Abstract
Chronic lymphocytic leukemia (CLL) is a clonal disease of a subset of human B lymphocytes. Although the cause of the disease is unknown, its development and evolution appear to be promoted by signals delivered when B-cell receptors (BCRs) engage (auto)antigens. Here, using a peptide phage display library of enhanced size and diverse composition, we examined the binding specificity of a recombinant monoclonal antibody (mAb) constructed with the heavy chain and light chain variable domains of a CLL BCR that does not exhibit somatic mutations. As determined by testing the peptides identified in the selected peptide phage pool, this CLL-associated unmutated mAb bound a diverse set of sequences, some of which clustered in families based on amino acid sequence. Synthesis of these peptides and characterization of binding with the CLL-associated mAb revealed that mAb-peptide interactions were generally specific. Moreover, the mAb-peptide interactions were of lower affinities (micromolar KD), as measured by surface plasmon resonance, than those observed with a CLL mAb containing somatic mutations (nanomolar KD) and with immunoglobulin heavy chain variable (IGHV)-mutated antibodies selected by environmental antigens. This information may be of value in identifying and targeting B lymphocytes expressing specific BCRs in CLL patients and healthy subjects with monoclonal B lymphocytosis.
INTRODUCTION
Chronic lymphocytic leukemia (CLL) is an incurable disease of unknown etiology (1,2). The blood and lymphoid tissues of CLL patients contain high levels of a single clone of B lymphocytes with unique surface membrane immunoglobulin (smIg) components of B-cell receptors (BCRs) for antigen, essentially the cell-surface version of a monoclonal antibody (mAb). On the basis of the biased use of specific immunoglobulin heavy chain variable (IGHV) genes by CLL B cells (3) and the not uncommon striking structural restriction in amino acid sequence (4–8), however, it seems likely that disease initiation and progression are influenced by (auto)antigen selection and drive of a specific set of human B lymphocytes (1,9,10). Studies to date suggest that this drive is more frequent and effective among CLL clones with unmutated IGHV genes (U-CLL) (11) that can bind multiple antigens than among CLL clones with mutated IGHVs (M-CLL) (12–15).
Furthermore, the categorization of CLL BCRs according to the degree of somatic nucleotide mutations in the expressed IGHV segment (3) can quite accurately predict a patient’s clinical course (16,17). Specifically, patients whose clones exhibit BCRs containing 2% or less difference from the germline (U-CLL BCRs) are much more likely to exhibit aggressive disease than those >2% difference (M-CLL BCRs). Therefore, characterization of BCR-antigen interactions is of general importance for understanding more and less aggressive forms of the disease. Current evidence suggests that U-CLL BCR combining sites can interact with a variety of antigens (12–15). Although these U-CLL BCR-antigen interactions are presumed to be of low affinity, specific measurements of affinity are not available.
Because of these observations, it is tantalizing to hypothesize that multispecific interactions between U-CLL BCRs and antigens drive the proliferation of leukemic B cells. Here we describe the use of peptide phage display containing a very large number of random epitopes to study recognition by a U-CLL BCR. Our motivation for this work was twofold. First, the identification of agents that can target U-CLL BCRs would be of potential use for diagnosis and treatment. Current CLL therapies are not targeted to BCRs with specific structures and, therefore, the specificity of such treatments could be enhanced by development of U-CLL BCR-binding peptides as targeting agents. Furthermore, such reagents could be used in flow cytometry assays to determine whether B lymphocytes bearing specific BCRs are present in blood and in what abundance. This might be especially helpful, not only in CLL, but also in healthy individuals with the condition known as monoclonal B lymphocytosis (18,19) that appears to be a prerequisite preamble to CLL (20). Second, the characterization of binding specificities offers insight into the behavior of a disease-associated germline-encoded antibody, and the extent that these antibodies resemble early stages of normal B-cell and antibody evolution. Peptides that are specific for U-CLL could serve as biological and structural probes to decipher aspects of U-CLL BCR recognition.
In this study, we identified a diverse panel of peptide-phage clones that bind an mAb derived from a U-CLL clone. Studies with synthetic peptides indicated that some were specific for this U-CLL BCR and that in general the affinities of these, measured by surface plasmon resonance, were much lower than that of an M-CLL mAb.
MATERIALS AND METHODS
Expression and Purification of U-CLL mAb 068 and M-CLL mAb169
Studies were approved by the Institutional Review Board of North Shore–LIJ Health System (Manhasset, NY, USA) and performed in accordance with the Helsinki agreement. In brief, ribonucleic acid (RNA) from patient’s blood mononuclear cells was reverse-transcribed into cDNA. The variable segments of rearranged immunoglobulin heavy (IGH) and immunoglobulin light (IGL) of U-CLL 068 and M-CLL 169 were expressed as human IgG1 as described previously (15).
