SUMMARY
Multipotent stromal cells (MSC) and their osteoblastic lineage cell (OBC) derivatives are part of the BM niche and contribute to hematopoietic stem cells (HSC) maintenance. Here, we show that myeloproliferative neoplasia (MPN) progressively remodels the endosteal BM niche into a self-reinforcing leukemic niche that impairs normal hematopoiesis, favors leukemic stem cell (LSC) function and contributes to BM fibrosis. We show that leukemic myeloid cells stimulate MSCs to overproduce functionally altered OBCs, which accumulate in the BM cavity as inflammatory myelofibrotic cells. We identify roles for TPO, CCL3 and direct cell-cell interactions in driving OBC expansion, and for changes in TGFβ, Notch and inflammatory signaling in OBC remodeling. MPN-expanded OBCs, in turn, exhibit decreased expression of many HSC retention factors and severely compromised ability to maintain normal HSCs, but effectively support LSCs. Targeting this pathological interplay could represent a novel avenue to treat MPN patients and prevent myelofibrosis.
INTRODUCTION
Hematopoietic stem cells (HSC) sustain the life-long production of all types of mature blood cells (Orkin and Zon, 2008). At steady state, HSCs primarily reside in the bone marrow (BM) cavity where they interact with different types of stromal cells expressing important regulatory molecules including SCF, CXCL12 (SDF1α) and TGFβ (Frenette et al., 2013). Although early mouse studies implicated mature bone-forming osteoblasts (Calvi et al., 2003), recent work has refined the identity of HSC-supportive cells to several populations of multipotent stromal cells (MSC) and their early osteoblastic lineage cell (OBC) derivatives. Both Runx2-expressing OBCs (Chitteti et al., 2010) and perivascular MSC-like cells expressing either Nestin (Nes) (Mendez-Ferrer et al., 2010), high levels of CXCL12 (so-called Cxcl12-abundant reticular cells or CAR cells) (Omatsu et al., 2010) or leptin receptor (Lepr) (Ding et al., 2012) have been shown to be important for HSC maintenance. However, not all MSC derivatives have HSC-supportive activity and, in fact, mature adipocytes negatively impact HSC function (Naveiras et al., 2009). Other known HSC-supporting BM niche cells include endothelial cells (EC) (Doan and Chute, 2012) and ectodermally derived non-myelinating Schwann cells (Yamazaki et al., 2011). Therefore, the emerging picture of the HSC BM niche is a complex array of regulatory cell types with a predominant role for MSCs and their early OBC derivatives in forming both perivascular end endosteal BM niches that maintain HSCs and regulate blood production.
Deregulation of HSC activity is an important step in the development of myeloid malignancies (Passegué et al., 2003). This is particularly the case for myeloproliferative neoplasms (MPN), a class of clonal disorders that are propagated by leukemic stem cells (LSC) arising from transformed HSCs and carrying oncogenic lesions such as BCR/ABL for chronic myelogenous leukemia (CML), or activating Jak2 mutations for polycythemia vera (PV), essential thrombocythosis (ET) and primary myelofibrosis (PMF) (Van Etten and Shannon, 2004; Levine and Gilliland, 2008). Deregulation of the BM microenvironment is another important factor in the development of myeloid malignancies (Lane et al., 2009). Genetic ablation of the retinoic acid receptor gamma (Rar-γ) or retinoblastoma (Rb) genes in BM stromal cells promote MPN development (Walkley et al., 2007a; 2007b), while inactivation of the miRNA-processing enzyme dicer in immature Osterix (Osx)-expressing osteoprogenitors causes a myelodysplastic syndrome (MDS) (Raaijmakers et al., 2010). In addition, changes in the signaling activity of OBCs can alter HSC numbers and cause lineage-specific defects in blood production (Schepers et al., 2012). Recent evidence indicates that myeloid malignancies also affect the function of the BM microenvironment. In particular, decreased expression of Cxcl12 by BM stromal cells is observed in chronic phase CML (Zhang et. al, 2012), thereby impairing support for normal HSCs, while severe osteoblastic defects are found in blast crisis CML leading to a major loss of bone (Frisch et al., 2012). However, much remains to be understood about how leukemic hematopoiesis impacts the BM microenvironment and, in turn, how changes in the activity of specific BM niche cells contribute to MPN pathogenesis. Here, we used an inducible Scl-tTA::TRE-BCR/ABL double transgenic mouse model of human chronic phase CML (Reynaud et al., 2011) to investigate the effect of MPN development on the endosteal BM niche.
RESULTS
Endosteal OBCs contain cells with HSC-supporting activity
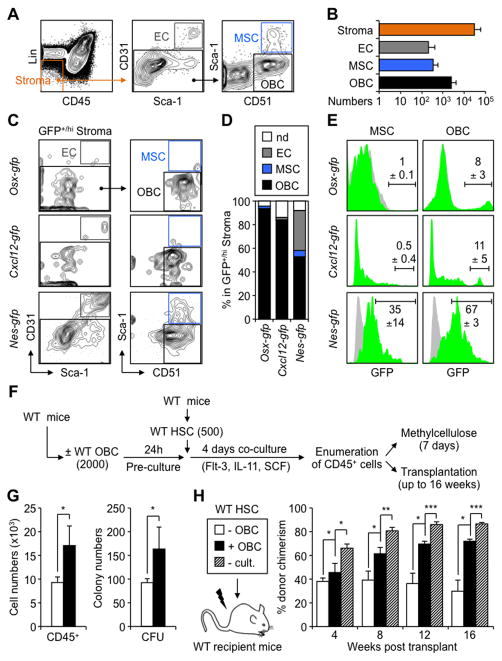
Several flow cytometry approaches have been developed to identify endosteum-associated BM stromal cells. Here, we used a previously described protocol to isolate ECs (Lin−/CD45−/CD31+/Sca-1+), MSCs (Lin−/CD45−/CD31−/CD51+/Sca-1+) and OBCs (Lin−/CD45−/CD31−/CD51+/Sca-1−) from hematopoietic cell-depleted, collagenase-treated crushed bones of wild type (WT) mice (Figures 1A and 1B) (Winkler et al., 2010). In vitro characterization of these populations showed the expected high frequency of colony forming-unit fibroblast activity (CFU-F) and PDGFRα levels in MSCs (Figures S1A and S1B). In contrast, OBCs had lower CFU-F frequencies and PDGFRα levels, while ECs lacked PDGFRα expression and were devoid of CFU-F activity. Consistent with their lineage relationship, both MSCs and OBCs produced alkaline phosphatase positive colonies (CFU-Alk) and von Kossa positive bone nodules (CFU-OB) upon osteoblastic differentiation, with MSCs giving rise to larger colonies than their OBC derivatives (Figure S1A). These results confirm reliable enrichment of endosteal MSCs and their OBC derivatives using this flow cytometry protocol.
Figure 1. HSC-supportive activity of endosteal OBCs.
(A) Flow cytometry approach used to identify endosteal BM stromal populations. (B) Average numbers of ECs, MSCs and OBCs contained in the endosteal (Lin−/CD45−) BM stromal fraction of wild type (WT) mice (n = 23 in 7 independent experiments). (C) Immunophenotype and (D) frequencies of GFP+ endosteal ECs, MSCs and OBCs in Osx-gfp, Cxcl12-gfp and Nes-gfp reporter mice (n = 2–4 per genotype; nd: not determined). (E) Frequency of GFP+/hi cells in endosteal MSCs and OBCs of Osx-gfp, Cxcl12-gfp and Nes-gfp reporter mice (green histograms). Grey histograms indicate background GFP fluorescence levels in control populations. (F) Schematic of the short-term co-culture of HSCs with or without OBCs, and follow up analyses. (G) Cell numbers and methylcellulose colony-forming unit (CFU) activity. (H) Transplantation in lethally irradiated WT CD45.1 recipients (n = 3–5 mice per group, with results replicated in another independent experiment). Mice were bled every 4 weeks and analyzed for the percentage of CD45.2+ donor-derived cells (- cult.: no culture).
Data are means ± SD; *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001. See also Figure S1.
