Abstract
Cystatin 9 (CST9) is a member of the type 2 cysteine protease inhibitor family, which has been shown to have immunomodulatory effects that restrain inflammation, but its functions against bacterial infections are unknown. Here, we report that purified human recombinant (r)CST9 protects against the deadly bacterium Francisella tularensis (Ft) in vitro and in vivo. Macrophages infected with the Ft human pathogen Schu 4 (S4), then given 50 pg of rCST9 exhibited significantly decreased intracellular bacterial replication and increased killing via preventing the escape of S4 from the phagosome. Further, rCST9 induced autophagy in macrophages via the regulation of the mammalian target of rapamycin (mTOR) signaling pathways. rCST9 promoted the upregulation of macrophage proteins involved in antiinflammation and antiapoptosis, while restraining proinflammatory-associated proteins. Interestingly, the viability and virulence of S4 also was decreased directly by rCST9. In a mouse model of Ft inhalation, rCST9 significantly decreased organ bacterial burden and improved survival, which was not accompanied by excessive cytokine secretion or subsequent immune cell migration. The current report is the first to show the immunomodulatory and antimicrobial functions of rCST9 against Ft. We hypothesize that the attenuation of inflammation by rCST9 may be exploited for therapeutic purposes during infection.
INTRODUCTION
Cystatin 9 (CST9) is a small, ~18-kDa human protein and a member of the type 2 cystatin superfamily that is compromised of 14 members (1). Cystatins are structurally conserved, low molecular weight endogenous cysteine protease inhibitors found in most body compartments and fluids. Characteristically, they are described to function intra- and/or extracellularly to inhibit their target cysteine enzymes (for example, cathepsins) to maintain a crucial protease-inhibitor balance, thus regulating damaging proteolytic activities (2,3). Distinct cystatins regulate specific cysteine proteases. For instance, cystatin C is typically secreted and acts as a strong inhibitor of papainlike proteases, while cystatin F primarily regulates intracellular cathepsin C (4,5). Sustaining the equilibrium between cysteine proteases and cystatins also serves to regulate immunomodulatory functions not related to their inhibition of proteases (6). Under pathophysiological conditions, dysregulated inflammatory responses can cause decreased levels of cystatins that are insufficient to regulate proteases (1,7–9). This, in turn, results in excessive cytokine production, unrestrained tissue breakdown (for example, degradation of the extracellular matrix [ECM]) and promotes excessive immune cell extravasation to inflammation sites exacerbating organ damage (1,3,7–9).
Although the biological roles of most cystatins have not yet been fully characterized, there are some data to suggest that exogenous restoration of selected cystatins can reestablish vital immunomodulatory capabilities to the host and, thereby, may exert therapeutic effects. For example, cystatin C (10,11), as well as cystatin B (8) and E/M (9) have been shown to have antitumor effects via significantly decreasing tumor proliferation and metastasis. In addition, cystatin C has been shown to be neuroprotective in neurodegenerative disorders (for example, Alzheimer’s disease) leading to its use as a potentially promising new cancer and neuroprotective therapy (12,13). Cystatin B has been shown to have antiviral properties as it decreased HIV replication in macrophages (14). Less is known regarding cystatin’s ability to modulate immune responses during bacterial infection.
The data presented in the current report demonstrate a hitherto unknown and surprising function for CST9 during Francisella tularensis (Ft) infection of the lung. The select agent Ft is a deadly gram-negative human inhalational pathogen and is the causative agent of pneumonic tularemia (15–17). Here we describe the ability of CST9 to increase the intracellular killing of Ft by macrophages (Ft’s primary target cell population) and simultaneously restrained inflammation ultimately resulting in host survival. We show that a single dose of human recombinant (r)CST9 given to S4-infected human monocyte-derived macrophages (MDM) prevents intracellular S4 from escaping the phagosome, and induces macrophage autophagy while regulating the phosphorylation events of a key kinases in the mTOR pathway that determine the induction of autophagy. Additionally, we show that rCST9 increased MDM proteins involved in anti-inflammation/antiapoptosis while restraining proinflammatory-associated proteins compared with untreated MDM. These findings were confirmed, in a murine model of pulmonary tularemia. Intranasal (i.n.) administration of rCST9 decreased bacterial burden in organs and increased survival without inducing exaggerated levels of cytokine secretion or immune cell migration. Finally, we show that rCST9 exerts antimicrobial activity by decreasing Ft viability and virulence.
MATERIALS AND METHODS
Synthesis and Purification of CST9 Protein Expression Constructs
Oligonucleotide primers (Sigma-Aldrich, St. Louis, MO, USA) based on the full-length (aa 1–159) homo sapiens cystatin 9 (GenBank accession number NM001008693) coding sequence were used for PCR amplification. PCR products derived from human genomic DNA were subsequently cloned into the bacterial protein expression plasmid pPROEX (Life Technologies, Grand Island, NY, USA) using introduced restriction enzyme sites. Sequence confirmed recombinant plasmids were transformed into E. coli BL21 DE3 competent cells (Protein Express, Cincinnati, OH, USA).
E. coli BL21 DE3 cells, transformed with CST9 expression constructs, were induced by the addition of 3 mmol/L IPTG to produce proteins as previously described (18). Recombinant proteins were eluted using a buffer containing 8 mol/L urea, 100 mmol/L NaH2PO4, 100 mmol/L Tris-HCl and 500 mmol/L imidazole (pH 8.0). Purified proteins were dialyzed overnight into 4°C PBS (Sigma-Aldrich) using slide-a-lyzer dialysis cassettes (Pierce, Waltham, MA, USA) with a molecular weight cutoff of 3.5 kDa. After dialysis, proteins were quantified using BCA (Pierce) and quantifed using SDS-PAGE electrophoresis (Bio-Rad, Hercules, CA, USA). The final protein concentration was 0.2 μg/μL as determined by the Bradford assay (Bio-Rad).
Bacterial Strains
Schu 4 was obtained from United States Army DPG, Life Sciences Division (Dugway, UT, USA); and live virus strain (LVS) (ATCC 29684) was obtained from Karen Elkins (CBER/FDA, Rockville, MD, USA). Bacterial colonies were expanded overnight in 10 mL of modified Mueller-Hinton II broth supplemented with IsoVitale X (BD, Franklin Lakes, NJ, USA) with shaking at 37°C. The overnight culture was pelleted by centrifugation and then resuspended in 10 mL of sterile phosphate-buffered saline (PBS). The bacterial concentration then was adjusted to 1 × 109 colony-forming units (CFU)/mL using a Klett photoelectric densitometer (Scienceware, Pequannock, NJ, USA) and then were diluted in sterile PBS to the desired concentrations. Bacterial ten-fold dilutions were plated on CHA plates to confirm experimental dosage.
Human MDM and Alveolar Macrophages (hAM)
Human monocytes were isolated from peripheral blood mononuclear cells obtained from healthy donors and alveolar macrophages were obtained from healthy human volunteers via bronchial alveolar lavages as per an approved UTMB IRB protocol. Monocytes were isolated using a Hypaque-Ficoll (Amersham Biosciences, Piscataway, NJ, USA) density gradient (800 × g at 21°C) followed by selection using a counterflow centrifugal elutriation (Beckman J2–21 M/E centrifuge with JE-B6 elutriator rotor, Beckman Instruments, Palo Alto, CA, USA). The purified monocytes were cultured in RPMI 1640, 10% FBS supplemented with GM-CSF (100 ng/mL) for 7 d to ensure differentiation into macrophages. The macrophages were removed from the culture plate using a nonenzymatic cell dissociation solution (Sigma-Aldrich). Differentiation of MDM was confirmed by flow cytometry of cell surface markers expression (CD11b, CD80) consistently showing purities of >95%.
Macrophage Phagocytosis and Intracellular Replication
MDM (5 × 105 cells per 200 μL) were given 50 pg of rCST9, then immediately infected with S4 (MOI 40:1). To examine phagocytosis and intracellular replication/killing of S4, at 1 h after infection, macro -phages were washed with 2 mL of PBS, then incubated for 30 min with serum-free medium containing 50 μg/mL of gentamicin (Sigma-Aldrich) to kill extra-cellular organisms. Cells were lysed by adding 0.1% SDS and lysates were plated on CHA plates and incubated at 37°C in 5% CO2 for 48 h. Single colonies were counted to quantify bacterial phagocytosis. To determine killing, additional macrophages were treated and infected in parallel. Except after the 30-min gentamicin incubation, cells were washed, then resuspended in serum-free medium containing 5 μg/mL of gentamicin. Cells were incubated overnight at 37°C/5% CO2; then cells were lysed, plated, and single colonies were counted as described above. These bacterial counts were compared with counts taken immediately following the 1-h time points to determine phagocytosis.