Phage Library Sorting and Analysis
Preparation of the pVIII peptide library was reported previously (21). Wells of Costar EIA/RIA high-binding plates (Corning, Big Flats, NY, USA) were coated with U-CLL mAb068 in phosphate-buffered saline (PBS, pH 7.4) for 1 h at room temperature. The well solutions were decanted and unbound well sites blocked by incubation with PBS containing 3% (w/v) human serum albumin (HSA) for 1 h at room temperature. After washing with PBS containing 0.05 % (v/v) Tween 20 (PBS-T), freshly amplified library phage at titers of ~1013 plaque-forming units (PFU)/mL in PBS/0.5 % HSA were added to wells and allowed to bind for 1 h. Next, wells were washed six times with PBS-T. Bound phages were eluted by the addition of 100 μL 100 mmol/L glycine hydrochloride (Gly·HCl) pH 2.0 for 5 min. The phage solution was neutralized in 30 μL of 2 mol/L tris(hydroxymethyl)aminomethane (Tris) pH 8.0 and then propagated overnight at 37°C in XL1-Blue E. coli (Stratagene, La Jolla, CA, USA). The cells were removed by centrifugation, the phage precipitated by addition of 0.5 mol/L NaCl and 4% (w/v) polyethylene glycol (PEG) 8000 and used directly for the subsequent round of selection.
Reactivity of output populations from the selection with U-CLL mAb068 or HSA were assessed using phage enzyme-linked immunosorbent assay (ELISA). mAb068 or HSA was immobilized into wells of Costar EIA/RIA plates and blocked as above. Freshly-amplified phage populations from each round of selection were incubated with the target for 1 h at room temperature, and the wells washed three times with PBS-T. An anti-M13/horseradish peroxidase conjugate (GE Healthcare, Piscataway, NJ, USA) was added as per the manufacturer’s protocol and allowed to incubate for 1 h. After washing with PBS-T, 100 μL of 3,3′,5,5′-tetramethylbenzidine substrate (SureBlue, Kirkegaard & Perry Laboratories, Gaithersburg, MD, USA) were added. The color was allowed to develop for 5 min, quenched with 1 mol/L HCl, and the absorbance at 450 nm determined. Analyses of individual clones from the R3 and R4 populations were similar, except that the phages were grown as small-scale cultures (1 mL) in 96-deep well plates. Following centrifugation, the culture supernatants were applied directly to the immobilized U-CLL mAb068 or HSA.
Peptide Synthesis and Purification
All peptide were produced by solid-phase peptide synthesis using standard Fmoc (N-[9-fluorenyl]methoxycarbonyl) chemistry at the Tufts University Biopolymer Facility (Boston, MA, USA). Following synthesis, simultaneous side chain deprotection and cleavage from resin were achieved by treating them with 95% trifluoroacetic acid, 2.5% 1,2-ethanedithiol, and 2.5% thioanisole for 3 h. The resin was removed by filtration, and the peptide precipitated by the addition of cold diethylether. The peptide was pelleted by centrifugation, the pellet washed twice with cold diethyl ether, and the peptides dissolved in water/acetonitrile and lyophilized. For peptides containing disulfide bonds, the crude peptide was dissolved in water at low concentrations (<0.5 mg/mL) and the pH adjusted to 7 to 8. Disulfide oxidation was achieved by bubbling air through this solution for 2 to 3 d. Oxidation progress was monitored by reverse-phase analytical high- performance liquid chromatography (HPLC) on a Vydac® C18 column (10 μm × 250 mm × 21.2 mm; Grace, Deerfield, IL, USA) with water/acetonitrile/0.1% TFA mobile phases. Once oxidation was complete, the solution was lyophilized. Crude peptides were purified by reverse phase (RP)-HPLC on a Vydac preparatory-scale C18 column (10 μm × 250 mm × 21.2 mm). Peptide purity was generally >95% as judged by analytical RP-HPLC, and the identity of all peptides was confirmed by matrix-assisted laser desorption/ionization (MALDI MS).
Surface Plasmon Resonance
All measurements were performed on a Biacore 3000 instrument at room temperature in running buffer (20 mmol/L phosphate buffer [pH 7.4], 130 mmol/L KCl, 3.4 mmol/L EDTA, 0.005% Tween 20 [v/v]). U-CLL mAb068 or M-CLL mAb169 were immobilized to CM5 chips (GE Healthcare) as specified by the manufacturer at loading levels of 17,000–22,000 response units (RUs). The peptides were dissolved in running buffer and sensorgrams obtained by flowing peptide solutions at various concentrations over the immobilized mAb surface at a flow rate of 40 μL/min. Sensorgrams were fit to a standard 1:1 Langmuir model using BIAevaluation 4.1 (Biacore) software. Adequacy of data fitting was assessed using the parameters χ2. When possible, mAb-coated surfaces were regenerated by 10 mmol/L KOH. Peptide and protein concentrations were determined by absorbance at 280 nm.
All supplementary materials are available online at www.molmed.org.