We then used GFP reporter mice to determine the relationship between endosteal subsets and BM niche cells with demonstrated HSC-supportive activity. Strikingly, we found the presence of Osx+ osteoprogenitors, CXCL12hi CAR cells and Nes+ MSC-like cells within the OBC fraction (Figures 1C and 1D), with frequencies ranging from ~10% in Osx-gfp and Cxcl12-gfp mice to ~70% in Nes-gfp mice (Figure 1E). As expected, we also found that ~35% of the MSC fraction was GFP+ in Nes-gfp mice (Mendez-Ferrer et al., 2010), while less than 1% was GFP+/hi in either Osx-gfp or Cxcl12-gfp mice (Figure 1E). Additional flow cytometry analyses of stromal vs. hematopoietic BM cells (Figure S1C) and immunofluorescence analyses of bone sections (Figure S1D) confirmed that most Osx-GFP+ cells were located at the bone surface, while Cxcl12-GFPhi and Nes-GFP+ cells were found both at the bone surface and throughout the BM cavity. These results indicate that the endosteal OBC fraction contains some of the previously-described HSC niche cells.
To confirm that endosteal OBCs are able to maintain HSCs, we performed short-term in vitro co-culture experiments where 500 wild type (WT) HSCs (Lin−/c-Kit+/Sca-1+/Flk2−/CD150+/CD48−) were grown for 4 days with or without 2,000 purified OBCs (Figure 1F). As expected, HSCs co-cultured with OBCs showed more in vitro hematopoietic expansion and higher myeloid differentiation potential in methylcellulose than HSCs cultured on plastic (Figure 1G). Mice transplanted with the progeny of 500 HSCs co-cultured with OBCs also displayed significantly higher levels of donor chimerism than mice receiving cells cultured without OBCs (Figure 1H). However, the chimerism level was still lower than in mice transplanted with 500 freshly-isolated WT HSCs, indicating that OBCs were primarily maintaining HSC function without increasing their numbers. In all cases, we observed similar multilineage reconstitution (data not shown). These results directly demonstrate that phenotypically defined endosteal OBCs have HSC-supporting activity.
MPN development causes endosteal OBCs expansion and myelofibrosis development
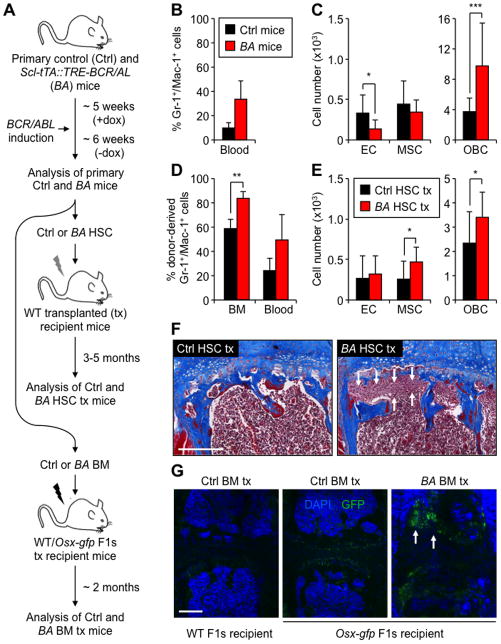
We then used our inducible Scl-tTA::TRE-BCR/ABL (BA) double transgenic mouse model of chronic phase CML (Reynaud et al., 2011) to address how MPN development affects the endosteal BM niche. We first quantified the numbers of endosteal stromal BM cells in age-matched littermate controls (Ctrl) and primary diseased BA mice, which develop severe granulocytic expansion within 5 to 6 weeks upon doxycycline withdrawal and BCR/ABL induction (Figures 2A and 2B). Remarkably, we found a large increase in OBC numbers in BA mice, with MSC numbers remaining largely unaffected and EC numbers actually decreasing (Figures 2C and S2A). This OBC expansion was associated with an accumulation of myelofibrotic cells with increased collagen deposition (Figure S2B), as previously reported in both primary BA mice (Reynaud et al., 2011) and human CML patients (Thiele and Kvasnicka, 2006). While no significant changes in bone mineral density (BMD) were detected (data not shown), microCT analyses of proximal tibiae revealed increased trabecular thickening of primary and secondary spongiosa with significant expansion of trabecular bone below the growth plate in BA mice (Figure S2C). Although trabecular number and spacing were unchanged, trabeculae were thicker and showed increased connectivity (Figure S2D). In contrast, cortical bone formation was unaffected (Figure S2C) and tartrate resistant acid phosphatase (TRAP) staining showed no qualitative differences in osteoclast numbers between Ctrl and BA mice (Figure S2E). To exclude the possibility that BCR/ABL was expressed in OBCs, we generated Scl-tTA::TetO-H2B-gfp double transgenic reporter mice (Figure S2F). While some ECs (Lécuyer and Hoang, 2004) and all hematopoietic BM cells including HSCs were GFP+, no cells within the OBC and MSC fractions expressed GFP. This finding indicates that the Scl/Tal-1 3′-enhancer element is not active in endosteal MSCs and OBCs, and rules out an intrinsic effect of BCR/ABL expression in these populations. These results demonstrate that MPN development is accompanied by a striking expansion of endosteal OBCs, an increase in trabecular bone formation and BM fibrosis.
Figure 2. MPN hematopoiesis induces expansion of endosteal OBCs.
(A) Overall experimental design. Primary diseased Scl-tTA::TRE-BCR/ABL (BA) and age-matched control (Ctrl) mice were analyzed ~6 weeks after doxycycline (dox) withdrawal (n = 9–14 per group in 4 independent experiments). Purified CD45.2 HSCs (500 to 4,000 cells) were transplanted into lethally-irradiated WT CD45.1 recipients (n = 10–13 mice per group in 6 independent experiments), and unfractionated CD45.2 BM cells (2×106 cells) into WT or Osx-gfp F1s recipients (n = 3–6 mice per group in 2 independent experiments). (B) Percentage of Gr-1+/Mac-1+ myeloid cells in the blood of primary Ctrl and BA mice. (C) Numbers of endosteal ECs, MSCs and OBCs in primary Ctrl and BA mice. (D) Percentage of donor-derived Gr-1+/Mac-1+ myeloid cells in the BM and blood of Ctrl and BA HSC tx WT mice. (E) Numbers of endosteal ECs, MSCs and OBCs in Ctrl and BA HSC tx WT mice. (F) Masson’s trichrome staining of sternums from the indicated mice. Arrows indicate areas with myelofibrotic cells. Scale bar, 250 μm. (G) Immunofluorescence analyses of sternums from Ctrl and BA BM tx WT and Osx-gfp mice stained for DAPI (blue) and GFP (green). Arrows indicate areas with GFP+ myelofibrotic cells. Scale bar, 250 μm.
Data are means ± SD; *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001. See also Figure S2.
To demonstrate that MPN development is directly responsible for the OBC expansion, we transplanted purified HSCs isolated from primary Ctrl and diseased BA mice into sub-lethally-irradiated WT recipients (Figure 2A). Mice that received BA HSCs all developed CML within 3 to 5 months following transplantation (Figure 2D). Remarkably, diseased BA HSC transplanted WT recipients also showed OBC expansion, which was now accompanied by an increase in MSC numbers, and BM fibrosis (Figures 2E and 2F). To test whether MPN-expanded OBCs are involved in myelofibrosis development, we transplanted unfractionated Ctrl or BA BM cells into lethally-irradiated Osx-gfp recipients, where ~10% of endosteal OBCs are marked by GFP expression (Figure 1E). Strikingly, diseased BA BM transplanted Osx-gfp recipients show accumulation of GFP+ cells in the BM cavity (Figure 2G), which reflected the contribution of MPN-expanded recipient-derived Osx-GFP+ OBCs to the myelofibrotic tissue. Collectively, these results indicate that MPN progressively alters the architecture of the BM microenvironment, and directly causes an expansion of endosteal OBCs that, in turn, contribute to BM fibrosis.
