Abstract
Endophytic fungi were isolated from healthy leaf and root samples of Taraxacum coreanum. Of the 72 isolates recovered, 39 were from leaves and 33 from roots with an isolation frequency of 54% and 46%, respectively. Based on ITS sequence analysis, 72 isolates were classified into 19 genera of which 17 were under the phylum Ascomycota and 2 were under Basidiomycota. Diverse genera were found and Alternaria, Cladosporium, Fusarium and Phoma were dominant. Out of 19 genera, Apodus, Ceriporia, Dothideales, Leptodontidium, Nemania, Neoplaconema, Phaeosphaeria, Plectosphaerella and Terfezia were new to Korea. Seventy two isolates were screened for antifungal activity, of which 10 isolates (14%) were found active at least against one of the tested fungi. Isolate 050603 had the widest antifungal spectra of activity, and isolates 050592 and 050611 were active against three plant pathogenic fungi.
Keywords: Antifungal activity, Endophytic fungi, ITS sequence, Taraxacum coreanum
Endophytes are microorganisms that live asymptomatically within plant tissues (Miguel et al., 2002). Fungal endophytes can colonize internal plant tissues without causing apparent harm to their host plant. Fungal endophytes are being recognized for their potential role as biological control agents and as metabolite sources of medical and agricultural importance. Most investigations into their occurrence have involved in trees and shrubs of north-temperate regions (Duran et al., 2005).
Endophytes were recognized for the first time in New Zealand and USA in the end of 1940's when livestock grazing on some pastures suffered from serious, unexplained symptoms. Thereafter, toxic compounds produced by endophytes were found from the grasses on these pastures (Bony et al., 2001). Recently number of studies has reported that, in addition to increasing plant resistance to herbivores, endophytes may also enhance plant competitive abilities, germination success, and resistance to drought and water stress. Endophytes, especially fungi, are increasingly recognized as important mediators of interaction between plants and their competitors, seed dispersers, herbivores and pathogens (Santos et al., 2003). Different studies have established that the composition of the endophytic communities varies among plants and plant organs (Santos et al., 2003). Investigations of species composition in woody plants have shown that a large number of endophytic species can be recovered from a single host species. Studies based largely on pure culture of surface-sterilized stems and leaves have revealed a broad diversity of fungal species existing subcuticularly or within the tissues of many kinds of healthy plants. Usually one and, at most a few species dominate the community, while the majority of the species are rare (Carroll, 1998).
The endophytic microorganisms are not considered as saprophytes since they are associated with living tissues, and may in some way contribute to the well being of the plant. That is, the plant is thought to provide nutrients to the microbe, while the microbe may produce factors that protect the host plant from attack by animals, insects and microbes (Yang et al., 1994). There are many reports demonstrating that many bioactive compounds could be produced by endophytic microorganisms (Huang et al., 2001). In search for bio-control agent, endophytic fungi bore antibiotics, volatile antibiotics, antioxidants, and showed activity against several plant pathogenic fungi (Strobel and Daisy, 2003).
The present study was conducted to isolate and identify fungal endophytes from living symptomless tissues of white dandelion (Taraxacum coreanum Nakai) which is a perennial compositae plant, whose roots are used in traditional medicine and young leaves are used as vegetables in Korea (Koo et al. 2004, Lee 1980) and to evaluate the antifungal activity of their secondary metabolites.
Materials and Methods
Sample collection
Healthy roots and leaves of white dandelion (Taraxacum coreanum) were collected from Chungnam province, Republic of Korea in 2005. Plants were identified based on their morphological characteristics. The fresh plant samples were brought to the laboratory in sterile polyethylene bags and stored in a refrigerator prior to isolation of endophytic fungi.
Isolation of endophytic fungi
To determine the distribution of endophytic fungi within white dandelion plant parts, the sampled leaves and roots were washed in running tap water and cut into small pieces of 0.5 cm. Tissue pieces were rinsed in 0.1% Tween 20 for 30 sec, then in 2% sodium hypochlorite for 5 min and then washed in sterile distilled water for 5 min. Next the tissue pieces were surface sterilized in 70% ethanol for 5 min and washed with sterile distilled water. The surface sterilized samples were allowed to dry on paper towel in a laminar air flow chamber. Ten fragments from each leaf and root were placed onto dichloran rose bengal cholamphenicol agar (DRBC) and potato dextrose agar (PDA) media. After incubation at 25℃ for 5, 10 and 25 days, individual hyphal tips of the developing fungal colonies were collected and placed onto PDA media and incubated for 8~10 days and checked for culture purity. Eventually pure cultures were transferred to PDA slant tubes and 20% glycerol stock solution.