RESULTS
A mAb That Corresponds to a U-CLL BCR
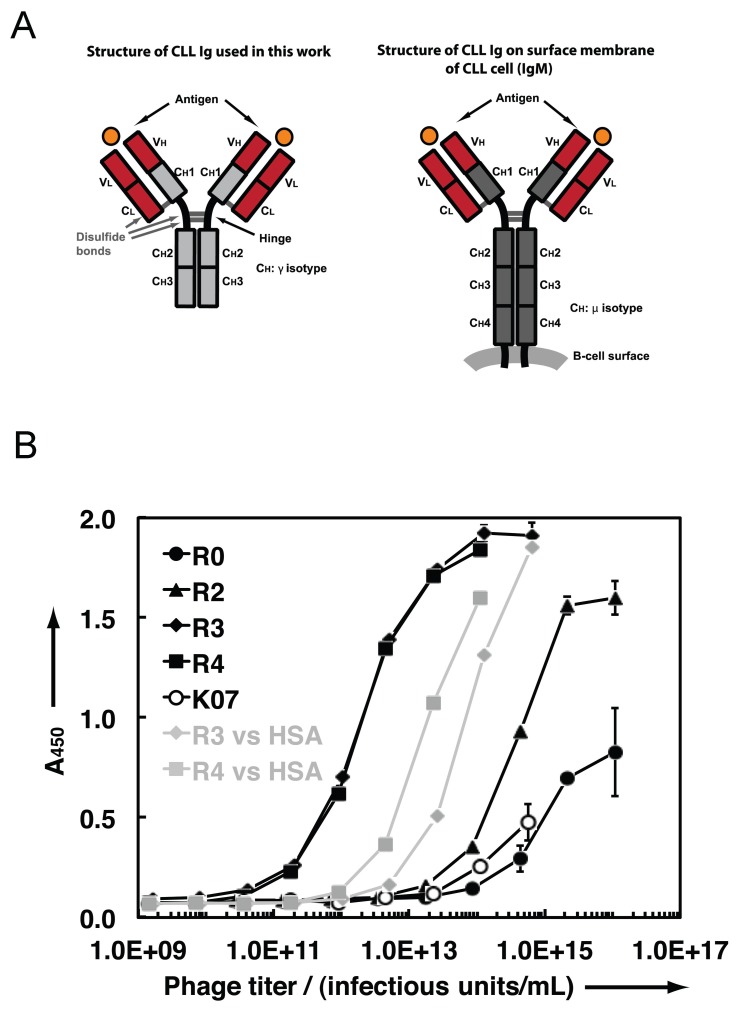
The amino acid sequences of the IGHV-D-J and IGLVκ-Jκ rearrangements determine the architecture of the antigen-binding domain of a BCR (Figure 1). For the following studies, the IGHV-D-J and IGKVκ-Jκ rearrangements from the leukemic cells of patient CLL068 were linked molecularly to an IGHCγ1 segment to produce an immunoglobulin G1 (IgG1) mAb. Although the native CLL 068 BCR/mAb is of the immunoglobulin M (IgM) isotype, structural studies with full-length mAbs and mAb fragments (for example, antigen-binding fragments, Fabs, or single-chain variable fragments (scFvs), indicate that overall combining site topology is usually independent of constant domains (22), although exceptions have been noted (23). Therefore, an IgG1 mAb containing the IGHV-D-J/IGLV/J rearrangements of a U-CLL BCR will likely be an effective mimic of the cell-surface combining site.
Figure 1.
(A) Domain architecture of an IgG1 mAb and the surface membrane IgM of a BCR. (B) Polyclonal phage ELISA of R0–R4 populations, as well as K07 control phage, against U-CLL mAb068. The R3 and R4 populations also were tested against wells containing the negative control protein HSA (gray). CH: immunoglobulin heavy chain constant region; CL: Immunoglobulin light chain constant region; CH1, CH2, CH3, and CH4: immunoglobulin constant region heavy chains 1, 2, 3, and 4, respectively; VL: immunoglobulin light chain variable region; VH: immunoglobulin heavy chain variable region.
The design and analysis of U-CLL mAb068 expressed with a human IgG1 domain has been described previously (15). The U-CLL mAb068 antigen-binding domain contains the IGHV1-69 germline segment, which is frequently selected among virus neutralizing antibodies and whose combining site interactions typically involve a hydrophobic second heavy chain complementarity-determining region (HCDR2) (24), linked to IGHD3-16 and IGHJ3 segments. It is estimated that the IGHV1-69 germline segment comprises less than 2% of productive rearrangements in normal blood B lymphocytes (25), perhaps a surprising observation considering its prevalence in CLL cells and viral responses. Both the IGHV-D-J and IGLVκ-Jκ rearrangements of U-CLL mAb068 are identical to germline segments (Supplementary Figure S1). In addition, these rearrangements exhibit characteristic junctional amino acids (4). Finally, the antigen-binding site represented by the U-CLL mAb068 IGHV-D-J and IGLVκ-Jκ rearrangement is shared by multiple CLL patients (4–8). Therefore understanding the antigen-binding characteristics of U-CLL mAb068 will be applicable to those of multiple patients.
Phage Display Selection of Peptides that Bind U-CLL mAb068
To explore the range of U-CLL mAb068 combining site reactivities, we screened a high complexity peptide phage display library against immobilized U-CLL mAb068. Previous attempts to isolate binding peptides that target U-CLL mAb068 from a commercially-available library containing random linear peptide segments displayed on the M13 minor coat protein pIII (the “PhD library”) yielded only a single sequence with low-level binding activity (sequence WPLWSIPPFSPR) (26). Here, we used a library in which random peptide segments (containing both linear and disulfide crosslinks) were expressed on the major M13 coat protein pVIII using a two-plasmid phagemid/helper phage system (27). The diversity of the displayed peptides (Table 1) includes random segments from 7 to 20 residues, in many cases containing cysteine residues at various spacings that are expected to form disulfide bonds upon phage assembly. Linear random sequences (X8) also were included, as were sequences containing a central reverse turn–promoting ~GP~ dipeptide. The pVIII display format is expected to display peptides on ~50 of the 2700 pVIII copies per M13 phage particle. The library complexity was 1011 clones (21).