MPN myeloid cells stimulate MSCs to overproduce OBCs
To understand how MPN development leads to OBC expansion, we developed a novel in vitro co-culture/imaging approach modeling the interactions between hematopoietic and stromal BM populations (Figure 3A). WT MSCs or OBCs were isolated from β-actin-gfp mice and co-cultured for up to 10 days with BM cells isolated from Ctrl or BA mice. A combination of manual and software-automated cell counting of fluorescence microscopy images obtained with an IN Cell Analyzer 2000 was then used to determine the number of MSC- or OBC-derived colonies and GFP+ cells per colony. As expected, MSCs co-cultured with BM cells showed more numerous colonies than MSC cultured without hematopoietic cells (0.6 ± 0.7 vs. 6.9 ± 3.2 colonies; n = 10; p ≤ 0.001). In these conditions, BA BM cells significantly increased the size of MSC-derived colonies compared to Ctrl BM cells (Figure 3B), without changing the total numbers of colonies (6.9 ± 3.2 vs. 6.5 ± 3.6 colonies; n = 10). In contrast, BA BM cells had no effect on the size (Figure 3B) or the number of OBC-derived colonies (3.7 ± 2.5 vs. 4.0 ± 1.0 colonies; n = 10), hence demonstrating a specific effect of leukemic hematopoiesis on MSCs. Fluidigm-based gene analyses uncovered no major differences in expression of the cell cycle machinery in MSCs and OBCs isolated from primary Ctrl and BA mice, besides a significant decrease in Ccnd2 (cyclinD2) and p27 expression in BA populations (Figure S3A). EdU incorporation experiments performed after 7 days co-culture showed that BA BM cells directly increased the proliferation rates of expanding MSC-derived cells (Figures 3C and S3B). Consistent with an enhanced OBC differentiation, MSC-derived cells co-cultured for 10 days with BA BM cells also showed a smaller, more rounded osteoblastic-like morphology, and increased expression of early osteoblastic differentiation markers as measured by qRT-PCR analyses (Figures S3C and S3D). However, upon differentiation, MPN-expanded OBCs did not maintain a proliferative state as shown in vivo by the absence of staining with the proliferation marker Ki67 of recipient-derived Osx-GFP+ OBCs (Figure S3E). These results indicate that MPN directly stimulates MSCs to overproduce OBC derivatives.
Figure 3. MPN myeloid cells stimulate MSCs to overproduce OBCs.
(A) Schematic of the in vitro co-culture/imaging approach. MSCs or OBCs isolated from β-actin-gfp mice were cultured with Ctrl or BA BM cells and imaged for the number of GFP+ cells per colony using an IN Cell Analyzer 2000. Representative images are of wells containing MSC- and OBC-derived GFP+ colonies after 10 days culture with Ctrl and BA BM cells. Scale bar, 1 mm. (B) Average numbers of GFP+ cells obtained per MSC or OBC colony after 10 days culture ± Ctrl or BA BM cells (n ≥ 3 per group, with at least 11 individual colonies scored per condition; nd: not determined). (C) Frequencies of EdU+ cells in re-sorted MSC-derived GFP+ cells cultured with Ctrl or BA BM cells for 7 days and pulsed with 10 μM EdU for 3 hours (n = 7–8 per group). (D) Frequencies of hematopoietic cells stained with the indicated markers in unfractionated and myeloid-enriched (my) Ctrl and BA BMs, and average numbers of GFP+ cells obtained per MSC colony after 10 days culture with Ctrl or BA myBM cells (n ≥ 3 per group, with at least 65 individual colonies scored per condition). (E) Schematic of the in vivo MPN regression experiment. Primary diseased BA and age-matched Ctrl mice were re-exposed for 2 or 4 months (m) to doxycline (+dox) to block BCR/ABL expression (n = 3–6 mice per group). (F) Percentage of Gr-1+/Mac-1+ myeloid cells in the BM of re-exposed mice and Masson’s trichrome staining of sternums from the indicated mice. Scale bar, 100 μm.
Data are means ± SD; *p ≤ 0.05, ***p ≤ 0.001. See also Figure S3.
We next tested the importance of leukemic myeloid cells for OBC expansion. We first confirmed that mature BA BM cells, rather than immature c-Kit+ stem and progenitor cells, were responsible for the increase in MSC colony size (data not shown). We then used magnetic bead depletion to obtain ≥ 95% Mac-1+ myeloid-enriched BM cells from both Ctrl and BA mice (Figure 3D). Strikingly, we found that myeloid-enriched Ctrl BM cells did not increase MSC colony size in contrast to myeloid-enriched BA BM cells, hence demonstrating the specific involvement of leukemic myeloid cells. To address the reliance of OBC expansion on leukemic myeloid cells, we re-exposed primary diseased BA mice to doxycycline to block BCR/ABL expression and eradicate leukemic myeloid cells (Figure 3E). Following normalization of myeloid counts, we observed a progressive decrease in OBC numbers with full recovery by 4 months post reinduction, and a quick disappearance of myelofibrosis as early as 2 months post re-induction (Figure 3F). These results are consistent with normal turnover rates of MSCs (Park et al., 2012), and demonstrate that leukemic myeloid cells are both necessary and sufficient to drive OBC expansion and to remodel the endosteal BM niche.
Both soluble factors and direct cell contact drive OBC expansion
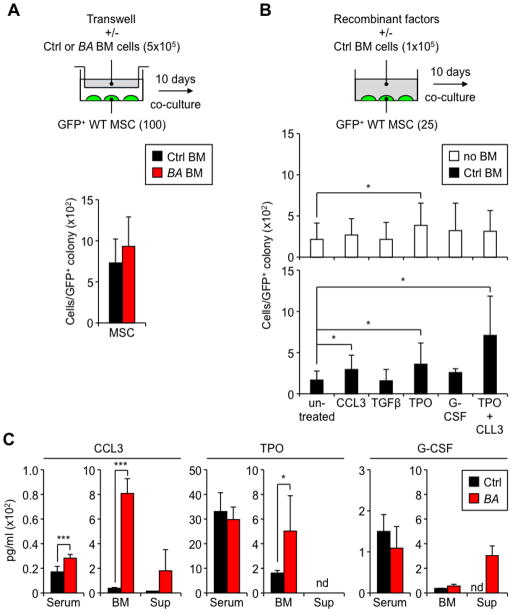
To understand how leukemic myeloid cells stimulate MSCs to overproduce OBC derivatives, we first used transwell plates to assess the relative contribution of secreted soluble factors vs. direct cell-cell interactions. Strikingly, we did not observe a significant increase in MSC colony size when BA BM cells were physically separated from MSCs (Figure 4A). Examination of MSCs co-cultured with myeloid-enriched BA BM cells also showed close contacts between the two cell types throughout the culture period (Figure S4A). These findings indicate that direct contacts or close proximity signals between leukemic myeloid cells and MSCs are important in driving OBC expansion.
Figure 4. MSC stimulation requires TPO, CCL3 and direct contact with MPN myeloid cells.
(A) Schematic and average numbers of GFP+ cells obtained per MSC colony after 10 days co-culture without direct contact with Ctrl or BA BM cells in 24-well transwell plates (n = 3 per group, with at least 30 individual colonies scored per condition). (B) Schematic and average numbers of GFP+ cells obtained per MSC colony after 10 days co-culture with the indicated recombinant cytokines (50 ng/ml except for TGFβ used at 10 ng/ml) ± Ctrl BM cells (n ≥ 6 per group, with at least 20 individual colonies scored per condition). (C) ELISA quantification of CCL3, TPO and G-CSF levels in the serum and BM plasma (BM) of primary Ctrl and BA mice (n = 6–8 per group), and supernatant (sup) from 10-day co-cultures of MSCs with Ctrl and BA BM cells (n = 3 per group; nd: not detectable).
Data are means ± SD; *p ≤ 0.05, ***p ≤ 0.001. See also Figure S4.