Intranasal Mouse Model of Treatment and Infection
Eight-week old female Balb/c mice weighing between 21 and 24 grams (Jackson Laboratories) were housed in an Association for Assessment and Accreditation for Laboratory Animal Care (AAALAC)-approved housing facility and permitted to adjust to their environment for 7 d prior to procedures receiving free access to food and water throughout the study. All procedures were approved by the University of Texas Medical Branch IACUC and performed humanely with minimal suffering. The animals were anesthetized with sodium pentobarbital and restrained vertically using sterilized commercial fishing line looped behind the upper incisors and connected to a support platform. Ten microliters of 50 pg of rCST9 were placed at the anterior of each nares (10 μL/nare) concurrent with 500 CFU/mouse of LVS and the animals inhaled the solutions naturally. Survival (n = 12 mice/group) was observed for up to 25 d after treatment and infection.
Bacterial Burden in Mouse Tissues and Lung Histology
The lungs, liver and spleen (n = 6 mice/group) were harvested at 48 h after treatment from rCST9-treated and/or LVS (500 CFU/mouse)–infected mice. A small section of the lungs from three mice was fixed and processed for hematoxylin and eosin (H & E) staining to determine the overall condition of the lungs. Tissues were homogenized, plated on CHA, then incubated at 37°C/5% CO2 for 48 h. Individual colonies were counted to determine CFU.
Cytokine Analysis
Cell culture supernatants and mouse BAL were analyzed by a species appropriate (mouse or human) enzyme-linked immunosorbent assay (ELISA) (Endogen, Pierce) to quantify secreted cytokines at designated times after rCST9 treatment and/or infection.
2D Gel Analysis
S4 or MDM were incubated for 4 h at 37°C/5% CO2 in presence or absence of CST9. The 2D gel analysis was performed as previously described (19). Briefly, the bacteria were lysed using lysis buffer (2 mol/L thiourea, 7 mol/L urea, 4% 3-[(3-cholamidopropyl) dimethylammonio] propanesulfonate, 2% dithiothreitol) supplemented with protein inhibitors. The immobilized pH gradient (IPG) strips (pH: 3–10, Bio-Rad) were rehydrated at 50 V for 12 h with 250 μL buffer consisting of 1 mmol/L thiourea, 8 mmol/L urea, 2% CHAPS, 1% dithiothreitol, and 0.2% ampholytes containing 100 μg protein sample and 0.002% bromophenol blue. Isoelectric focusing on the strips was performed at 500 V for 1 h, 1,000 V for 1 h, 8,000 V for 2 h, and then 8,000 V for a total of 50,000 V. The IPG strips then were suspended in equilibration buffer (50 mmol/L Tris-HCl, pH 6.8, 6 mmol/L urea, 20% glycerol) and incubated for 15 min each with 2% dithiothereitol/2% SDS (alkylating conditions) and 2.5% iodoacetamide/2% SDS (alkylating conditions). Followed by equilibrated strips being subjected to second-dimension electrophoresis using 8% to 10% linear gradient precast Tris-HCl gels (Bio-Rad) on a PROTEAN plus Dodeca Cell System at 75 V for 1 h and then at 120 V. Gels were fixed (10% methanol/7% acetic acid), then stained with SYPRO Ruby (Bio-Rad). Destaining of the gels was accomplished by using 10% ethanol and imaged by Separations Technology Core part of Biomolecular Resource Facility at UTMB, using high-resolution Typhoon Trio (GE Health-care) imaging system. The comparative analysis on different gels was done using ImageMaster 2D Platinum 7.0 software (GE Healthcare). Proteins spots showing the most differences in area and intensity were selected to be identified by MS analysis as performed by UTMB Proteomics Core Facility and the Biomolecule Analysis Core Facility at UTEP (20).
Bacterial Viability Assays
Live S4 (1 × 104 CFU/mL) was incubated with 50 pg of rCST9 for 4 h at 37°C. S4 alone was used as a control. One hundred microliters of each sample was plated on CHA plates and incubated for 48 h; then colonies were counted to determine the affect rCST9 had on bacterial viability. Further, a portion (200 μL) of the S4 organisms preincubated rCST9 were added to 5 × 105 hMDM/sample to determine intracellular killing/replication following a 6-h incubation via CFU. Additionally, S4 was incubated with rCST9 (50 pg) for 4 h; then Balb/c mice were challenged i.n. with LVS (500 CFU/mouse), and survival was observed.
Fluorescent Microscopy
MDM and hAM grown on coverslips, then given 50 pg of rCST9 and/or S4 (MOI 40:1). As previously described (21), cells were fixed with 2% formaldehyde for 5 min and then permeabilized with 0.2% Triton × 100 for 7 min. If applicable, coverslips were blocked for 30 min with 10% nonfat dried milk, then incubated overnight with anti-LC3-I and LC3-II antibody (Novus Biologicals, Littleton, CO, USA) to label autophagy markers. Coverslips were washed, then incubated with the appropriate secondary antibody conjugated to a fluorescent tag (Life Technologies) for 2 h. After final washes, the coverslips were mounted with Vectashield Mounting Solution (Vector Laboratories, Burlingame, CA, USA) containing DAPI to stain the cell nuclei side up on a microscope slide and visually examined using a Nikon Eclipse Ti immunofluorescent microscope (Nikon Instruments, Melville, NY, USA).
Transmission Electron Microscopy (TEM)
S4 was incubated with 50 pg of rCST9 for 4 h; or MDM were exposed to S4 (MOI 40:1) and/or rCST9 for 2 or 20 h; then samples were processed for TEM as previously described (19). Briefly, hMDM or S4 were fixed in a mixture of 2.5% formaldehyde and 0.1% glutaraldehyde in 0.05 mmol/L cacodylate buffer (pH 7.2) containing 0.03% trinitrophenol and 0.03% CaCl2, washed in 0.1 mmol/L cacodylate buffer (pH 7.2), and postfixed in 1% OsO4 in the same buffer. Treated and/or infected hMDM were stained en bloc with 1% uranyl acetate in 0.1 mmol/L maleate buffer, dehydrated in ethanol and embedded in Poly/Bed 812 epoxy resin (Polysciences, Warrington, PA, USA). Sections were cut on an Ultracut S ultramicrotome (Reichert; Leica Microsystems GmbH, Wetzlar, Germany), then stained with 2% aqueous uranyl acetate and lead citrate and examined with a Philips 201 or CM 100 electron microscope at 60 kV.
Migration Assay
A modified Boyden chamber assay (Cell Biolabs Inc., San Diego, CA, USA) cell migration assay was used as described previously (22). Briefly, rCST9 (50 pg) in the presence or absence of human vascular endothelial growth factor (VEGF) was added to the lower chamber and lymphocytes were added to the upper chamber to determine immune cell migration through the membrane. Following a 4-h incubation, the membrane between the two compartments was fixed and stained, and the cells that migrated to the lower side of the membrane was quantified.
Western Blot Analysis
Western blots were performed as we described previously (23). Briefly, MDM were given 50 pg of rCST9 and/or S4 (MOI 40:1). MDM alone served as negative controls. Total cell protein was collected at selected times after treatment and/or infection. Cell lysates were analyzed for the amount of protein in each sample by the Bradford assay (Bio-Rad, Hercules, CA, USA), then boiled in loading buffer (4% SDS, 20% glycerol, 125 mmol/L Tris-HCl [pH 6.8], and 10% 2-mercaptoethanol); then 20 μg of protein was loaded on an 8% to 16% Tris-glycine gradient gel (Novex, San Diego, CA, USA). Electrophoresed proteins were transferred to a nitrocellulose membrane (Novex) and membranes were blocked with 10% nonfat dried milk for 30 min prior to incubation with rabbit polyclonal anti-LC3-I and LC3-II (Novus Biologicals) at 2 ug/mL, 1:1000 rabbit monoclonal anti-P70S6 kinase (K), anti-phospho-P70S6 kinase (Thr389), or anti-phospho-inositide kinase-3 (PI3-K) class III (C3) (Cell Signaling Technology, Danvers, MA, USA) overnight. Blots were washed, followed by the addition of peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma-Aldrich) at a dilution of 1:10,000 for 3 h. Blots were washed and then incubated for 1 min in enhanced chemiluminescence reagents (ECL kit; Amersham, Little Chalfont, Buckinghamshire, England). Processed blots were placed on X-ray film (Kodak, Rochester, NY, USA) for empirically optimized exposures.