Table 1.
Random peptide sequences in phage display library.a
| CX5C | X2CX4CX2 | X4CX10CX4 |
| CX5CX | X2CX5CX2 | X5CX8CX5 |
| CX5CX2 | X2CX6CX2 | X5CX9CX4 |
| XCX5C | X2CX7CX2 | X6CX6CX6 |
| X2CX5C | X2CX8CX2 | X6CX7CX5 |
| X2CX9CX2 | X7CX4CX7 | |
| X2CX2CX2 | X2CX10CX2 | X7CX5CX6 |
| X2CX2CX3 | ||
| X2CX3CX2 | X4CX2GPX4CX4 | X8 |
All random segments produced as N-terminal pVIII fusions; X = any amino acid as encoded by the combinatorial codon NNS (N = A/T/C/G, S = C/G).
Four rounds of selection were performed against U-CLL mAb068. Phage ELISA of the unselected library population (R0) and the output populations from rounds 2, 3 and 4 (R2–R4) indicate that the selection resulted in enhanced binding activity for wells containing U-CLL mAb068. To examine the specificity of the later rounds, the R3 and R4 populations were tested for binding to wells containing the negative control protein, human serum albumin (HSA) (Figure 1B). Although some binding activity of the R3 and R4 populations was observed against HSA, the higher binding signals against U-CLL mAb068 at lower phage titers relative to HSA indicated that U-CLL mAb068-specific clones were present in the R3 and R4 population. Furthermore, a negative control phage bearing no peptide (K07) showed minimal binding to U-CLL mAb068. Sequence analysis indicated the R3 population clones were highly diverse in sequence.
Analysis of Individual Phage Clones
Individual clones from the R3 and R4 population were screened for reactivity with U-CLL mAb068 using high-throughput phage ELISA. We identified 36 clones that had strong positive ELISA signals for U-CLL mAb068 (A450 >1.0) and had specificity over control wells containing HSA (an [A450 (mAb068)]/[A 450 (HSA)] ratio of 13-fold was used as a cutoff). These clones corresponded to 23 unique amino acid sequences (Table 2). Many of the sequences contained the cysteine residues at various spacings ranging from 2 to 10, although a CX6C pattern was more commonly observed. Four copies of the linear sequence NMSIATVP also were present in the dataset. To further evaluate selectivity for binding the CLL-associated mAb, we tested all 23 of these sequences for binding to an unrelated mAb and for their capacity to retain U-CLL mAb068 binding activity under high salt conditions (750 mmol/L NaCl). Eighteen of the 23 sequences fulfilled the criteria of being specific for U-CLL mAb068 and retaining binding activity in high salt (underlined in Table 2 and shown in Supplementary Table S1). Since these clones did not bind an unrelated IgG1 mAb, we conclude that they have specificity for U-CLL mAb068 and that the peptides bind the variable domains of the CLL mAb. Furthermore, the ability of these clones to bind under high salt conditions suggests the interactions between the peptides and U-CLL mAb068 involve specific contacts and are not due to general electrostatic or hydrophobic binding.
Table 2.
Isolated phage clones from selection with U-CLL mAb068.
| Clone(s) | (Frequency)a | Sequenceb | [A450 (mAb068)]/[A450 (HSA)]c |
|---|---|---|---|
| B12, D7, E4, F9 | (4) | NMSIATVP | 28 |
| C2, F3, F7, H11 | (4) | ASCYIDDGIRYCTG | 28 |
| A6, B1, D2, E12 | (4) | VSCNPFSWLPRCYA | 19 |
| A5, D4, D5 | (3) | DYCNLSVSWCYP | 26 |
| F1, F5 | (2) | STCWLMFSMCRV | 24 |
| A4, B6 | (2) | TPCEGSWFFCTL | 23 |
| E2 | LGCDVIYWSSGWGCKF | 31 | |
| B3 | GSCSLSIVTIPCGV | 30 | |
| D3 | VTCAAFSLFPFCYN | 28 | |
| E7 | CHPGWPCFW | 28 | |
| D8 | GGCNLSVLWCLD | 27 | |
| F10 | NECILVNGDRWCIS | 27 | |
| E6 | FPCWALPSQYLCRP | 27 | |
| D11 | TACNLSVSWCEF | 20 | |
| G11 | TNCGFLYLGCVI | 18 | |
| E9 | FDCGDGSWRYYLDCVR | 18 | |
| B7 | TSCRVLWIMTLCNA | 18 | |
| B10 | EVCNNKLWPCFW | 16 | |
| A8 | GGCGWLWVSCVI | 14 | |
| A2 | CSPAWPCWW | 14 | |
| F2 | LPCAGRSFPCYW | 14 | |
| C8 | DRCLLWVSLCMA | 13 | |
| E1 | CIPMWPCWW | 13 |
Identified clones and observed frequencies in cases where multiples were observed.