We then directly tested the effect of candidate factors known to increase osteoblastic differentiation, like IL-6 and IL-1β (Erices et al., 2002; Sonomoto et al., 2012), or previously implicated in either leukemia-induced stromal changes, like CCL3 (MIP-1α) and G-CSF (Frisch et al. 2012; Zhang et al., 2012), or myelofibrosis development, like TGFβ and thrombopoietin (TPO) (Varricchio et al., 2009). Although both IL-6 and IL-1β levels were increased in the serum of primary diseased BA mice (Figure S4A) (Reynaud et al., 2011; Zhang et al., 2012), neither of these cytokines, nor TNFα, were able to increase MSC colony size when added individually to the cultures (Figure S4B). In addition, OBC expansion and myelofibrosis were still observed in primary BA mice lacking either Il-6 or Il-1r1 genes (Figures S4C and S4D), confirming that MPN-mediated remodeling of the endosteal BM niche was largely independent of the aberrant secretion of these pro-inflammatory cytokines by leukemic myeloid cells. Among the other factors tested, only TPO increased MSC colony size by itself (Figure 4B). Strikingly, addition of Ctrl BM cells significantly potentiated the effect of TPO and CCL3 on MSC colony size, but did not promote OBC expansion with either TGFβ or G-CSF (Figure 4B). We also confirmed elevated levels of CCL3 and TPO in the BM plasma of primary diseased BA mice, and detected a trending increase in CCL3 and G-CSF levels in the supernatant of MSC co-cultures with BA BM cells (Figure 4D). These results suggest that TPO and CCL3, in conjunction with direct interactions between MSCs and leukemic myeloid cells, drive the overproduction of OBC derivatives during MPN development.
Molecular features of MPN-expanded OBCs
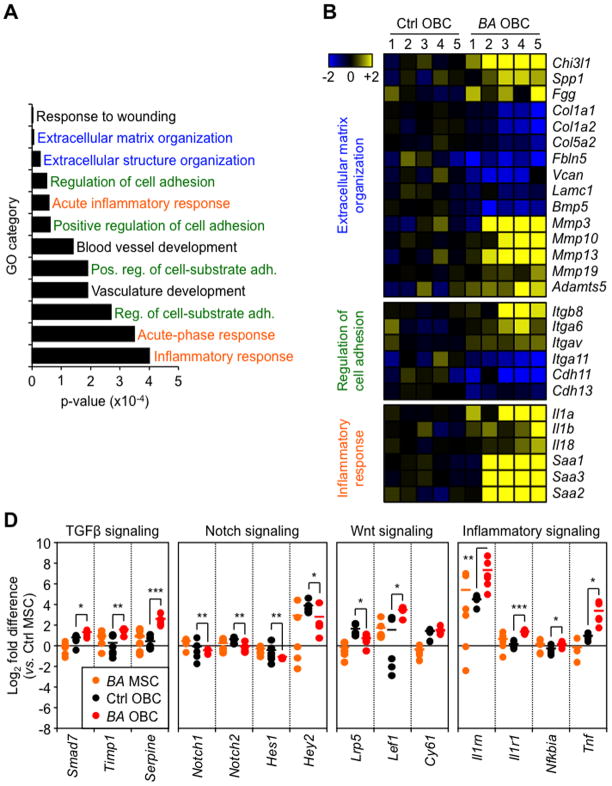
To determine whether MPN-expanded OBCs are functionally altered, we compared the gene expression profiles of OBCs isolated from primary Ctrl and diseased BA mice using Affymetrix Gene ST 1.0 microarrays. Consistent with their identity, both Ctrl and BA OBCs expressed high levels of osteoblast and chondrocyte differentiation markers, low levels of MSC and adipocyte differentiation genes, and were not contaminated by adherent myeloid cells (Figures S5A). Statistical analysis of microarrays (SAM) uncovered 610 genes that were differentially expressed between Ctrl and BA OBCs (≥ 1.5 fold; p ≤ 0.05), with 437 up-regulated and 173 down-regulated genes (Tables S1 and S2). Gene ontology (GO) analyses of these genes identified processes such as extracellular matrix organization, regulation of cell adhesion and inflammatory responses as being significantly enriched in BA OBCs (Figure 5A and Table S3). Examination of individual genes showed changes in extracellular matrix components that are relevant for myelofibrosis development, such as increased expression of chitinase (Chi3l1), Spp1 and fibrinogen (Fgg), and decreased expression of several types of collagens (Col), versican proteoglycans (Vcan), laminin glycoproteins (Lamc1) and integrin ligands (Fbln5), and increased expression of a whole range of matrix metallopeptidases (Mmp) and ADAM metallopeptidases (Adamts) that are likely important for tissue remodeling (Figure 5B). BA OBCs also displayed changes in integrin (Itg) and cadherin (Cdh) expression levels, as well as a striking up-regulation of the IL-1 super-family of pro-inflammatory cytokines and many serum amyloids (Saa) (Figure 5B). These results were validated by subsequent qRT-PCR and Fluidigm-based gene expression analyses (Figures S5B and S5C). Importantly, many of the genes up-regulated in primary BA OBCs were also up-regulated in WT MSC-derived cells cocultured with BA BM cells (Figure S3A), thus confirming that these molecular changes were a direct consequence of exposure to leukemic myeloid cells. Taken together, these results confirm the myelofibrotic, pro-inflammatory nature of MPN-expanded OBCs.
Figure 5. Molecular features of MPN-expanded OBCs.
(A) OBCs were purified from individual primary Ctrl and BA mice (n = 5 per group) and used for microarray analyses. Histogram shows the gene ontology (GO) results for the biological processes significantly affected in BA OBCs. (B) Microarray results detailing the genes involved in extracellular matrix organization, regulation of cell adhesion and inflammatory response. Data are expressed as log2 fold relative to the average expression level in Ctrl OBCs (set to 0). (C) Fluidigm-based gene expression analyses of TGFβ, Notch, Wnt and inflammation pathway components in MSCs and OBCs isolated from primary Ctrl and BA mice (n = 4–6 pools of 100 cells per population). Data are expressed as log2 fold relative to the average level in Ctrl MSCs (set to 0). Bars indicate average levels, and °/* statistical differences between Ctrl vs. BA MSCs and OBCs, respectively.
Data are means ± SD; *p ≤ 0.05, ** p ≤ 0.01, ***p ≤ 0.001. See also Figure S5
We then took advantage of our Fluidigm-based gene expression analyses to investigate changes in signaling activity occurring in MPN-expanded OBCs (Figure 5D and Table S4). We found evidence of elevated TGFβ signaling in BA OBCs as shown by increased expression of the TGFβ targets Smad7, Timp1 and Serpine. We also observed attenuated Notch signaling in BA OBCs as evidenced by decreased expression of both Notch1 and Notch2 receptors and Notch targets Hes1 and Hey2. In contrast, we found no consistent trend in the status of the Wnt pathway with decreased expression of the Lrp5 co-receptor, increased expression of Lef1 transcription factor and unchanged levels of the Wnt target Cy61. Moreover, we confirmed active inflammatory signaling in BA OBCs, with increased expression of IL-1 receptor (Il-1r1) and receptor antagonist (Il-1rn), and up-regulation of both TNFα (Tnf) and IκBα (Nfkbia). These results indicate that changes in TGFβ, Notch and inflammatory signaling are associated with the remodeling of MPN-expanded OBCs into inflammatory myelofibrotic cells.