Statistical Analyses
Where applicable, numerical results are reported as mean ± SEM of two-to-three independent experiments. Analysis of numerical data was determined by one-way analysis of variance (ANOVA) and Student t test using Prism software (Graph Pad, San Diego, CA, USA). Survival data were analyzed by log-rank analyses with Welch corrections using Prism software (v 4.0 GraphPad). Differences were considered statistically significant when the P value was <0.05.
RESULTS
rCST9 Modulates Macrophage Responses to Ft
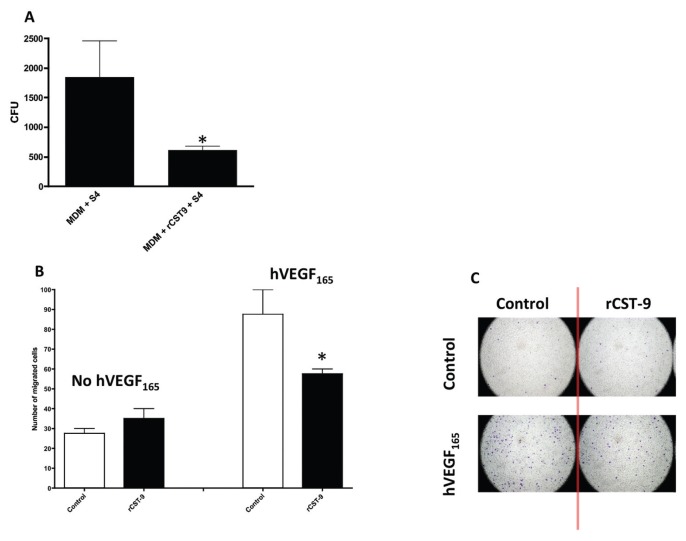
We accessed the ability of rCST9 to modulate macrophage responses to Ft. MDM were treated with an approximate physiological dose (24,25) of our human rCST9 protein (50 pg), then immediately infected with S4 (MOI 40:1). At 24 h after treatment and/or infection, rCST9 significantly increased intra-macrophage killing of S4 compared with S4-infected MDM alone (Figure 1A). Interestingly, S4 killing by MDM was not accompanied by marked levels of cytokine secretion (for example, IL-8, IL-6, IFN-γ, IL-1β, TNF-α; data not shown). Correlating with these results, we found that rCST9 significantly decreased leukocyte migration through the endothelium in the presence of VEGF compared with VEGF alone (Figures 1A, 1B). VEGF is a powerful chemoattractant for immune cells (26).
Figure 1.
rCST9 modulates MDM responses Ft S4. (A) MDM (5 × 105 in 200 μL media/sample) were treated concurrently with 50 pg of rCST9 then immediately infected with S4 (MOI of 40:1). At 24 h after infection and/or treatment, bacterial killing was significantly higher in rCST9-treated MDM, as compared with S4-infected MDM alone (P < 0.05). (B) rCST9 significantly decreased leukocyte migration in the presence of VEGF compared with VEGF alone (P < 0.05), as shown in a modified Boyden chamber assay. (C) Cells were stained to visualize and quantify migrated cells in the modified Boyden chamber assay. Data are presented as mean ± SEM. Asterisks signify significant differences of P < 0.05.
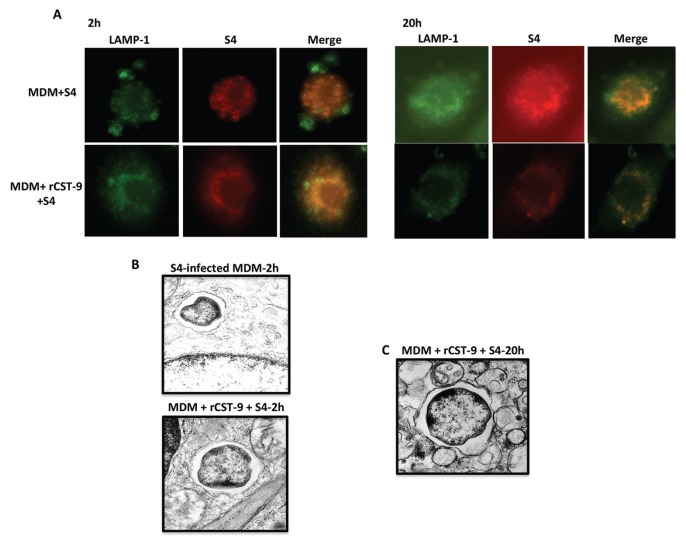
These findings indicate that rCST9 is not inducing conventional innate immune responses. Therefore, we next addressed the mechanisms by which this protein induces intramacrophage killing of S4. It has been reported that Ft must escape the phagosome of macrophages to replicate in the cytosol (27,28). Thus, the fate of intra-cellular S4 in rCST9-treated macro phages evaluated whereby MDM were simultaneously given 50 pg of CST9 and S4 (MOI 40:1) or S4 alone. Following a 2- or 20-h incubation period, the intracellular localization and replication of Ft was examined via fluorescence microscopy. The results showed that S4 (red) colocalized (yellow) with the lysosome associated membrane protein marker 1 (LAMP)-1 (green) at the early stages (2 h) of infection in all tested conditions (Figure 2A). However, at 20 h after infection, S4-infected MDM alone showed minimal colocalization with LAMP-1 with extensive cytosolic replication as indicated by the dispersed red labeling of S4 throughout the cell consistent with phagosomal escape (see Figure 2A). Alternatively, in rCST9-treated MDM, intracellular S4 was either cleared (as indicated by the diminishing red stain of S4) (see Figure 2A) or remained colocalized to LAMP-1 with minimal cytosolic replication at 20 h after infection compared with the 2-h time point of these same experimental conditions and S4-infected MDM controls (see Figure 2A).
Figure 2.
rCST9 confined S4 to MDM phagosome and decreased intracellular replication. rCST9 (50 pg) treated MDM (105) were infected with S4 (MOI 40:1). S4-infected MDM alone served as controls. (A) All tested conditions show that the majority of S4 (red) colocalized with LAMP-1 (green) at 2 h. At 20 h, S4 showed minimal LAMP-1 colocalization and increased cytosolic replication in infected MDM alone. However, the majority of S4 remained colocalized with LAMP-1 with minimal cytosolic replication in MDM treated with rCST9. (B) TEM analysis showed S4 in phagosomes at 2 h after infection with or without rCST9 treatment. (C) However, S4 remained in the phagosomes of MDM at 24 h after infection and/or rCST9 treatment. The images are representative of at least three replicate studies.
We further confirmed that rCST9 decreased the escape of Ft from the phagosomal macrophages as determined by TEM, which showed S4-containing phagosomes at 2 h after infection in rCST9-treated MDM and S4-infected MDM alone (Figure 2B). By 20 h after infection, in rCST9-treated MDM, the majority of S4 remained in phagosomes (see Figure 2B) with few cytosolic bacteria.