The underlined sequences specifically bound to U-CLL mAb068, but not an unrelated mAb, and retained binding activity in 750 mmol/L NaCl.
Ratio of ELISA signals for wells coated with U-CLL mAb068 over wells coated with HSA negative control. In cases where a sequence was observed more than once, the average is shown. For all others, results for a single replicate are shown. In general, data among independent experiments were in good agreement.
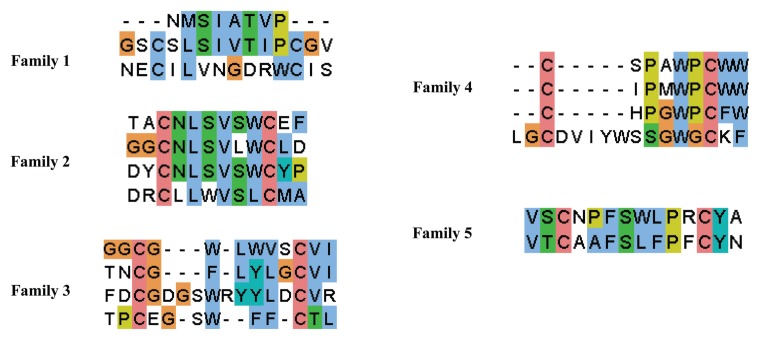
To gain further insight into binding selectivity, we grouped the 18 mAb068-specific sequences into families based on amino acid composition. We found that the clones clustered into five distinct families of varying conservation (Figure 2). Clone E6 was not closely related to any other clone and therefore was not classified according to any family. Members of families 1, 2 and 4 showed high conservation, suggesting that these clones bind in a similar manner. However, families 3 and 5 contained more disparate primary sequences. Overall, the diversity in the sequences, from content of hydrophilic/hydrophobic residues to spacing between cysteine residues, indicates that U-CLL mAb068 is able to bind a number of sequences.
Figure 2.
Family analysis of the 18 sequences that had specificity for U-CLL mAb068.
Phage display studies with the linear PhD library resulted in strong consensus sequences against mAbs associated with the less aggressive M-CLL BCR-containing B-cells (26). Previous studies suggest that greater antigen-binding promiscuity of U-CLL than M-CLL BCR combining sites may lead to the higher activation of U-CLL clones (28) and their more aggressive clinical properties (16,17). Of note, selection of phage peptides against antibodies does not necessarily provide information about binding promiscuity. For example, an affinity-matured HIV-1 antibody has been reported to display binding promiscuity for several disparate peptide sequences from synthetic and phage displayed libraries (29). Nonetheless, it is interesting to note that distinct families of sequences were obtained from selection against U-CLL mAb068.
In Vitro Characterization of U-CLL mAb068-Binding Peptides
We synthesized several peptides corresponding to sequences from the U-CLL mAb068 selection isolated here: peptide D4-Btn belonging to the well conserved family 2; peptide D2-Btn from the less conserved family 5 with longer length; and peptide E6-Btn with no significant similarity to other selectants (Table 3). We also produced two peptides identified in our previous study using the commercially available 12-mer PhD library (26): peptide PhD-068-4-Btn isolated with U-CLL mAb068 and peptide PhD-169-8-Btn, isolated by an IGHV-mutated CLL mAb with different IGH and IGL variable regions (M-CLL mAb169). All peptides were purified by HPLC in good yield. In all of the above cases, a C-terminal biotin was included for detection. Disulfide bonds in D4-Btn, D2-Btn and E6-Btn were formed by air oxidation; MALDI-MS and HPLC analysis indicated disulfide bond formation was complete in all cases.
Table 3.
Peptides synthesized for in vitro analysis.a
| Peptidea | Sequence | KDb | Selection target (comment) |
|---|---|---|---|
| D4-Btn | DYCNLSVSWCYP-K (Btn) | ND | U-CLL mAb 068 (this work; family 2) |
| D4-3K | DYCNLSVSWCYP-GSGKKK | NB | |
| D2-Btn | VSCNPFSWLPRCYA-K (Btn) | ND | U-CLL mAb 068 (this work; family 5) |
| D2-3K | VSCNPFSWLPRCYA-GSGKKK | 58 μmol/L | |
| E6-Btn | FPCWALPSQYLCRP-K (Btn) | ND | U-CLL mAb 068 (this work) |
| E6-3K | FPCWALPSQYLCRP-GSGKKK | 111 μmol/L | |
| PhD-068-4-Btn | WPLWSIPPFSPR-K (Btn) | weak | U-CLL mAb 068 (reference [5]) |
| PhD-068-4-3K | WPLWSIPPFSPR-KKKK | weak | |
| PhD-169-8-Btn | DNYAAALAQRAR-K (Btn) | ND | M-CLL mAb 169 (reference [5]) |
| PhD-169-8-4K | DNYAAALAQRAR-KKKK | 31 nmol/L |
All peptides synthesized as C-terminal amides.
ND, not determined; NB, no binding.