MPN-expanded OBCs have compromised HSC-supportive activity
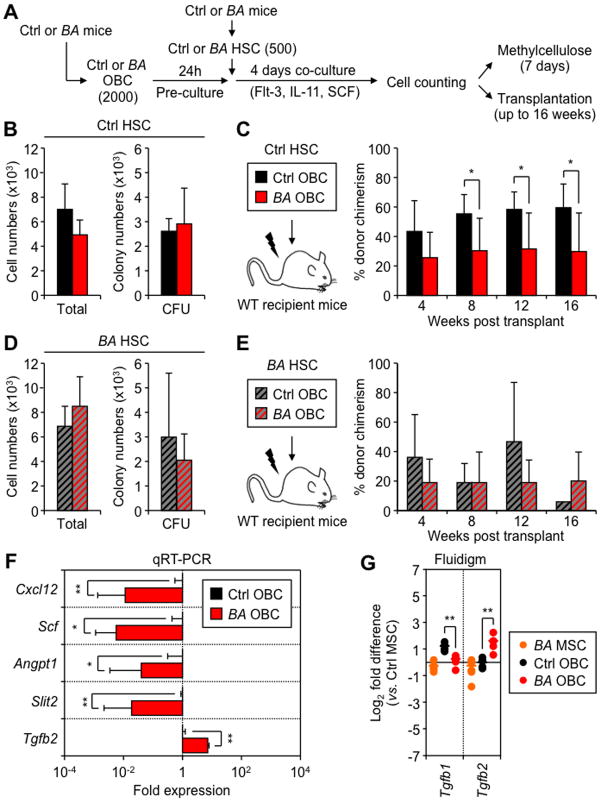
Finally, we investigated whether MPN-expanded OBCs had altered HSC-supportive activity towards either normal HSCs or transformed LSCs. We performed short-term in vitro co-culture experiments where Ctrl or BA HSCs were grown for 4 days with Ctrl or BA OBCs (Figure 6A). Although Ctrl HSCs co-cultured with either OBC population displayed similar in vitro hematopoietic expansion and myeloid differentiation activity (Figures 6B), transplantation experiments showed significantly impaired donor chimerism in mice injected with the progeny of Ctrl HSCs co-cultured with BA OBCs (Figures 6C). Defective reconstitution was not the result of changes in lineage distribution (Figure S6A), but due to reduced numbers of engrafted donor-derived Ctrl HSCs (Figure S6B). BA HSCs also displayed similar hematopoietic expansion and myeloid differentiation activity when co-cultured with either OBC populations (Figure 6D). However, in sharp contrast to their normal counterparts, the engraftment ability of BA HSCs was largely independent the OBC population on which they were co-cultured (Figure 6E and S6C). In both cases, we observed either impaired engraftment or CML development eventually leading to an early death, which are consistent with previous findings showing poor engraftment from freshly isolated BA HSCs (Zhang et al., 2012). These results indicate that MPN-expanded OBCs are severely compromised in their ability to maintain normal HSCs, while transformed LSCs are essentially resistant to the detrimental effect of BA OBCs.
Figure 6. Impaired HSC-supportive activity of MPN-expanded OBCs.
(A) Schematic of the short-term co-culture of Ctrl or BA HSCs with Ctrl or BA OBCs, and follow up analyses. Cell numbers and methylcellulose CFU activity for the progeny of Ctrl HSCs (B) or BA HSCs (D) co-cultured with Ctrl or BA OBCs. Transplantation experiments for the progeny Ctrl HSCs (C) or BA HSCs (E) co-cultured with Ctrl or BA OBCs (n = 3–5 mice per group, with results replicated in another independent experiment). (F) qRT-PCR-based gene expression analyses of HSC regulatory molecules in OBCs isolated from individual primary Ctrl and BA mice (n = 3 per group). Data are expressed as fold relative to the average expression level in Ctrl OBCs (set to 1). (G) Fluidigm-based gene expression analyses of members of the TGFβ family in MSCs and OBCs isolated from primary Ctrl and BA mice (n = 4–6 pools of 100 cells per population). Data are expressed as log2 fold relative to the average level in Ctrl MSCs (set to 0). Bars indicate average levels, and * statistical differences between Ctrl vs. BA OBCs.
Data are means ± SD; *p ≤ 0.05; **p ≤ 0.01. See also Figure S6.
We then used our microarray results as well as confirmatory qRT-PCR and Fluidigm-based gene expression analyses to address the underlying mechanism (Figures 6F, 6G and S6D). We found a broad down-regulation of many HSC retention factors in BA OBCs compared to Ctrl OBCs, including Lepr, Cxcl12, N-cadherin (Cdh2), Scf, Angpt1 and Slit2. We also observed a striking shift in the expression pattern of TGFβ molecules in BA OBCs, with down-regulation of the quiescence-enforcing Tgfb1 and massive up-regulation of the myeloid-promoting Tgfb2. These molecular changes are likely to contribute to the impaired ability of MPN-expended OBCs to maintain normal HSCs, and to favor MPN development. Collectively, these data identify how an MPN-remodeled endosteal BM niche can favor LSC function by impairing normal hematopoiesis and promoting myeloid differentiation.
DISCUSSION
Transgenic mouse models of human MPNs provide an ideal platform for understanding how leukemic hematopoiesis disrupts the normal mechanisms controlling HSC function and blood production. Previously, we found that aberrant secretion of pro-inflammatory cytokines by leukemic myeloid cells establishes a feed-forward loop that drives myeloid differentiation and CML development by influencing fate decisions in the leukemic multipotent progenitor (MPP) compartment (Reynaud et al., 2011). Here, we show that leukemic myeloid cells also remodel the endosteal BM niche into a self-reinforcing leukemic niche that impairs normal hematopoiesis, favors LSC function and contributes to myelofibrosis development (Figure 7). These results expand our understanding of the effects of leukemic hematopoiesis on the BM microenvironment, and the contribution of the endosteal BM niche to MPN pathogenesis. Moreover, they identify a novel mechanism for the BM fibrosis and loss of normal hematopoiesis that often accompanies MPN development in humans. Altogether, they uncover a close interrelationship between leukemic hematopoiesis and leukemic microenvironment, which could be exploited to develop new therapeutic strategies.
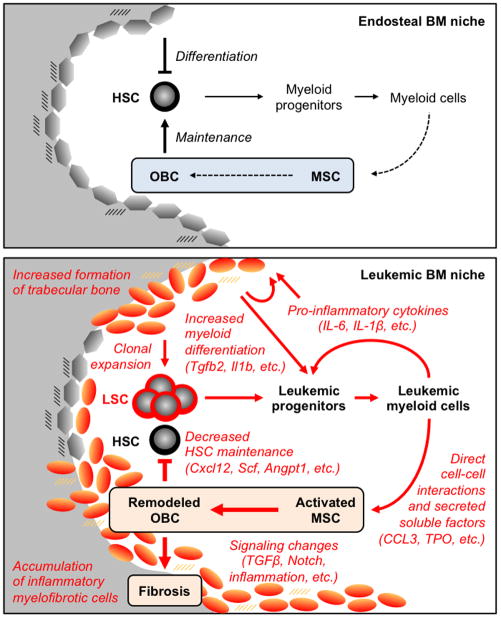
Figure 7. Model for the leukemic BM niche.
The endosteal BM niche contributes to HSC maintenance and regulated production of myeloid cells (top panel). The normal composition of the BM milieu likely contributes to regulated production of bone-lining OBCs from MSCs. During MPN development, transformed HSCs with LSC properties overproduce leukemic myeloid cells that secrete high levels of pro-inflammatory cytokines, thus creating a paracrine feedback loop that drives myeloid differentiation (bottom panel). Leukemic myeloid cells also directly stimulate MSCs to overproduce functionally altered OBCs, which accumulate in the BM cavity as inflammatory myelofibrotic cells. These MPN-expanded OBCs are severely compromised in their ability to maintain normal, but not transformed, HSCs and promote myeloid differentiation. Our results demonstrate that MPN development remodels the endosteal BM niche into a self-reinforcing leukemic niche that impairs normal hematopoiesis, favors LSC function and contribute to BM fibrosis. See also Figure S7.