Together these data show that rCST9 decreased phagosomal escape and subsequent cytosolic replication of S4. This is one possible mechanism by which rCST9 induced intramacrophage killing of S4 that was independent of exaggerated cytokine secretion.
rCST9 Induced Autophagy in Macrophages
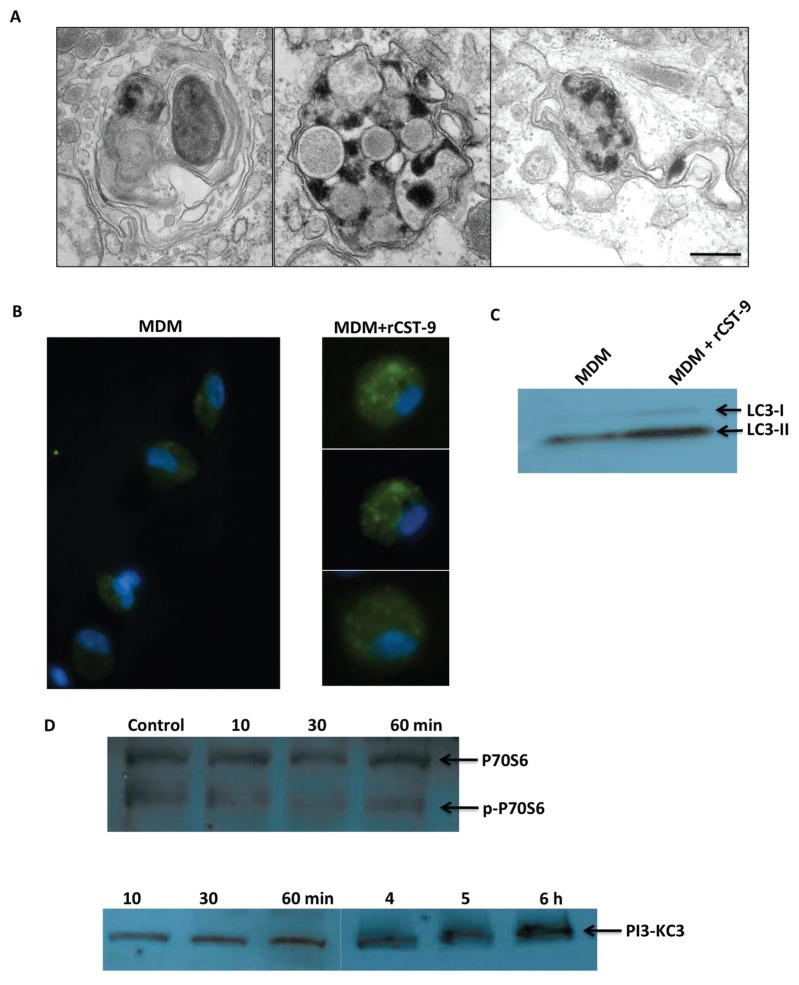
In addition to confining S4 to the phagosome, TEMs also showed evidence of autophagy in S4-infected MDM treated with rCST9 (Figure 3A) as indicated by the double membrane autophagic vacuoles (AV) compared with no AVs observed in S4-infected MDM alone. To confirm rCST9-induced autophagy, we treated primary human alveolar macrophages (hAM) with 50 pg of rCST9 overnight, then stained for LC3-1 and LC3-II. LC3 is a specific marker for AVs and is present as LC3-I under normal circumstances. However, during certain pathophysiologic conditions, LC3-I is cleaved into the AV membrane-associated LC3-II, which appears as small spots within the cell. Immunofluorescent microscopy results showed that rCST9 induced LC3-II labeled AV in hAM compared with untreated hAM (Figure 3B). Further, Western blot analysis showed a marked increase in the conversion of LC3-I to LC3-II in MDM treated with rCST9 compared with MDM alone (Figure 3C).
Figure 3.
rCST9 induced autophagy. (A) MDM showed evidence of autophagy (that is, double membrane AV) in MDM infected with S4 (MOI 40:1) at 20 h after rCST9 (50 pg) treatment and S4 infection as per TEM analysis. (B) Immunofluorescent microscopy showed LC3-II protein (small green spots) in human alveolar macrophages at 20 h after rCST9 (50 pg) treatment compared with untreated macrophages. Images shown in A and B are representative of experiments repeated three times using duplicate conditions. (C) Western blot analysis revealed marked conversion of LC3-I to LC3-II in rCST9 (50 pg) treated MDM compared with MDM alone. (D) rCST9 (50 pg) induced the dephosphorylation of P70S6 kinase at 30 and 60 min after treatment. PI3-KC3 phosphorylation of was detected at 10, 30 and 60 min as well as 4, 5 and 6 h in rCST9-treated MDM. Experiments were performed three times.
Because autophagy was induced in macrophages by rCST9, we sought to determine if rCST9 modulates the mTORC1 signaling pathway, which is a negative regulator of autophagy. The phosphorylation of P70S6 kinase is a key indicator of mTOR complex 1 (C1) pathway activation. Alternatively, the activation of class III PI3-K induces autophagy, indicating mTOR complex 2 (C2) activity. Therefore, we treated MDM with 50 pg of rCST9 for 10, 30 or 60 min or 4, 5 or 6 h. MDM alone served as negative controls. Total proteins from whole cell lysates were analyzed for P70S6 and p-P70S6 by Western blot. Results showed that rCST9 induced the dephosphorylation of P70S6 kinase overtime in MDM thus inhibiting mTORC1 signaling pathway while increasing P70S6 kinase levels (Figure 3D). Further promoting autophagy, rCST9 induced the phosphorylation of PI3-KC3 by 30 min after treatment that was detectable up to 6 h after treatment (see Figure 3D).
On the basis of these results, rCST9 induced autophagy, inhibited mTORC1, but induced mTORC2 pathway activation in primary macrophages.
rCST9 Demonstrated Antimicrobial Activity and Induced Antibacterial Resistance in MDM
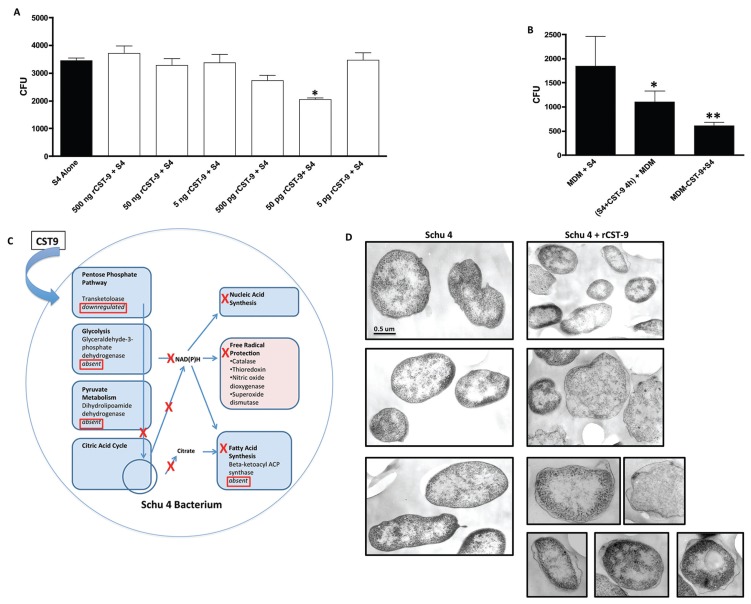
Because the rCST9 induced intra-macrophage killing of S4 that did not induce inflammatory responses, we hypothesized that rCST9 also may affect S4 directly. To test this hypothesis, S4 (104 CFU/sample) was cultured with various doses of rCST9 for 4 h and then plated to quantify viability. Results showed that rCST9 significantly decreased the replication of S4 in a dose-dependent manner establishing 50 pg of rCST9 as the most effective dose (P < 0.05; Figure 4A). A portion of the S4 incubated with rCST9 (50 pg) was washed to remove the bacterial broth, resuspended in RPMI media, then added to MDM (MOI 40:1) to determine intramacrophage killing. The results showed that MDM incubated with rCST9 killed S4 significantly better than infected MDM alone, but not as effectively as MDM given rCST9 and S4 concurrently (P < 0.05 or 0.01 respectively; Figure 4B).
Figure 4.
rCST9 directly affected protein changes in S4 and MDM. (A) S4 (1 × 104 CFU/sample) was incubated with various dilutions rCST9 for 4 h, then plated to determine bacteria viability via replication. 50 pg of rCST9 significantly decreased S4 viability compared with all other tested dilutions (P < 0.05). (B) A portion of the S4 incubated with 50 pg of rCST9 was given to MDM (MOI 40:1) where it was killed significantly better (P < 0.05) than infected MDM alone but not as effectively as rCST9 and S4 given concurrently to MDM. (C) The schematic showing that MS analysis identified numerous proteins involved in bacterial metabolic pathway that were either downregulated or eliminated by CST9. (D) TEM analysis of untreated S4 revealed an intact, uninterrupted cell wall. However, the membranes of S4 incubated with 50 pg of rCST9 for 4 h had the periplasmic space disrupted, and in some areas, the bacterial membrane was discontinuous. Where applicable data are presented as mean ± SEM and asterisk signifies significant differences of *P < 0.05 or **P < 0.01 as indicated. Scale bar, 0.5 μm.