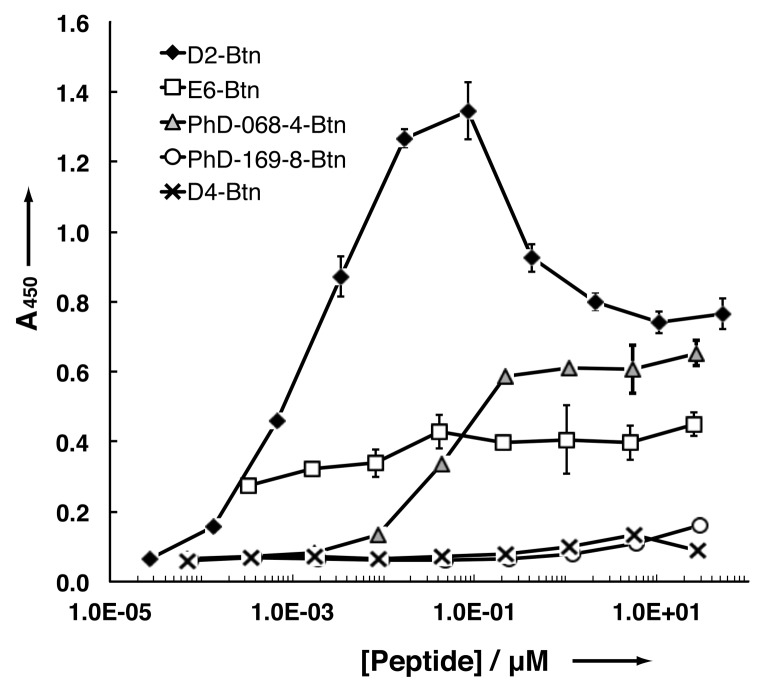
ELISAs in which the binding of peptides to immobilized U-CLL 068 mAb was detected with streptavidin-horse-radish peroxidase (SA-HRP) were performed (Figure 3). Peptide D2-Btn bound U-CLL mAb068 with the highest ELISA signal and the lowest half-maximal binding titer. A drop-off in the ELISA signal was evident at concentrations higher than 100 nmol/L, possibly due to the prozone effect. Peptide E6-Btn also exhibited some ELISA signal, albeit weaker in comparison to D2-Btn and with less dose responsiveness. As expected, PhD-068-4-Btn exhibited a binding ELISA signal, but this had higher half-maximal binding titer than D2-Btn. Peptide D4-Btn from conserved family 2, did not show any binding signal. Thus, binding did not correlate directly with conservation among families shown in Figure 2. As expected, PhD-169-8-Btn (selected against M-CLL mAb169 (26)), did not bind U-CLL mAb068.
Figure 3.
ELISA of biotinylated peptides against U-CLL mAb068.
To further explore the binding properties of these peptides, we examined the binding of these peptides to U-CLL mAb068 and/or M-CLL mAb169 by surface plasmon resonance (SPR). To do this, we synthesized several variants with a C-terminal polylysine segment (E6-3K, D2-3K, D4-3K, PhD-068-4-4K, PhD-169-8-4K). The polylysine segment was included to improve solubility and minimize aggregation or nonspecific hydrophobic effects for in vitro assays.
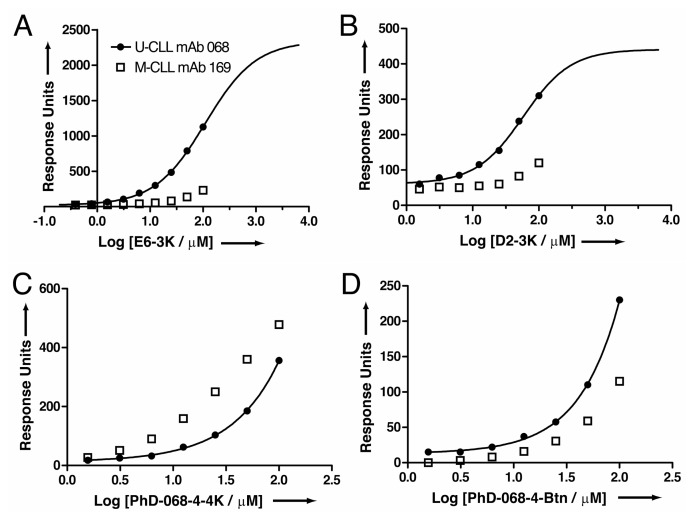
Because many binding sensorgrams did not obey 1:1 Langmuir binding, we performed equilibrium analysis rather than kinetic analysis in those cases. We found that E6-3K had a strong concentration-dependent binding signal to U-CLL mAb068 but not M-CLL mAb169 (Figure 4A). These data are consistent with the biotinylated peptide ELISA results in Figure 3, and indicate that E6-3K bears specificity for the U-CLL mAb068 over M-CLL mAb169. Peptide D2-3K also bound U-CLL mAb068, but the amplitude of the equilibrium signal (in response units) was generally lower than E6-3K at equivalent peptide concentrations (Figure 4B). Furthermore, D2-3K also showed specificity for U-CLL mAb068 over M-CLL mAb169. Aggregation at high concentrations prevented determination of the saturation point for binding U-CLL mAb068 for both peptides. However, fitting of the partial sigmoids led to KD estimates in the micromolar range (~110 μmol/L for E6-3K and ~60 μmol/L for D2-3K). Both PhD-068-4-4K and PhD-068-4-Btn had robust SPR binding signals for U-CLL mAb068, whereas peptide D4-3K did not (not shown); these were consistent with the biotinylated peptide binding ELISA findings. However, thermodynamic analysis indicated these interactions were weaker than E6-3K and that there was little specificity for U-CLL mAb068 over M-CLL mAb169 (Figures 4C, 4D).
