cell activation factor of the TNF family). APRIL and BAFF can activate macrophages via shared receptors such as TACI and initiate inflammatory changes leading to the induction of mediators of pro-inflammatory cytokines and matrix degrading enzymes (34, 35). Adverse adiposity distribution particularly accumulation of visceral fat mass is linked to insulin resistance. One possible hypothesis is that chronic state of inflammation largely attributed to macrophage infiltration of visceral adipose tissue could play a major causative role in the pathogenesis of MetS. Our results suggest a role for TNSRSF13B in the etiology of several phenotypes that may be genetically linked via a network through inflammatory pathways. These include VF, TAF, REE, WHR, and some lipid densities subclasses.
Based on our filtering criteria focusing on the biology of MetS, our other prioritized genes include LPP, MCF2L2, TBL1XR1, ATPB11 and NLG1 at 3q27 and HS3ST3A1, COX10, PMP22, RICH2, TRPV2 at 17p12. Some of these genes have been implicated in functions that may potentially be related to the pathogenesis of MetS but more research is needed to elucidate their precise biological role. We also noticed that none of the SNPs we found significantly associated with MetS traits in our cohort have been identified in previous GWAS for related phenotypes. The genes some of these SNPs are mapped to, including IGF2BP2 and TNFRSF13B (31, 32, 36), have been implicated in diabetes related phenotypes. In conclusion, our QTL-based association and PBMC expression genetic analyses guided by biology-focused approach revealed novel genes that could account for the complexity of MetS phenotypes both in relation to their informativeness as well as the epistatic and pleiotropic influences of its identified QTLs. They also account at least in part for potential biologic pathways underlying the MetS phenotypes. Whereas IGF2BP2 could relate to aberrations in glucose/insulin responsiveness, TNFRSF13B could mediate its association with adverse adiposity as well as aberrations of plasma lipids/lipoprotein sizes profile.
Acknowledgements
Dr. Ahmed Kissebah, the senior author of this work, was fully involved in the preparation of this report up to his death on 17 May 2012. We are saddened by his loss and deeply respectful of his long and distinguished career in obesity research.
This work is supported by grants from the NIH (DK071895-03 and DK65598-01) for A.H.K and NIH (MH059490) to J.B. and by TOPS (Take Off Pounds Sensibly) Club, Inc., G.E.S. and R.M.A were former employees of MCW. No potential conflicts of interests are relevant to this article. Dr. Yi Zhang is the guarantor of this work, had full access to all the data, and takes full responsibility for the integrity of the data and the accuracy of the data analysis.
Y.Z. analyzed data, contributed to discussion, and wrote the manuscript. J.W.K. performed statistical genetic analyses, contributed to discussion and co-wrote the manuscript. M.O. provided laboratory assistance in genotyping, contributed to discussion and data analysis. O.A. and R.M.A. assisted in the phenotyping procedures. U.B. facilitated the genotyping procedures. G.E.S. supervised the biologic phenotyping procedures. A.C. contributed to the initial linkage findings, J.E.C. and M.A.C. did cytokine and transcriptome procedures. T.D.D. prepared the genotyping raw data for genetic analysis procedures. H.H.H.G. assisted in the transcriptome data analysis. D.L.R. did the lipoprotein particle sizing. A.K. and J.B. supervised and directed the overall statistical genetic analyses and contributed to discussion. Acknowledgement also includes the contribution of Roland James (Medical College of Wisconsin), the project manager of TOPS Center for Obesity and Metabolic Research and all his effort in recruitment, phenotyping and database management, Jacqueline Marks (Medical College of Wisconsin) for her supervision of all biochemical procedures, Melena Zelembaba and Regina Cole (Medical College of Wisconsin) for their technical assistance in the Affymetrix chips genotyping procedures, and Ruth Gielow (TOPS Club, Inc.) and her assistants in the recruitment process and finally to all the members of TOPS Club, Inc. and their families who volunteered for this study.
Footnotes
Disclosure The authors declared no conflict of interest.