Because rCST9 decreased S4 viability, we sought to determine how rCST9 was directly affecting S4. Therefore, we incubated S4 with medium containing CST9 for 4 h and determined S4 protein changes by 2-DE gel analysis untreated. S4 alone served as a negative control. The gels were analyzed via computerized comparison to identify the maximum differences in spot area (mm2) and intensity, and then selected spots were analyzed by MS. The most significant changes in spot intensity with high peptide counts are reported herein. S4 incubated with CST9 markedly upregulated the bacterial cell wall enzyme, d-alanyl-d-alanine carboxypeptidase (29,30) (Table 1) that has been shown to render gram-negative bacteria more susceptible to killing by compromising the cell wall (29). CST9 also eliminated or markedly decreased dihydrolipoamide dehydrogenase (pyruvate metabolism), β-ketoacyl-ACP synthase (fatty acid synthesis), aldo/keto reductase (enzymes that utilize NADPH) and GAPDH (glycolysis) (Table 1). These results were placed into a functional pathway format (Ingenuity Pathways Analysis) to map the disruptive effects of the CST9 on S4 metabolic pathways (Figure 4C). Further confirming the direct effects of rCST9 on S4, TEM analysis revealed intact, uninterrupted cell walls while S4 incubated with rCST9 displayed disruption of the periplasm and loss of cell wall integrity as seen by damaged, discontinuous cell walls (Figure 4D). Collectively, these data demonstrate that rCST9 decreased S4’s viability and virulence via directly modulating proteins involved in S4 metabolic pathway and cell wall synthesis.
Table 1.
S4 protein changes induced by rCST9.
| Spot no. | Protein name | Accession no. | Peptide count | Protein score |
|---|---|---|---|---|
| 3 | Dihydrolipoamide hydrogenase | gi118498063 | 14 | 724 |
| 4 | β-ketoacyl-ACP synthase | gi87931425 | 8 | 617 |
| 5 | Aldo/keto reductase | gi118497417 | 12 | 548 |
| 6 | GAPDH | gi282159610 | 11 | 543 |
| 7 | 30S ribosomal protein S2 | gi118496840 | 16 | 375 |
| 8 | Transketolase | gi187931433 | 9 | 347 |
| 11 | Sigma-54 modulation protein | gi89256500 | 8 | 522 |
| 12 | Peptidly-prolyl cis-trans isomerase | gi254367995 | 4 | 250 |
| 13 | d-alanyl-d-alanine carboxylpeptidase | gi118497499 | 16 | 807 |
Because the treatment of MDM with rCST9 significantly enhanced S4 killing, we next investigated protein changes in rCST9-treated MDM after 4 h of incubation. Proteomics analysis was employed as described above. There was an upregulation of various multifaceted proteins in rCST9-treated MDM such as those involved in mediating cytoskeletal actin organization (31) as well as phagosome maturation (32) (that is, cofilin, profilin, myosin light chain kinase, calmodulin, GADPH) and regulation of phagocytosis (macrophage capping protein) compared with untreated MDM (Table 2). There was also an upregulation of proteins in rCST9-treated MDM that have been shown to temper inflammation that is, glutaredoxin-1, whose expression is key in the activation of alveolar macrophages by regulating the production of inflammatory mediators through control of S-glutathionylation–sensitive signaling pathways such as NF-κB (33). Further, we observed a moderate upregulation of vimentin in rCST9-treated MDM, which can suppress the production of reactive oxygen intermediates (ROIs) (34) but is also involved in wound healing and remodeling during pulmonary injury (35). Alternatively, S100-A10 and cathepsin B proteins were downregulated in rCST9-treated MDM (Table 2). The up-regulation of these two proteins has been associated with pathogenic inflammation and various disease processes.
Table 2.
MDM protein changes induced by rCST9.
| Spot no. | Proteins upregulated by rCST9 | Accession no. | Protein score |
|---|---|---|---|
| 6 | Macrophage capping protein | P40121 | 792 |
| 10 | GAPDH | P04406 | 629 |
| 12 | Calmodulin | P62158 | 158 |
| 13 | Myosin light chain kinase | P60660 | 315 |
| 28 | Cofilin | P23528 | 158 |
| 29 | Profilin | P07737 | 397 |
| 30 | Glutaredoxin-1 | P35754 | 250 |
| 31 | Vimentin | P08670 | 250 |
|
| |||
| Proteins downregulated by rCST9 | |||
|
| |||
| 19 | S100-A10 | P60903 | 0.997 |
| 27 | Cathepsin B | P07858 | 0.02 |
These data show that rCST9 has direct, multifaceted effects on both the macrophages and S4. Thus, rCST9 serves to divert MDM defenses away from potentially damaging inflammation while weakening the virulence of S4.
rCST9 Protections against Pulmonary Tularemia
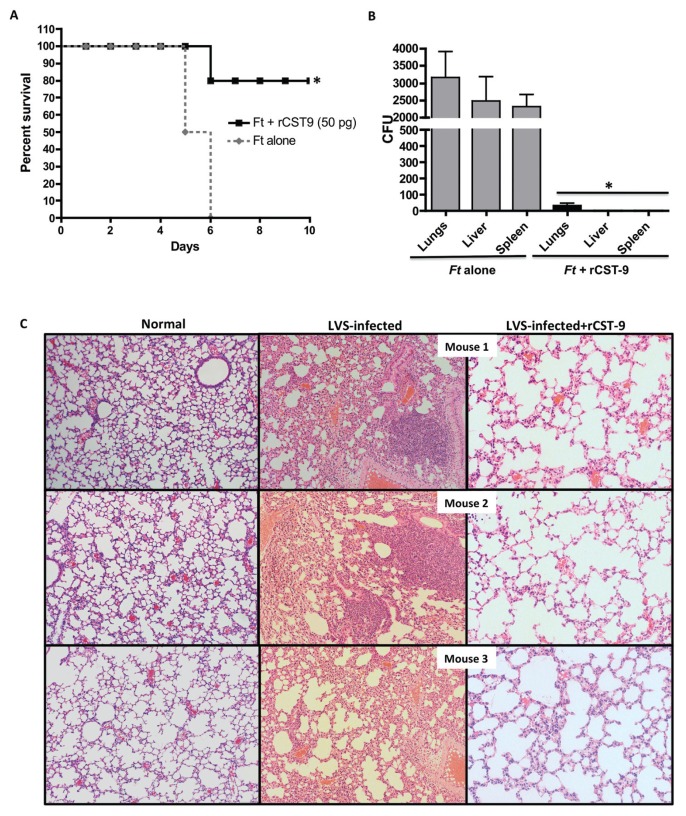
Because rCST9 directly decreased Ft viability and induced intramacrophage killing, we investigated rCST9’s ability to improve survival in a Ft LVS model of inhalation. Note that the lethality of our LVS in a mouse model of inhalation is nearly equivalent to S4, in that death is eminent by d 8 after i.n. challenge with as few as 50 LVS organisms/mouse (36). Therefore, we used LVS in our inhalation mouse model. Mice were i.n. infected with 500 CFU of LVS, then given 50 pg of rCST9/mouse via the same route. Mock-treated and infected mice were used as controls. Survival rates of mice given rCST9 and LVS had significantly higher survival (80%) compared with infected mice alone, which succumbed to the infection by d 6 (Figure 5A). LVS-infected mice became increasingly hunched and lethargic with unkempt fur by d 4 after infection. Conversely, rCST9-treated/infected mice were active and groomed throughout the study with no visible signs of illness. To this end, rCST9-treated mice which cleared the infection as survivors were euthanized at 25 d after treatment and/or infection and no LVS was detected in the lungs, liver or spleen via CFU quantification (data not shown).
Figure 5.
rCST9 afforded protection against pulmonary tularemia. (A) Balb/c mice (n = 12 mice/group) were given 50 pg of rCST9 i.n. concurrently with LVS (500 CFU/mouse; 100 × LD50s). rCST9-treated/infected mice had significantly higher survival compared with infected mice alone (P < 0.01). Data are presented as mean ± SEM. (B) Parallel groups of mice (n = 6/group) were euthanized 48 h after treatment and/or infection to collect lungs, liver and spleen to determine LVS burden. Bacterial loads in all three organs of rCST9-treated/infected mice were significantly lower compared with infected mice alone (P < 0.01). (C) Ft-infected mice treated with rCST9 showed moderate cellularity and maintained the architecture of the alveolar space similar to uninfected/untreated (normal) controls. LVS-infected mice alone showed a loss of alveolar architecture and hypercellularity. Magnification, 10×.