Previous lineage tracking studies have identified various populations of perivascular MSC-like cells and endosteal osteoprogenitor cells as important BM niche cells for HSCs (Frenette et al., 2013). In parallel, several flow cytometry approaches have been developed to isolate and interrogate the function of different endosteal BM stromal subsets. However, it remains unclear how these two methodologies overlap in identifying BM niche cells important for HSC activity. Here, we used a fractionation method developed by the Levesque group to isolate BM stromal populations enriched for endosteal MSCs (Lin−/CD45−/CD31−/CD51+/Sca-1+) and their OBC derivatives (Lin−/CD45−/CD31−/CD51+/Sca-1−), which represents a mixture of both immature and mature osteoblasts (Winkler et al., 2010). We show that such phenotypically-defined OBC population contains cells known from lineage tracking studies to be important for HSC maintenance, including Osx+ osteoprogenitor cells, some CXCL12hi CAR cells and a large fraction of Nes+ MSC-like cells. We also found that the phenotypically-defined endosteal MSC fraction contains Nes+ MSC-like cells, and are now interested in extending these cross-identification approaches to the recently described perivascular Lepr+, Mx1+ or Prx1+ MSC-like cells (Ding et al., 2012; Park et al., 2012; Greenbaum et al., 2013). Using a series of in vitro co-culture assays followed by imaging and transplantation approaches, we confirm the ability of endosteal MSCs to give rise to OBCs and, most importantly, directly demonstrate the HSC-supportive capability of endosteal OBCs. Our results indicate that HSC niche cells are also present at the bone surface, a finding that reinforces a role for the endosteal BM niche in controlling HSC maintenance and blood production.
Recent studies indicate that dysfunctional BM microenvironments contribute to the development of myeloid malignancies and that, in turn, leukemic hematopoiesis can create dysfunctional BM microenvironments. Here, we asked whether MPN development could impact on the activity of the endosteal BM niche. Through analyses of primary BA mice and LSC-transplanted WT mice, we show that MPN development results in a massive expansion of endosteal OBCs associated with increased trabeculation and trabecular thickening. These features contrast with the severe osteoblastic defects and bone loss reported in a CML blast crisis mouse model (Frisch et al. 2012), and are likely to reflect fundamental and clinically relevant differences between chronic MPNs and transformed acute myeloid leukemias (AML). We confirm that MPN-mediated OBC expansion is not restricted to BCR/ABL expression since also observed in junB-deficient mice (Figure S7)), which develop a CML-like MPN with high penetrance (Santaguida et al., 2009). Moreover, using a combination of in vitro and in vivo approaches, we demonstrate that MPN-mediated OBC expansion is driven by MSCs that are stimulated by leukemic myeloid cells to proliferate and overproduce OBC derivatives. This involvement of MSCs is in line with their high expansion capability as compared to the restricted growth potential of OBCs (Park et al., 2012). This function of leukemic myeloid cells is also consistent with the newly described role of monocytes/macrophages in regulating the activity of BM niche cells and maintenance of HSC (Frenette et al., 2013). While we did not characterize MPN myeloid cells beyond their broad expression of Mac-1, it is possible that they could contain the leukemic and aberrantly behaving equivalents of the recently described CD169+ macrophages (Chow et al., 2011) or F4/80+ osteomacs lining the endosteum (Chang et al., 2008; Winkler et al., 2010). Altogether, our results show that MPN development alters the normal activity of MSCs and their OBC derivatives leading to a major remodeling the endosteal BM niche.
Many factors and signaling pathways are known to stimulate MSC growth and to control osteoblastic differentiation (Long, 2012). Here, we took advantage of our in vitro co-culture/imaging approach to demonstrate the importance of direct cell-cell interactions between leukemic myeloid cells and expanding MSCs, and to screen for the involvement of candidate factors known to affect OBC production or myelofibrosis development. Although BA myeloid cells exhibit aberrant secretion of pro-inflammatory cytokines known to increase osteoblastic differentiation, we show that neither IL-6, nor IL-1β or TNFα are involved in MSC stimulation. We also exclude a direct role for G-CSF and TGFβ, despite their importance for osteoblastic differentiation and myelofibrosis. On the other hand, we show that both TPO, the physiologic regulator of platelet production and a strong inducer of myelofibrosis (Varricchio et al., 2009), and CCL3, an inflammatory chemokine that regulates MSC migration and osteoblastic differentiation (Sordi et al., 2005; Vallet et al., 2011), synergistically expand OBCs especially when BM cells are directly in contact with the cultured MSCs. Since both TPO and CCL3 levels are elevated in the BM plasma of BA mice, they are likely part of the mechanism by which leukemic myeloid cells stimulate MSCs to overproduce OBC derivatives during MPN development. However, it is clear that other soluble or membrane-bound factors expressed by leukemic myeloid cells also contribute to this process. Our results support the idea that a complex interplay in the BM cavity between secreted soluble factors and direct interactions between myeloid cells and MSCs controls the rate of OBC production in both normal and disease conditions. They suggest that distinct deregulations in this finely tuned equilibrium could result in OBC expansion during MPN development and OBC loss during AML development.
MPNs, unlike AML, often progress to BM fibrosis prior to leukemic transformation, and the development of myelofibrosis can have severe consequences even at the pre-leukemic stage (Abdel-Wahab and Levine, 2009). Here, we show that MPN-expanded OBCs accumulate in the BM cavity as inflammatory myelofibrotic cells (Figure 7). We find clear evidence of myelofibrosis in both primary BA mice and LSC-transplanted WT mice, and demonstrate that MPN-expanded OBCs can directly contribute to the myelofibrotic tissue in LSC-transplanted Osx-gfp reporter mice. Furthermore, we show that OBCs, either directly isolated from primary diseased BA mice or generated in vitro by co-culture of WT MSCs with BA BM cells, display the molecular hallmarks of inflammatory myelofibrotic cells, including changes in the expression of genes involved in cellular adhesion, extracellular matrix remodeling and inflammation. Strikingly, many of the up-regulated genes in MPN-expanded OBCs are also elevated in human patients with rheumatoid and osteoid arthritis (Okamoto et al., 2008). This convergence suggests a tantalizing, but still un-explored, link between chronic inflammatory diseases and MPN-associated myelofibrosis. Since MPN-expanded OBCs also express higher levels of pro-inflammatory factors, they may directly contribute to the overproduction of leukemic myeloid cells, thus enforcing a vicious cycle that perpetuates MPN development and OBC expansion (Figure 7). Pathway-directed gene expression analyses implicate increased TGFβ and inflammatory signaling, and attenuated Notch signaling as potential contributors to the remodeling of MPN-expanded OBCs into inflammatory myelofibrotic cells. It is therefore tempting to speculate that pro-inflammatory factors secreted by either leukemic myeloid cells or MPN-expanded OBCs, together with tissue remodeling factors produced by the expanded OBCs could create positive feedback loops driving this process, similar to the interplay between IL-1β and serum amyloids described in rheumatoid arthritis (Okamoto et al., 2008).
Myelofibrosis is a prominent feature of PV and PMF MPNs (Abdel-Wahab and Levine, 2009) and is observed in ~30% of chronic phase CML patients, where it is often associated with poor-prognosis disease and progression to blast crisis (Thiele and Kvasnicka, 2006). Myelofibrosis in PV and PMF MPNs has been linked to defective megakaryopoiesis, and impaired megakaryopoiesis can result in the development of PV- and PMF-like MPNs in mice (Varricchio et al., 2009). In contrast, we never observed any defects in megakaryopoiesis in CML mouse models, including BA and junB-deficient mice (data not shown). Here, we identify a novel route for the pathogenesis of BM fibrosis associated with CML-like MPNs, which involves leukemic myeloid cells driving the expansion and remodeling of endosteal OBCs (Figure 7). We show that both OBC expansion and myelofibrosis development depend on the presence of leukemic myeloid cells, and can be fully reverted upon blockade of BCR/ABL expression and recovery of normal hematopoiesis. This is consistent with the reduction in fibrosis observed in CML patients successfully treated with tyrosine kinase inhibitors (Thiele and Kvasnicka, 2006), which reinforces the idea that a similar mechanism could be operating in humans. It will now be important to directly address its contribution to fibrosis occurring in CML patients, as well as in PV and PMF patients since overproduction of leukemic myeloid cells is also observed in these other MPN diseases (Abdel-Wahab and Levine, 2009). Our results implicate remodeling of the endosteal BM niche by leukemic myeloid cells as an important new mechanism for BM fibrosis associated with MPN development.