Figure 4.
Equilibrium SPR analysis of peptide binding to U-CLL mAb068 or M-CLL mAb 169. (A) E6-3K; (B) D2-3K; (C) PhD-068-4-4K; (D) PhD-068-4-Btn.
Thus, U-CLL mAb068 participated in a broad range of low-affinity interactions. The disparity in the off-phage and on-phage binding behaviors may be due to polyvalency effects on the phage surface (~50 copies of each peptide are displayed for the peptide isolated in this report; ~5 copies/phage for peptides from the PhD library in the previous report) (26).
By contrast, PhD-169-8-4K showed a strong, robust binding signal to M-CLL mAb169 (Supplementary Figure S2). The binding behavior of this peptide obeyed 1:1 Langmuir binding kinetics, and both kinetic and equilibrium analyses were in agreement, resulting in KD values of 32 nmol/L and 31 nmol/L. Furthermore, this interaction was highly specific because no significant binding was observed against U-CLL mAb068 (not shown). It is interesting to note that high affinity peptides could be readily isolated against M-CLL mAb 169, but not against U-CLL mAb 068 using the PhD library.
DISCUSSION
Thus we have selected and characterized peptides that bind an antibody from a CLL B-cell clone with a surface membrane Ig coded by germline IGHV-D-J and IGLVκ-Jκ segments, finding that this U-CLL mAb recognizes a diverse set of peptide sequences, some of which cluster into conserved families. These U-CLL mAb-peptide interactions are generally of modest micromolar affinities as determined by surface plasmon resonance and contrast with the high affinity (nanomolar) interaction of an M-CLL mAb with a phage-derived peptide.
These findings indicate that some U-CLL mAbs bind with relative selectivity to peptides of defined and discrete structure, suggesting that selection for reactivity with these epitopes occurred in normal B cells giving rise to CLL in patients. These epitopes may be environmental or self in nature, depending on the type of CLL mAb (1,9,10). Consistent with this are several recent studies analyzing the reactivities of U-CLL and M-CLL mAbs with foreign antigens, self- antigens and peptides. Hoogeboom et al. have shown that a specific stereotyped BCR expressing a mutated IGHV3-07 reacts with β-(1,6)-glucan, a major antigenic determinant of yeasts and filamentous fungi (30), and Binder et al. (31) and Dühren-von Minden et al. (32) have identified peptides bound by an array of CLL mAbs (both U-CLL and M-CLL) that recognize autoepitopes in human Igs which may, after BCR binding, deliver survival signals to CLL B cells and to their precursors. The data reported here provide biochemical information about the level of specificity and the strength of some of these interactions. This information may be of value in identifying and targeting B lymphocytes expressing specific BCRs in CLL patients and in healthy individuals with monoclonal B lymphocytosis.
Supplemental Data
ACKNOWLEDGMENTS
This work was supported in part by grants from the United States National Institutes of Health (CA81554 to N Chiorazzi and CA155472 to JR Lai), and by startup funds from the Albert Einstein College of Medicine (to JR Lai). The Karches Foundation, the Nash Family Foundation, the Marks Foundation, the Jerome Levy Foundation, the Leon Levy Foundation and the Mildred and Frank Feinberg Foundation also provided support (to KR Rai and N Chiorazzi). We thank Gregory Weiss (University of California – Irvine) for providing the pVIII library, Samantha Wilner for technical assistance with peptide purification and Huiyong Cheng for assistance with SPR experiments.
Footnotes
Online address: http://www.molmed.org
DISCLOSURE
The authors declare that they have no competing interests as defined by Molecular Medicine, or other interests that might be perceived to influence the results and discussion reported in this paper.