Additional groups of mice were treated and infected as described above but were euthanized at 48 h after infection to determine lung histology, cytokine secretion in the lungs, dendritic cell and neutrophil migration to the lung and bacterial burden in the lungs, liver and spleen. Cytokine secretion and subsequent dendritic cell and neutrophil migration was minimal in rCST9-treated/infected mice (data not shown). However, the bacterial burden in the lungs, liver and spleen was significantly lower in rCST9-treated/infected mice at 48 h compared with infected mice alone (Figure 5B; P < 0.05). Correlating with decreased bacterial burden, the lung histology of rCST9-treated/infected mice maintained the architecture of the alveolar space similar to uninfected/untreated control lungs with minimal to modest cellularity (Figure 5C). Conversely, there was marked disruption of alveolar architecture likely caused by hypercellularity in the lungs of LVS-infected mice (see Figure 5C).
DISCUSSION
Unique and unexpected characteristics of several cystatins have started to emerge to show that these cysteine protease inhibitors function outside its inhibitor role. Our study showed that a single dose of exogenous rCST9 plays a novel, multifaceted role in the protection against pulmonary tularemia that did not include the induction of conventional innate immune responses. We found that rCST9 enhanced antimicrobial resistance in macrophages, directly and indirectly modulated inflammation as a result of infection in vitro and in vivo, and decreased Ft viability and virulence.
Cystatins are naturally occurring proteins, however, decreased levels of cystatins and increased activity of cysteine proteases are observed in many disease conditions such as Alzheimer’s disease (10–13), organ failure (2,8,9) and cancer (7–9,24,37). Activated macrophages release a variety of cysteine proteases that can result in inflammatory cytokine secretion, degradation and destruction of the extracellular matrix (ECM) allowing for excessive immune cell infiltration that results in tissue damage. Cystatins selectively regulate particular cysteine proteases, thus stabilizing the ECM, resulting in the direct and indirect restraint of inflammatory responses. Therefore, selected cystatins are now being used as biomarkers to determine the disease severity and to predict patient outcomes (7,8,38–41). Subsequently, the replenishment of selected cystatins has been shown to be a promising treatment, specifically in certain types of cancer, to regulate tissue-damaging inflammation and maintain the integrity of the endothelium to prevent tumor metastasis (2,7,9,37). However, little is known about the role of cystatins during bacterial infection. Therefore, to our knowledge, we are the first to show that rCST9 protected against a pulmonary Ft infection.
Ft’s ability to evade host recognition by delaying the upregulation of inflammatory mediators during the first 24 h after infection following by unrestrained systemic inflammation contributes to its status as one of the deadliest human pathogens (15). This delay in host recognition is not well defined, but gives the pathogen opportunity to invade alveolar macrophages and then to replicate to high numbers. Ft then disseminates out of the lungs, resulting in the development of severe sepsis as characterized by deregulated inflammation (42). Our findings showed that rCST9 prevented Ft from escaping the phagosome of macrophages subsequently reducing cytosolic replication, which is lethal to the pathogen (43–45). It is reasonable to hypothesize that the lack of cytosolic Ft replication in rCST9-treated MDM likely prohibited the activation of the cysteine protease caspase-1 within the inflammasome that results in inflammatory cytokine release, including IL-1β, and cellular apoptosis (46–48). Our working hypothesis, therefore, is that inhibiting cytosolic S4 replication via confining the pathogen to phagosomes may explain why we did not detect substantial cytokine secretion from rCST9-treated macrophages or in the lungs of Ft LVS-infected mice. This would result in rCST9 having an indirect modulatory effect on inflammation and the subsequent reduction in immune cell migration to the lungs in vivo. However, rCST9 did directly modulate cell migration in vitro while in the presence of the strong cell migration chemoattractant, VEGF. Although a certain level of inflammation is required for protection against infections, failure to control the extent and intensity of the inflammatory cascade can result in immunopathology that damages the host and worsens an infection and/or disease. Similar to our findings, other studies have shown that muted or restrained immune responses enhanced the host’s ability to successfully fight against invading pathogens. For instance, Malik et al. (49) reported that cysteine proteinase knockout mice (specifically matrix metallopeptidase-9) resolved a pulmonary S4 infection, which correlated with decreased proinflammatory cytokines production, the diminishment of neutrophil infiltration and reduction of bacterial burden in tissues, (49). Likewise, Bosio et al. (50) showed that excessive neutrophil infiltration in B-cell-deficient mice infected with Ft was detrimental and induced extensive tissue damage.
While examining the intracellular fate of Ft S4 via TEM, we observed a number of double membrane AV in rCST9-treated/infected macrophages that suggested the development of autophagy. Regulated autophagy usually is associated with cell survival, cell cycle progression, metabolism and apoptosis suppression (51–53). More specifically, autophagy is a cellular process mediated by PI3-KC3, which shuttles cytoplasmic content, such as damaged organelles, to lysosomes for degradation and rids the cell protein aggregates that leads to apoptosis (51). We were able to visualize AV-associated LC3-II protein by immunofluorescent microscopy in rCST9-treated human alveolar macrophages, which was confirmed by the conversion levels of LC3-I to LC3-II. rCST9 induced autophagy in macrophages by the activation of PI3-KC3 while promoting the dephosphorylation of a key mTORC1 kinase, P70S6, that is known to inhibit autophagy. PI3-KC3 positively regulates autophagy in addition to functions in cell trafficking, endocytic pathway activation and cytoskeletal changes, which occurs through the mTORC2 pathway (54,55). Alternatively, the phosphorylation of P70S6 kinase is a key activator of the mTORC1 that is a negative regulator of autophagy but positive regulator of apoptosis (13,55). Our findings are similar to a study showing that cystatin C is neuroprotective by the induction of autophagy in murine primary cortical neurons and neuronal cell lines via inhibition of the mTOR signaling pathway (13).
To further delineate the immunomodulatory effects of rCST9 on macrophages, proteomic analysis identified proteins that were differentially expressed by rCST9-treated macrophages. We found that rCST9 upregulated actin dynamic proteins, including cofilin and profilin, that are associated with cytoskeletal stabilization, antiapoptosis and cell survival and play an important role in the regulation of the actin coat of phagosomes (32). Moreover, actin dynamics have been linked to the activation of PI3-KC3, resulting in the induction mTORC2 pathway (13,54,55).
Additional evidence of rCST9 diverting macrophage from potentially harmful inflammation responses was the upregulation of glutaredoxin-1. This protein indirectly controls NF-κB signaling pathways, which regulate the production of inflammatory mediators (33). Interestingly, there was a moderate but notable upregulation of vimentin in rCST9-treated MDM. Vimentin is another multi-faceted protein that, among its various functions, has been shown to suppress the production of ROIs, thus attenuating their organ-damaging effects as seen during sepsis and septic shock (34,35). Conversely, rCST9 downregulated S100-A10, a protein also known as an alarmin. Upon release of S100-A10 from macro -phages and dying cells, alarmins act as chemoattractants and immune activators. During pathological circumstances, such as Ft-induced sepsis, alarmins can stimulate immune cells to secrete excessive levels of cytokines and ROIs, turning advantageous immune responses into damaging inflammation (42). Sharma et al. (42) found that the upregulation of alarmins in systemic organs correlated with the progression of Ft-induced sepsis. Similarly, rCST9 downregulated another protein associated with proinflammatory responses known as the cysteine protease, cathepsin B, which is normally located in cell lysosomes where it is tightly regulated by specific cystatins (2,5,6). Among many functions, cathepsin B degrades proteins shuttled to the lysosome and activates procollagenase leading to collagen degradation (56). Cathepsin B is found in abundance in activated macrophages where it is involved in apoptosis and inflammation (56). Consequently, significant upregulation of this proteolytic enzyme plays a significant role in many diseases, including cancer, bronchial disease and arthritis (56). A recent study showed that cathepsin B significantly upregulated HIV-infected macrophages and inhibited cystatin B and C (14). Therefore, maintaining a cysteine protease-cystatin balance is crucial for normal cell function and preservation, as well as protecting cells during pathophysiological conditions.