Loss of normal hematopoiesis is another event occurring in human MPNs, and during the development of most leukemias, for reasons that still remain largely unknown. Here, we show that leukemic hematopoiesis turns the endosteal BM niche into a leukemic niche, which promotes LSC function and impairs the maintenance of normal HSCs (Figure 7). We find altered expression of many HSC regulatory genes in MPN-expanded OBCs, including a broad down-regulation of essential HSC retention factors (i.e., Cxcl12, Scf, Lepr, Angpt1, Cdh2, Slit2, Tgfb1) and up-regulation of factors promoting myeloid differentiation (i.e., Il1b, Tgfb2). We directly show that MPN-expanded OBCs exhibit reduced ability to maintain HSCs, which mainly affects normal HSCs, with minimal effects on transformed LSCs. The molecular changes we uncovered are likely responsible for the compromised HSC-supportive activity of the remodeled OBCs, especially with respect to decreased Cxcl12 expression, which could directly contribute to loss of normal HSCs through increased mobilization to the periphery (Zhang et al. 2012; Greenbaum et al., 2013). Molecular changes in MPN-expanded OBCs are also likely to promote MPN development by favoring myeloid differentiation and overproduction of leukemic myeloid cells from LSCs. The fact that LSC maintenance is unaffected by the remodeled OBCs could be, in large part, due to their different requirement in adhesion molecules for homing and retention in the BM compared to normal HSCs (Gordon et al. 1987; Krause et al., 2006). The loss of normal HSCs also likely contributes to the clonal dominance of BCR/ABL-expressing LSCs in transplanted mice (Reynaud et al., 2011) and CML patients (Holyoake et al., 1999). Moreover, such differential retention provided by a remodeled BM microenvironment could explain clonal dominance in situations where transformed HSCs have impaired functions such as in MDS and BM failure syndromes (Levine and Gilliland, 2008). Taken together, our results unveil novel features of MPN pathogenesis and uncover how leukemic myeloid cells create a self-reinforcing leukemic BM niche that promotes MPN development at the expense of normal hematopoiesis. Targeting this pathological interplay could represent a novel therapeutic avenue to treat MPN patients and prevent myelofibrosis development.
EXPERIMENTAL PROCEDURES
Extended Experimental Procedures can be found in Supplemental Information.
Mice
All the genetic models were previously published. Irradiation and transplantation procedures were performed as previously described (Reynaud et al., 2011). All mice were maintained at UCSF in accordance with IACUC approved protocols.
Bone analyses
Masson’s trichrome staining was performed as described (Reynaud et al., 2011). Bones for immunofluorescence imaging were stained with a rabbit anti-Ki67 primary (SP6 Neomarkers) followed by a goat anti-rabbit-A594 secondary (Invitrogen) antibody, and counterstained with DAPI. Images were taken on an IN Cell Analyzer 2000 (GE Healthcare).
Stromal and hematopoietic cell analyses
Staining and enrichment procedures for flow cytometry were performed as described (Reynaud et al., 2011; Schepers et al., 2012). Myeloid-enriched BM cells were obtained by depleting BM of CD19, B220, CD3, CD4, CD8, CD5 and Ter-119 positive cells using purified rat anti-mouse antibodies and anti-rat Dynal beads. TPO, CCL3 and G-CSF protein levels were measured by ELISA according to the manufacturer’s instructions (R&D Systems and Raybiotech).
Cell culture experiments
Short-term co-culture experiments were performed as previously described (Schepers et al., 2012), with each well containing the progeny of 500 HSCs treated as follows: ~ 5% of the cells were used to measure cell numbers and plated at various dillutions in methylcellulose, and 95% of the cells were transplanted in lethally-irradiated mice with helper BM cells. Co-culture/imaging experiments were carried out in α-modified Eagle medium (αMEM) containing 10% FBS, 1x penicillin/streptomycin and 50 mM 2-mercaptoethanol. Unfractionated or myeloid-enriched BM cells were seeded with stromal BM cells isolated from β-actin-gfp mice in 96-well or 24-well transwell plates. Cells were co-cultured without medium replacement, and recombinant cytokines were refreshed every 2–3 days. EdU labeling experiments were performed after 7 days of co-culture. Stromal-derived GFP+ cells were imaged on an IN Cell Analyzer 2000 every 2–3 days, and automated counting was used to enumerate the total number of GFP+ cells per condition after 10 days of co-culture. The number of colonies (≥ 20 cells) was determined manually and used to calculate the average number of cells per colony.
Gene expression analyses
Analyses using the Fluidigm 96.96 Dynamic Array IFC platform and quantitative RT-PCR analyses were performed as previously described (Reynaud et al., 2011; Santaguida et al., 2009). For qRT-PCR and microarray analyses, RNA was purified using the Arcturus PicoPure RNA kit, amplified using the Nugen Pico WTA kit, and cleaned using the Qiagen QiaQuick PCR purification kit according to manufacturers’ instructions. For microarray analyses, sense-strand cDNA targets were generated using the Nugen WT-Ovation Exon Module, fragmented and biotinylated using the Nugen Encore Biotin Module and hybridized on Affymetrix Gene ST 1.0 microarrays according to manufacturers’ instructions.
Statistical analyses
p values were calculated using the unpaired Student’s t-test and differences with p ≤ 0.05 were considered statistically significant.
Supplementary Material
HIGHLIGHTS.
Leukemic myeloid cells stimulate MSCs to overproduce functionally altered OBCs
Direct contact with MPN cells and secretion of TPO and CCL3 drive OBC expansion
Remodeled OBCs accumulate as inflammatory myelofibrotic cells in the BM cavity
Remodeled OBCs have compromised ability to support HSCs but not LSCs
Acknowledgments
We thank Dr. Paul Frenette (Einstein College of Medicine) for Nes-gfp mice, Dr. Takashi Nagasawa (Kyoto University) for Cxcl12-gfp mice, Dr. Zena Web (UCSF) for Osx-gfp mice, the UCSF Mouse Pathology and Genomics core facilities, the San Francisco VA Histomorphometry core, Michael Kissner for management of our Flow Cytometry core facility, Daniel Carlin for help with the microarray analyses, Zhiqiang Cheng for help with TRAP staining, Stephanie Leong for help with the mouse colonies, and all members of the Passegué laboratory for critical insights and suggestions. This work was supported by NWO Rubicon and KWF fellowships to KS, NIH F32 HL106989 to EMP, NIH U01 HL100402 to AJW, NIH K08 AR056299 to ECH, and CIRM New Faculty Award and NIH R01 HL092471 to EP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdel-Wahab OI, Levine RL. Primary myelofibrosis: update on definition, pathogenesis, and treatment. Annu Rev Med. 2009;60:233–245. doi: 10.1146/annurev.med.60.041707.160528. [DOI] [PubMed] [Google Scholar]
- Calvi LM, Adams GB, Weibrecht KW, Weber JM, Olson DP, Knight MC, Martin RP, Schipani E, Divieti P, Bringhurst FR, et al. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- Chang MK, Raggatt LJ, Alexander KA, Kuliwaba JS, Fazzalari NL, Schroder K, Maylin ER, Ripoll VM, Hume DA, Pettit AR. Osteal tissue macrophages are intercalated throughout human and mouse bone lining tissues and regulate osteoblast function in vitro and in vivo. J Immunol. 2008;181:1232–1244. doi: 10.4049/jimmunol.181.2.1232. [DOI] [PubMed] [Google Scholar]
- Chitteti BR, Cheng YH, Streicher DA, Rodriguez-Rodriguez S, Carlesso N, Srour EF, Kacena MA. Osteoblast lineage cells expressing high levels of Runx2 enhance hematopoietic progenitor cell proliferation and function. J Cell Biochem. 2010;111:284–294. doi: 10.1002/jcb.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow A, Lucas D, Hidalgo A, Méndez-Ferrer S, Hashimoto D, Scheiermann C, Battista M, Leboeuf M, Prophete C, van Rooijen N, et al. Bone marrow CD169+ macrophages promote the retention of hematopoietic stem and progenitor cells in the mesenchymal stem cell niche. J Exp Med. 2011;208:261–271. doi: 10.1084/jem.20101688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan PL, Chute JP. The vascular niche: home for normal and malignant hematopoietic stem cells. Leukemia. 2012;26:54–62. doi: 10.1038/leu.2011.236. [DOI] [PubMed] [Google Scholar]