REFERENCES
- 1.Hetherington MM, Cecil JE. Gene-environment interactions in obesity. Forum Nutr: 2010;63:195–203. doi: 10.1159/000264407. [DOI] [PubMed] [Google Scholar]
- 2.Catenacci VA, Hill JO, Wyatt HR. The obesity epidemic. Clin Chest Med. 2009;30(3):415–44. vii. doi: 10.1016/j.ccm.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Day C. Metabolic syndrome, or What you will: definitions and epidemiology. Diab Vasc Dis Res. 2007 Mar;4(1):32–38. doi: 10.3132/dvdr.2007.003. [DOI] [PubMed] [Google Scholar]
- 4.Carmelli D, Cardon LR, Fabsitz R. Clustering of hypertension, diabetes, and obesity in adult male twins: same genes or same environments? Am J Hum Genet. 1994;55(3):566–573. [PMC free article] [PubMed] [Google Scholar]
- 5.Gibson F, Froguel P. Genetics of the APM1 locus and its contribution to type 2 diabetes susceptibility in French Caucasians. Diabetes. 2004;53(11):2977–2983. doi: 10.2337/diabetes.53.11.2977. [DOI] [PubMed] [Google Scholar]
- 6.Curran JE, Jowett JB, Elliott KS, et al. Genetic variation in selenoprotein S influences inflammatory response. J. Nat Genet. 2005;37(11):1234–1241. doi: 10.1038/ng1655. [DOI] [PubMed] [Google Scholar]
- 7.Kraja AT, Vaidya D, Pankow JS, et al. A bivariate genome-wide approach to metabolic syndrome: STAMPEED consortium. Diabetes. 60(4):1329–1339. doi: 10.2337/db10-1011. 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zabaneh D, Balding DJ. A genome-wide association study of the metabolic syndrome in Indian Asian men. PLoS One. 2010;5(8):e11961. doi: 10.1371/journal.pone.0011961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang K, Li WD, Zhang CK, et al. A genome-wide association study on obesity and obesity related traits. PLoS One. 2011;6(4):e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu YZ, Pei YF, Liu JF, et al. Powerful bivariate genome-wide association analyses suggest the SOX6 gene influencing both obesity and osteoporosis phenotypes in males. PLoS One. 2009;4(8):e6827. doi: 10.1371/journal.pone.0006827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kissebah AH, Sonnenberg GE, Myklebust J, et al. Quantitative trait loci on chromosomes 3 and 17 influence phenotypes of the metabolic syndrome. Proc Natl Acad Sci U S A. 2000;97(26):14478–83. doi: 10.1073/pnas.97.26.14478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grayson BL, Wang L, Aune TM. Peripheral blood gene expression profiles in metabolic syndrome, coronary artery disease and type 2 diabetes. Genes Immun. 2011;12(5):341–351. doi: 10.1038/gene.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheyssac C, Dina C, Leprêtre F, et al. EIF4A2 is a positional candidate gene at the 3q27 locus linked to type 2 diabetes in French families. Diabetes. 2006;55(4):1171–1176. doi: 10.2337/diabetes.55.04.06.db05-1298. [DOI] [PubMed] [Google Scholar]
- 14.Svendsen OL, Haarbo J, Heitmann BL, Gotfredsen A, Christiansen C. Measurement of body fat in elderly subjects by dual-energy x-ray absorptiometry, bioelectrical impedance, and anthropometry. Am J Clin Nutr. 1991;53(5):1117–1123. doi: 10.1093/ajcn/53.5.1117. [DOI] [PubMed] [Google Scholar]
- 15.Peiris AN, Hennes MI, Evans DJ, Wilson CR, Lee MB, Kissebah AH. Relationship of anthropometric measurements of body fat distribution to metabolic profile in premenopausal women. Acta Med Scand Suppl. 1988;723:179–188. doi: 10.1111/j.0954-6820.1987.tb05942.x. [DOI] [PubMed] [Google Scholar]
- 16.Bergman RN. Toward physiological understanding of glucose tolerance. Minimal-model approach. Diabetes. 1989;38:1512–1527. doi: 10.2337/diab.38.12.1512. [DOI] [PubMed] [Google Scholar]
- 17.Rainwater DL, Moore PH, Jr, Shelledy WR, Dyer TD, Slifer SH. Characterization of a composite gradient gel for the electrophoretic separation of lipoproteins. J Lipid Res. 1997;38(6):1261–1266. [PubMed] [Google Scholar]