The modulation abilities of rCST9 extended from the host to the pathogen directly whereby CST9 demonstrated an antibacterial activity that resulted in significant retardation of S4 growth/viability. Proteomic analysis showed that numerous key participants in bacterial metabolic pathways were decreased and/or eliminated in S4 exposed to CST9. In contrast, and most interesting, was the marked upregulation of a bacterial cell wall enzyme known as d-alanyl-d-alanine carboxypeptidase (29,30). The synthesis of bacterial cell walls, including glycopeptides and antigens, consists of complex mechanisms involving as many as 30 to 50 enzymes depending on the organism (29). The final steps in cell wall synthesis are catalyzed by a glycopeptide transpeptidase and a d-alanine carboxypeptidase. Excess expression of d-alanyl-d-alanine carboxypeptidase interferes with cell wall synthesis and makes the bacteria more susceptible to killing. As a result, they are the targets of various antibiotics such as vancomycin and penicillin (29,30). To this end, we found that rCST9 caused disruption of the cell wall and periplasmic space of S4. The periplasmic space of gram-negative bacteria is known to contain proteins that participate in acquiring nutrients for bacterial survival, enzymes involved in cell wall synthesis and proteins that modify lethal compounds that could harm the bacteria. Thus, it is conceivable that the overexpression of d-alanyl-d-alanine carboxypeptidase, disruption of the cell wall, as well as inhibiting S4 metabolic pathway proteins, contributed to the killing of the pathogen by macrophages and in vivo. Our findings are consistent with a recent report showing that recombinant cystatin 8 substantially decreased E. coli CFU in an effect that was dose- and time-dependent (57). Investigation of the antibacterial mechanism of cystatin 8 found that it killed E. coli by permeabilizing and disrupting its membrane, while also inhibiting macro-molecular synthesis to block bacterial growth (57).
A striking observation from the current set of studies was the finding that rCST9 affords significant protection to mice with pulmonary tularemia. Correlating with our macrophage data, the induction of a vigorous immune response was not detected in Ft-infected mice receiving rCST9, but a significant decrease in bacterial burden in the lungs and distal organs was observed. Moreover, rCST9 treatment preserved the alveolar architecture with moderate cellularity compared with the hypercellularity observed in the lungs of infected mice. Therefore, taking into consideration our macrophage findings, we conclude that exogenous rCST9 functioned in a multifold fashion to protect against inhaled Ft by (1) decreasing Ft virulence and preventing the pathogen’s escape from the macro phage phagosomes, (2) rendering the pathogen more susceptible to killing, (3) restraining potentially tissue-damaging cytokines, (4) deceasing immune cell infiltration, (5) decreasing dissemination to distal organs and (6) increasing host survival.
CONCLUSION
In summary, our study provides the first evidence showing the multilevels of protection provided by rCST9 against a pulmonary Ft infection. It is evident from our findings that the precise mechanism and function of rCST9 is complex and requires further investigation currently underway in our laboratory. However, our study has revealed novel immunomodulatory functions of rCST9 in vitro and in vivo during an Ft infection. The complexity of the immune system is associated with the convergence of numerous proteins and signaling pathways that can result in dysregulated inflammation as seen in Ft infections and other invading pathogens. Exogenous rCST9 functioned to target multiple immune mediators effective against Ft simultaneously. Thus, we hypothesize that rCST9 has the potential to reveal key host proteins in signaling pathways that could be targeted selectively to divert from excessive, damaging inflammation to attenuated, protective immune responses. Future studies may expose a role for the use of rCST9 as a prophylactic for patients at risk of developing an infection and/or sepsis.
ACKNOWLEDGMENTS
This work was supported by Eaves-Pyles’ NIH/NIAID (R21-A106877402). We would like to thank Istvan Boldogh (Department of Microbiology and Immunology, University of Texas Medical Branch) for his advice, suggestions and input on the in vivo studies presented herein and Bill Calhoun (Department of Internal Med-Pulmonary, University of Texas Medical Branch) for his contribution to the autophagy results. We are gratefuly to the Biomolecule Analysis Core Facility at the BBRC/Biology/UTEP (NIH grants G12M0007592, 5G12RR008124-16A1 and 5G12RR008124-16A1S1) for proteomic analysis.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Ochieng J, Chaudhuri G. Cystatin superfamily. J Health Care Poor Underserved. 2010;21:51–70. doi: 10.1353/hpu.0.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loffek S, Schilling O, C-W Franzke. Biological role of matrix metalloproteinases: a critical balance. Eur Respir J. 2011;38:191–208. doi: 10.1183/09031936.00146510. [DOI] [PubMed] [Google Scholar]
- 3.Zavasnik-Bergant T. Cystatin protease inhibitors and immune functions. Front Biosci. 2008:4625–37. doi: 10.2741/3028. [DOI] [PubMed] [Google Scholar]
- 4.Kopitar-Jerala. The role of cystatins in cells of the immune system. FEBS Letters. 2006;580:6295–301. doi: 10.1016/j.febslet.2006.10.055. [DOI] [PubMed] [Google Scholar]
- 5.Bobek LA, Levine MJ. Cystatins—inhibitors of cysteine proteinases. Crit Rev Oral Biol Med. 1992;4:307–32. doi: 10.1177/10454411920030040101. [DOI] [PubMed] [Google Scholar]
- 6.Vray B, Hartmann S, Hoebeke J. Immunomodulatory properties of cystatins. Cell Mol Life Sci. 2002;59:1503–12. doi: 10.1007/s00018-002-8525-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poteryaeva ON, et al. Cysteine proteinase inhibitor level in tumor and normal tissues in control and cured mice. Drugs Exp Clin Res. 2000;26:301–6. [PubMed] [Google Scholar]
- 8.Yang F, et al. Cystatin B inhibition of TRAIL-induced apoptosis is associated with the protection of FLIPLfrom degradation by the E3 ligase itch in human melanoma cells. Cell Death Differ. 2010;17:1354–67. doi: 10.1038/cdd.2010.29. [DOI] [PubMed] [Google Scholar]
- 9.Briggs JJ, et al. Cystatin E/M suppresses legumain activity and invasion of human melanoma. BMC Cancer. 2010;10:17. doi: 10.1186/1471-2407-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirttilä TJ, et al. Cystatin C modulates neurodegeneration and neurogenesis following status epilepticus in mouse. Neurobiol Dis. 2005;20:241–53. doi: 10.1016/j.nbd.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Gauthier S, Kaur G, Mi W, Tizon B, Levy E. Protective mechanisms by cystatin C in neurodegenerative diseases. Front Biosci. 2011;3:541–54. doi: 10.2741/s170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esposito E, Cuzzocrea S. New therapeutic strategy for Parkinson’s and Alzheimer’s disease. Curr Med Chem. 2010;17:2764–74. doi: 10.2174/092986710791859324. [DOI] [PubMed] [Google Scholar]
- 13.Tizon B, et al. Induction of autophagy by cystatin C: a mechanism that protects murine primary cortical neurons and neuronal cell lines. PLoS One. 2010;5(3):e9819. doi: 10.1371/journal.pone.0009819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rivera-Rivera L, Perez-Laspiur J, Colón K, Meléndez LM. Inhibition of interferon response by cystatin B: implication in HIV replication of macrophage reservoirs. J Neurovirol. 2012;18:20–9. doi: 10.1007/s13365-011-0061-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis DT, et al. Tularemia as a biological weapon: Medical and public health management. JAMA. 2001;285:2763–73. doi: 10.1001/jama.285.21.2763. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs RF. Tularemia. Ad Ped Infect Dis. 1977;12:55–69. [PubMed] [Google Scholar]
- 17.Sjöstedt A, Tärnvik A, Sandström G. Francisella tuarensis: Host-parasite interaction. FEMS Immun Med Micro. 1996;13:181–4. doi: 10.1111/j.1574-695X.1996.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 18.Eaves-Pyles TD, Wong HR, Odoms K, Pyles RB. Salmonella flagellin-dependent proinflammatory responses are localized to the conserved amino and carboxyl regions of the protein. J Immunol. 2001;167:7009–16. doi: 10.4049/jimmunol.167.12.7009. [DOI] [PubMed] [Google Scholar]
- 19.Dhiman M, et al. Enhanced nitrosative stress during Trypanosoma cruzi infection causes nitrotyrosine modification of host proteins: implications in Chagas’ disease. Am J Pathol. 2008;173:728–40. doi: 10.2353/ajpath.2008.080047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bayer-Santos E, et al. Proteomic analysis of Trypanosoma cruzi secretome: characterization of two populations of extracellular vesicles and soluble proteins. J Proteome Res. 2013;12:883–97. doi: 10.1021/pr300947g. [DOI] [PubMed] [Google Scholar]