REFERENCES
- 1.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 2.Zenz T, Mertens D, Kuppers R, Dohner H, Stilgenbauer S. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat Rev Cancer. 2010;10:37–50. doi: 10.1038/nrc2764. [DOI] [PubMed] [Google Scholar]
- 3.Fais F, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–25. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghiotto F, et al. Remarkably similar antigen receptors among a subset of patients with chronic lymphocytic leukemia. J Clin Invest. 2004;113:1008–16. doi: 10.1172/JCI19399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tobin G, et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood. 2004;104:2879–85. doi: 10.1182/blood-2004-01-0132. [DOI] [PubMed] [Google Scholar]
- 6.Stamatopoulos K, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood. 2007;109:259–70. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 7.Agathangelidis A, et al. Stereotyped B-cell receptors in one-third of chronic lymphocytic leukemia: a molecular classification with implications for targeted therapies. Blood. 2012;119:4467–75. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Messmer BT, et al. Multiple distinct sets of stereotyped antigen receptors indicate a role for antigen in promoting chronic lymphocytic leukemia. J Exp Med. 2004;200:519–25. doi: 10.1084/jem.20040544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiorazzi N, Ferrarini M. B cell chronic lymphocytic leukemia: lessons learned from studies of the B cell antigen receptor. Annu Rev Immunol. 2003;21:841–94. doi: 10.1146/annurev.immunol.21.120601.141018. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson FK, Caligaris-Cappio F. Chronic lymphocytic leukemia: revelations from the B-cell receptor. Blood. 2004;103:4389–95. doi: 10.1182/blood-2003-12-4312. [DOI] [PubMed] [Google Scholar]
- 11.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: a bird of a different feather. J Clin Oncol. 1999;17:399–408. doi: 10.1200/JCO.1999.17.1.399. [DOI] [PubMed] [Google Scholar]
- 12.Broker BM, et al. Chronic lymphocytic leukemic cells secrete multispecific autoantibodies. J Autoimmun. 1988;1:469–81. doi: 10.1016/0896-8411(88)90068-6. [DOI] [PubMed] [Google Scholar]
- 13.Sthoeger ZM, et al. Production of autoantibodies by CD5-expressing B lymphocytes from patients with chronic lymphocytic leukemia. J Exp Med. 1989;169:255–68. doi: 10.1084/jem.169.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borche L, Lim A, Binet JL, Dighiero G. Evidence that chronic lymphocytic leukemia B lymphocytes are frequently committed to production of natural autoantibodies. Blood. 1990;76:562–9. [PubMed] [Google Scholar]
- 15.Herve M, et al. Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J Clin Invest. 2005;115:1636–43. doi: 10.1172/JCI24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damle RN, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 17.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig VH genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 18.Rawstron AC, et al. Inherited predisposition to CLL is detectable as subclinical monoclonal B-lymphocyte expansion. Blood. 2002;100:2289–90. doi: 10.1182/blood-2002-03-0892. [DOI] [PubMed] [Google Scholar]
- 19.Ghia P, et al. Monoclonal CD5+ and CD5-B-lymphocyte expansions are frequent in the peripheral blood of the elderly. Blood. 2004;103:2337–42. doi: 10.1182/blood-2003-09-3277. [DOI] [PubMed] [Google Scholar]
- 20.Landgren O, et al. B-Cell clones as early markers for chronic lymphocytic leukemia. N Engl J Med. 2009;360:659–67. doi: 10.1056/NEJMoa0806122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murase K, et al. EF-Tu binding peptides identified, dissected, and affinity optimized by phage display. Chem Biol. 2003;10:161–8. doi: 10.1016/s1074-5521(03)00025-5. [DOI] [PubMed] [Google Scholar]
- 22.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302:575–81. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 23.Torres M, May R, Scharff MD, Casadevall A. Variable-region-identical antibodies differing in isotype demonstrate differences in fine specificity and idiotype. J Immunol. 2005;174:2132–42. doi: 10.4049/jimmunol.174.4.2132. [DOI] [PubMed] [Google Scholar]
- 24.Morris L, et al. Isolation of a human anti-HIV gp41 membrane proximal region neutralizing antibody by antigen-specific single B cell sorting. PLoS ONE. 2011;6:e23532. doi: 10.1371/journal.pone.0023532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brezinschek HP, et al. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J Clin Invest. 1997;99:2488–501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seiler T, et al. Characterization of structurally defined epitopes recognized by monoclonal antibodies produced by chronic lymphocytic leukemia B cells. Blood. 2009;114:3615–24. doi: 10.1182/blood-2009-01-197822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Houten NE, Zwick MB, Menendez A, Scott JK. Filamentous phage as an immunogenic carrier to elicit focused antibody responses against a synthetic peptide. Vaccine. 2006;24:4188–200. doi: 10.1016/j.vaccine.2006.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stevenson FK, Krysov S, Davies AJ, Steele AJ, Packham G. B-cell receptor signaling in chronic lymphocytic leukemia. Blood. 2011;118:4313–20. doi: 10.1182/blood-2011-06-338855. [DOI] [PubMed] [Google Scholar]
- 29.Keitel T, et al. Crystallographic analysis of anti-p24 (HIV-1) monoclonal antibody cross-reactivity and polyspecificity. Cell. 1997;91:811–20. doi: 10.1016/s0092-8674(00)80469-9. [DOI] [PubMed] [Google Scholar]
- 30.Hoogeboom R, et al. A mutated B cell chronic lymphocytic leukemia subset that recognizes and responds to fungi. J Exp Med. 2013;210:59–70. doi: 10.1084/jem.20121801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Binder M, et al. CLL B-cell receptors can recognize themselves: alternative epitopes and structural clues for autostimulatory mechanisms in CLL. Blood. 2013;121:239–41. doi: 10.1182/blood-2012-09-454439. [DOI] [PubMed] [Google Scholar]
- 32.Dühren-von Minden M, et al. Chronic lymphocytic leukaemia is driven by antigen-independent cell-autonomous signalling. Nature. 2012;489:309–12. doi: 10.1038/nature11309. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.