- Erices A, Conget P, Rojas C, Minguell JJ. Gp130 activation by soluble interleukin-6 receptor/interleukin-6 enhances osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. Exp Cell Res. 2002;280:24–32. doi: 10.1006/excr.2002.5627. [DOI] [PubMed] [Google Scholar]
- Frenette PS, Pinho S, Lucas D, Scheiermann C. Mesenchymal stem cell: keystone of the hematopoietic stem cell niche and a stepping-stone for regenerative medicine. Annu Rev Immunol. 2013;31:285–316. doi: 10.1146/annurev-immunol-032712-095919. [DOI] [PubMed] [Google Scholar]
- Frisch BJ, Ashton JM, Xing L, Becker MW, Jordan CT, Calvi LM. Functional inhibition of osteoblastic cells in an in vivo mouse model of myeloid leukemia. Blood. 2012;119:540–550. doi: 10.1182/blood-2011-04-348151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon MY, Dowding CR, Riley GP, Goldman JM, Greaves MF. Altered adhesive interactions with marrow stroma of haematopoietic progenitor cells in chronic myeloid leukaemia. Nature. 1987;328:342–344. doi: 10.1038/328342a0. [DOI] [PubMed] [Google Scholar]
- Greenbaum A, Hsu YM, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holyoake T, Jiang X, Eaves C, Eaves A. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood. 1999;94:2056–2064. [PubMed] [Google Scholar]
- Krause DS, Lazarides K, von Andrian UH, Van Etten RA. Requirement for CD44 in homing and engraftment of BCR-ABL-expressing leukemic stem cells. Nat Med. 2006;12:1175–1180. doi: 10.1038/nm1489. [DOI] [PubMed] [Google Scholar]
- Lane SW, Scadden DT, Gilliland DG. The leukemic stem cell niche: current concepts and therapeutic opportunities. Blood. 2009;114:1150–1157. doi: 10.1182/blood-2009-01-202606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E, Hoang T. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp Hematol. 2004;32:11–24. doi: 10.1016/j.exphem.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Levine RL, Gilliland DG. Myeloproliferative disorders. Blood. 2008;112:2190–2198. doi: 10.1182/blood-2008-03-077966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F. Building strong bones: molecular regulation of the osteoblast lineage. Nat Rev Mol Cell Biol. 2011;13:27–38. doi: 10.1038/nrm3254. [DOI] [PubMed] [Google Scholar]
- Mendez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma’ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveiras O, Nardi V, Wenzel PL, Hauschka PV, Fahey F, Daley GQ. Bone-marrow adipocytes as negative regulators of the haematopoietic microenvironment. Nature. 2009;460:259–263. doi: 10.1038/nature08099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto H, Cujec TP, Yamanaka H, Kamatani N. Molecular aspects of rheumatoid arthritis: role of transcription factors. FEBS J. 2008;275:4463–4470. doi: 10.1111/j.1742-4658.2008.06582.x. [DOI] [PubMed] [Google Scholar]
- Omatsu Y, Sugiyama T, Kohara H, Kondoh G, Fujii N, Kohno K, Nagasawa T. The essential functions of adipo-osteogenic progenitors as the hematopoietic stem and progenitor cell niche. Immunity. 2010;33:387–399. doi: 10.1016/j.immuni.2010.08.017. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park D, Spencer JA, Koh BI, Kobayashi T, Fujisaki J, Clemens TL, Lin CP, Kronenberg HM, Scadden DT. Endogenous bone marrow MSCs are dynamic, fate-restricted participants in bone maintenance and regeneration. Cell Stem Cell. 2012;10:259–272. doi: 10.1016/j.stem.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E, Jamieson CHM, Ailles LE, Weissman IL. Normal and leukemic hematopoiesis: are leukemias a stem cell disorder or a reacquisition of stem cell characteristics? Proc Natl Acad Sci USA. 2003;100:11842–11849. doi: 10.1073/pnas.2034201100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers MH, Mukherjee S, Guo S, Zhang S, Kobayashi T, Schoonmaker JA, Ebert BL, Al-Shahrour F, Hasserjian RP, Scadden EO, et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukaemia. Nature. 2010;464:852–857. doi: 10.1038/nature08851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Pietras E, Barry-Holson K, Mir A, Binnewies M, Jeanne M, Sala-Torra O, Radich JP, Passegué E. IL-6 controls leukemic multipotent progenitor cell fate and contributes to chronic myelogenous leukemia development. Cancer cell. 2011;20:661–673. doi: 10.1016/j.ccr.2011.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida M, Schepers K, King B, Sabnis AJ, Forsberg EC, Attema JL, Braun BS, Passegué E. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell. 2009;15:341–352. doi: 10.1016/j.ccr.2009.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers K, Hsiao EC, Garg T, Scott MJ, Passegué E. Activated Gs signaling in osteoblastic cells alters the hematopoietic stem cell niche in mice. Blood. 2012 Aug 15; doi: 10.1182/blood-2011-11-395418. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonomoto K, Yamaoka K, Oshita K, Fukuyo S, Zhang X, Nakano K, Okada Y, Tanaka Y. IL-1β induces differentiation of human mesenchymal stem cells into osteoblasts via the Wnt5a/Ror2 pathway. Arthritis Rheum. 2012;64:3355–3363. doi: 10.1002/art.34555. [DOI] [PubMed] [Google Scholar]
- Sordi V, Malosio ML, Marchesi F, Mercalli A, Melzi R, Giordano T, Belmonte N, Ferrari G, Leone BE, Bertuzzi F, et al. Bone marrow mesenchymal stem cells express a restricted set of functionally active chemokine receptors capable of promoting migration to pancreatic islets. Blood. 2005;106:419–27. doi: 10.1182/blood-2004-09-3507. [DOI] [PubMed] [Google Scholar]
- Thiele J, Kvasnicka HM. Myelofibrosis in chronic myeloproliferative disorders--dynamics and clinical impact. Histol Histopathol. 2006;21:1367–1378. doi: 10.14670/HH-21.1367. [DOI] [PubMed] [Google Scholar]
- Vallet S, Pozzi S, Patel K, Vaghela N, Fulciniti MT, Veiby P, Hideshima T, Santo L, Cirstea D, Scadden DT, et al. A novel role for CCL3 (MIP-1α) in myeloma-induced bone disease via osteocalcin downregulation and inhibition of osteoblast function. Leukemia. 2011;25:1174–81. doi: 10.1038/leu.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Etten RA, Shannon KM. Focus on myeloproliferative diseases and myelodysplastic syndromes. Cancer Cell. 2004;6:547–552. doi: 10.1016/j.ccr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Varricchio L, Mancini A, Migliaccio AR. Pathological interactions between hematopoietic stem cells and their niche revealed by mouse models of primary myelofibrosis. Expert Rev Hematol. 2009;2:315–334. doi: 10.1586/ehm.09.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Olsen GH, Dworkin S, Fabb SA, Swann J, McArthur GA, Westmoreland SV, Chambon P, Scadden DT, Purton LE. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007a;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Shea JM, Sims NA, Purton LE, Orkin SH. Rb regulates interactions between hematopoietic stem cells and their bone marrow microenvironment. Cell. 2007b;129:1081–1095. doi: 10.1016/j.cell.2007.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler IG, Sims NA, Pettit AR, Barbier V, Nowlan B, Helwani F, Poulton IJ, van Rooijen N, Alexander KA, Raggatt LJ, et al. Bone marrow macrophages maintain hematopoietic stem cell (HSC) niches and their depletion mobilizes HSC. Blood. 2010;116:4815–4828. doi: 10.1182/blood-2009-11-253534. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
- Zhang B, Ho YW, Huang Q, Maeda T, Lin A, Lee S, Hair A, Holyoake TL, Huettner C, Bhatia R. Altered Microenvironmental Regulation of Leukemic and Normal Stem Cells in Chronic Myelogenous Leukemia. Cancer Cell. 2012;21:577–592. doi: 10.1016/j.ccr.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.