- 21.Gentry M, et al. Role of primary human alveolar epithelial cells in host defense against Francisella tularensis infection. Infect Immun. 2007;75:3969–78. doi: 10.1128/IAI.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coletta C, et al. Hydrogen sulfide and nitric oxide are mutually dependent in the regulation of angiogenesis and endothelium-dependent vasorelaxation. Proc Natl Acad Sci U S A. 2012;109:9161–6. doi: 10.1073/pnas.1202916109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Popov V, Patel J, Eaves-Pyles T. Burkholderia mallei and Burkholderia pseudomallei stimulate differential inflammatory responses from human alveolar type II cells (ATII) and macrophages. Front Cell Infect Microbiol. 2012;2:165. doi: 10.3389/fcimb.2012.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Magister S, Kos J. Cystatins in immune system. Cancer. 2013;4:45–56. doi: 10.7150/jca.5044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turk V, Stoka V, Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front Biosci. 2008;13:5406–20. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- 26.Chang SH, et al. VEGF-A induces angiogenesis by perturbing the cathepsin-cysteine protease inhibitor balance in venules, causing basement membrane degradation and mother vessel formation. Cancer Res. 2009;69:4537–44. doi: 10.1158/0008-5472.CAN-08-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–17. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindgren H, et al. Factors affecting the escape of Francisella tularensis from the phagolysosome. J Med Microbiol. 2004;53:953–8. doi: 10.1099/jmm.0.45685-0. [DOI] [PubMed] [Google Scholar]
- 29.Izaki K, Matsuhashi M, Strominger JL. Glycopeptide transpeptidase and D-alanine carboxypeptidase: penicillin-sensitive enzymatic reactions. PNAS. 1966;55:656–63. doi: 10.1073/pnas.55.3.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers R, Yogum DJ, Waxman JRR, Stominger JL. Mechanism of penicillin action: Penicillin and substrate bind covalently to the same active site serine in two bacterial D-alanine carboxypeptidases. PNAS. 1979;76:2730–4. doi: 10.1073/pnas.76.6.2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez OC, et al. Conserved micro-tubule-actin interactions in cell movement and morphogenesis. Nat Cell Biol. 2003;5:599–609. doi: 10.1038/ncb0703-599. [DOI] [PubMed] [Google Scholar]
- 32.Yuan A, Cia CP. Co-loss of profiling I, II and cofilin with actin from maturing phagosomes in Dictyostelium discoideum. Protoplasma. 1999;209:214–225. [Google Scholar]
- 33.Chung S, Sundar IK, Yao H, Ho YS, Rahman I. Glutaredoxin 1 regulates cigarette smoke-mediated lung inflammation through differential modulation of I{kappa}B kinases in mice: impact on histone acetylation. Am J Physiol Lung Cell Mol Physiol. 2010;299:L192–203. doi: 10.1152/ajplung.00426.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mor-Vaknin N, et al. Vimentin suppresses the production of reactive oxygen species and the antimicrobial response via p47phox [abstract] Arthritis Rheum. 2011;63(Suppl 10):1003. [Google Scholar]
- 35.Rogel MR, et al. Vimentin is sufficient and required for wound repair and remodeling in alveolar epithelial cells. FASEB J. 2011;25:3873–83. doi: 10.1096/fj.10-170795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pyles RB, Jezek GE, Eaves-Pyles TD. Toll-like receptor 3 agonist protection against experimental Francisella tularensis respiratory tract infection. Infect Immun. 2010;78:1700–10. doi: 10.1128/IAI.00736-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mohamed MM, Sloane BF. Cysteine cathepsins: multifunctional enzymes in cancer. Nature Cancer. 2006;6:764–75. doi: 10.1038/nrc1949. [DOI] [PubMed] [Google Scholar]
- 38.Cantres-Rosario Y, et al. Cathepsin B and cystatin B in HIV-seropositive women are associated with infection and HIV-1-associated neurocognitive disorders. AIDS. 2013;27:347–56. doi: 10.1097/QAD.0b013e32835b3e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krawczeski CD, et al. Serum cystatin C is an early predictive biomarker of acute kidney injury after pediatric cardiopulmonary bypass. Clin J Am Soc Nephrol. 2010;5:1552–7. doi: 10.2215/CJN.02040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassinger AB, et al. Predictive power of serum cystatin C to detect acute kidney injury and pediatric-modified RIFLE class in children undergoing cardiac surgery. Pediatr Crit Care Med. 2012;13:435–40. doi: 10.1097/PCC.0b013e318238b43c. [DOI] [PubMed] [Google Scholar]
- 41.Qing X, et al. Cystatin C and asymptomatic coronary artery disease in patients with metabolic syndrome and normal glomerular filtration rate. Cardiovasc Diabetol. 2012;11:108. doi: 10.1186/1475-2840-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma J, Mares CA, Li Q, Morris EG, Teale JM. Features of sepsis caused by pulmonary infection with Francisella tularensis Type A strain. Microb Pathog. 2011;51:39–47. doi: 10.1016/j.micpath.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clemens DL, Lee BY, Horwitz MA. Virulent and avirulent strains of Francisella tularensis prevent acidification and maturation of their phagosomes and escape into the cytoplasm in human macrophages. Infect Immun. 2004;72:3204–17. doi: 10.1128/IAI.72.6.3204-3217.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bönquist L, Lindgren H, Golovliov I, Guina T, Sjöstedt A. MglA and Igl proteins contribute to the modulation of Francisella tularensis live vaccine strain-containing phagosomes in murine macrophages. Infect Immun. 2008;76:3502–10. doi: 10.1128/IAI.00226-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Checroun C, Whrly TD, Rishcher DR, Hayes SF, Celli J. Autophagy-mediated reentry of Francisella tularensis in to the endocytic compartment after cytoplasmic replication. Proc Natl Acad Sci U S A. 2006;103:14578–83. doi: 10.1073/pnas.0601838103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weiss DS, Henry T, Monack DM. Francisella tularensis: Activation of the inflammasome. Ann NY Acad Sci. 2007;1105:219–37. doi: 10.1196/annals.1409.005. [DOI] [PubMed] [Google Scholar]
- 47.Jones JW, Broz P, Monack DM. Innate immune recognition of Francisella tularensis: activation of type-I interferons and the inflammasome. Front Micro. 2011;2:1–10. doi: 10.3389/fmicb.2011.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mariathasan S, Weiss DS, Dixit VM, Monack DM. Innate immunity against Francisella tularensis is dependent on the ASC/caspase-1 axis. J Exp Med. 2005;202:1043–9. doi: 10.1084/jem.20050977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malik M, et al. Matrix metalloproteinase 9 activity enhances host susceptibility to pulmonary infection with type A and B strains of Francisella tularensis. J Immunol. 2007;178:1013–20. doi: 10.4049/jimmunol.178.2.1013. [DOI] [PubMed] [Google Scholar]
- 50.Bosio CM, Elkins KL. Susceptibility to secondary Francisella tularensis live vaccine strain infection in B-cell-deficient mice is associated with neutrophilia but not with defects in specific T-cell-mediated immunity. Infect Immun. 2001;69:194–203. doi: 10.1128/IAI.69.1.194-203.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leavy O. Mucosal immunology: autophagy helps man the barriers. Nat Rev Immunol. 2013;13:470–1. doi: 10.1038/nri3489. [DOI] [PubMed] [Google Scholar]
- 52.Patel AS, Morse D, Choi AM. Regulation and functional significance of autophagy in respiratory cell biology and disease. Am J Respir Cell Mol Biol. 2013;48:1–9. doi: 10.1165/rcmb.2012-0282TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong L, Devenish RJ, Prescott M. Autophagy as a macrophage response to bacterial infection. IUBMB Life. 2012;64:740–7. doi: 10.1002/iub.1070. [DOI] [PubMed] [Google Scholar]
- 54.Jaber N, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A. 2012;109:2003–8. doi: 10.1073/pnas.1112848109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lamouille S, Connolly E, Smyth JW, Akhurst RJ, Derynck R. TGF-β-induced activation of mTOR complex 2 drives epithelial-mesenchymal transition and cell invasion. J Cell Sci. 2012;125:1259–73. doi: 10.1242/jcs.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mort JS, Buttle DJ. Cathepsin B. Int J Biochem Cell Biol. 1997;29:715–20. doi: 10.1016/s1357-2725(96)00152-5. [DOI] [PubMed] [Google Scholar]
- 57.Wang L, et al. Antimicrobial activity and molecular mechanism of the CRES protein. PLoS One. 2012;7:e48368. doi: 10.1371/journal.pone.0048368. [DOI] [PMC free article] [PubMed] [Google Scholar]