- 18.Dupont NC, Wang K, Wadhwa PD, Culhane JF, Nelson EL. Validation and comparison of luminex multiplex cytokine analysis kits with ELISA: determinations of a panel of nine cytokines in clinical sample culture supernatants. J Reprod Immunol. 2005;66(2):175–191. doi: 10.1016/j.jri.2005.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burdick JT, Chen W-M, Abecasis GR, Cheung VG. In silico method for inferring missing genotypes in pedigrees. Nat Genet. 2006;38:1002–1004. doi: 10.1038/ng1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Göring HH, Curran JE, Johnson MP, et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat Genet. 2007;39(10):1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- 22.Kent JW, Jr, Comuzzie AG, Mahaney MC, et al. Intercellular adhesion molecule-1 concentration is genetically correlated with insulin resistance, obesity, and HDL concentration in Mexican Americans. Diabetes. 2004;53(10):2691–2695. doi: 10.2337/diabetes.53.10.2691. [DOI] [PubMed] [Google Scholar]
- 23.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50(Pt 2):181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 24.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 25.Moskvina V, Schmidt KM. On multiple testing correction in genome-wide association studies. Genet Epidemiol. 2008;32:567–573. doi: 10.1002/gepi.20331. [DOI] [PubMed] [Google Scholar]
- 26.Whitlock MC. Combining probability from independent tests: The weighted Z-method is superior to Fisher’s approach. J Evol Biol. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- 27.Ochiai Y, Serizawa M, Yanai K, et al. Search for type 2 diabetes susceptibility genes on chromosomes 1q, 3q and 12q. J Hum Genet. 2008;53(4):314–324. doi: 10.1007/s10038-008-0254-6. [DOI] [PubMed] [Google Scholar]
- 28.Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC. A family of insulin like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol. 1999;19(2):1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Cui H, Sandstedt B, Nordlinder H, Larsson E, Ekström TJ. Expression levels of the insulin-like growth factor-II gene (IGF2) in the human liver: developmental relationships of the four promoters. J Endocrinol. 1996;149(1):117–124. doi: 10.1677/joe.0.1490117. [DOI] [PubMed] [Google Scholar]
- 30.Heald AH, Kärvestedt L, Anderson SG, et al. Low insulin like growth factor-II levels predict weight gain in normal weight subjects with type 2 diabetes. Am J Med. 2006;119(2):167, e9–15. doi: 10.1016/j.amjmed.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 31.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cnop M, Landchild MJ, Vidal J, et al. The concurrent accumulation of intra-abdominal and subcutaneous fat explains the association between insulin resistance and plasma leptin concentrations : distinct metabolic effects of two fat compartments. Diabetes. 2002;51(4):1005–1015. doi: 10.2337/diabetes.51.4.1005. [DOI] [PubMed] [Google Scholar]
- 34.Lee SM, Kim EJ, Suk K, Lee WH. BAFF and APRIL induce inflammatory activation of THP-1 cells through interaction with their conventional receptors and activation of MAPK and NF-κB. Inflamm Res. 2011;60(9):807–815. doi: 10.1007/s00011-011-0336-3. [DOI] [PubMed] [Google Scholar]
- 35.Lee SM, Kim WJ, Suk K, Lee WH. Cell to Cell Interaction Can Activate Membrane-bound APRIL Which Are Expressed on Inflammatory Macrophages. Immune Netw. 2010;10(5):173–180. doi: 10.4110/in.2010.10.5.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Grassi MA, Tikhomirov A, Ramalingam S, Below JE, Cox NJ, Nicolae DL. Genome-wide meta-analysis for severe diabetic retinopathy. Hum Mol Genet. 2011;20(12):2472–2481. doi: 10.1093/hmg/ddr121. [DOI] [PMC free article] [PubMed] [Google Scholar]