DNA extraction
The isolates were grown in V8 broth medium for 7 days at 25℃ and 130 rpm. Mycelia were collected from the cultures by filtration and transferred to sterile plastic tubes. These samples were frozen at -70℃ for minimum 1 hour and then freeze dried in a freeze dryer (Eyela FDU-1000, Japan) and stored in hygrometer (Sanpla corporation). The frozen mycelia were ground in 1.5 eppendorf tube. Extraction buffer 200 mM Tris-HCL (pH 8.0), 200 mM NaCl, 30 mM EDTA, 0.5% SDS and proteinase K were added to each tube and incubated at 37℃ for 1 hr. Samples were extracted with 2 × CTAB solution (2% CTAB (w/v), 100 mM Tris-HCL (pH 8.0), 20 mM EDTA (pH 8.0), 1.4M NaCl, 1% PVP (Polyvinylpyrrolidone) and chloroform:isoamyl alcohol (24 : 1), followed by centrifuge for 10 min. Genomic DNA was washed with 70% ethanol, dried and re-suspended in 50 microgram sterile distilled water. RNase was added to each sample to remove RNA, followed by incubation at 37℃ for 2 hrs. Size and extracted DNA were examined by electrophoresis through a 0.7% agarose gel in TBE buffer stained with ethidium bromide and visualized with UV light. DNA were stored at 4℃ until used.
Polymerase chain reaction (PCR)
The DNA of all fungal samples were amplified by polymerase chain reaction (PCR) using two different ITS primers. The fungal ITS region was amplified using the universal primers ITS1 and ITS5. Amplifications reactions were performed in a total volume of 50 µl containing 10 × buffer 5 µl, 10 × BSA buffer 5 µl, dNTP 5 µl, 1 µl of each primer, 0.3 µl of Taq polymerase, 1 µl of genomic DNA and 31.7 µl of distilled water. PCR amplification was carried out in i-cycler (BIO-RAD, USA) for 30 cycles of 94℃ for 1 min denaturing, 55℃ for 1 min annealing and 72℃ for 1.30 min extension. Initial denaturing at 94℃ was extended to 5 min and the final extension was at 72℃ for 10 min.
Electrophoresis
Amplification products were separated by electrophoresis in gels containing 1.0% agarose. The electrophoresis was run in 0.5 × TBE-buffer and the amplification products were visualized by ethidium bromide staining under UV light. The lengths of the amplification products were estimated by comparing to 20, 50 and 100-bp DNA ladder. The PCR product was added to 100 µl of direct purification buffer in eppendorf tube and purified using the wizard PCR prep. DNA.
DNA sequencing and data analysis
DNA sequence was done using the ITS1 and ITS5 primer. Analyses of sequences were performed with the basic sequence alignment BLAST program run against the database (National Center for Biotechnology Information website, http://www.ncbi.nlm.nih.gov). Then phylogenetic tree were calculated and neighbour-joining algorithms were done, using PHYDIT program version 3.0. The nucleotide sequences obtained in this study have been submitted to the GenBank and assigned accession numbers.
Test organisms for antifungal activity
Seven phytopathogenic fungi were used to evaluate antifungal activity of endophytic fungi isolated from white dandelion plant. They were Alternaria panax, Botrytis cinerea, Colletrotichum gloeosporioides, Fusarium oxysporum, Phytophthora capsici, Pythium ultimum and Rhizoctonia solani. The phytopathogenic fungi were collected from the culture stock of Plant Pathology laboratory, Chungnam National University, Daejeon, Republic of Korea.
Screening for antifungal activity
Fungal mycelial-disks prepared from growing margin of cultures of test fungal isolates were placed in the centre of PDA plates and three endophytes disks were placed at 30 mm distance from the disk in the center. The plates were incubated at 25℃ for 7~10 days. Antifungal activity was indicative as mycelial growth of test fungus prohibited in the direction of active endophytic fungus. The level of inhibition was calculated by subtracting the distance (mm) of fungal growth in the direction of an antagonist colony from the fungal growth radius. The width of inhibition zones between the pathogen and the endophytes was evaluated as > 8 mm (+++, strong inhibition), 2~8 mm (++, moderate inhibition) and < 2 mm (+, weak inhibition).
Results and Discussion
Fungal isolation, identification and diversity
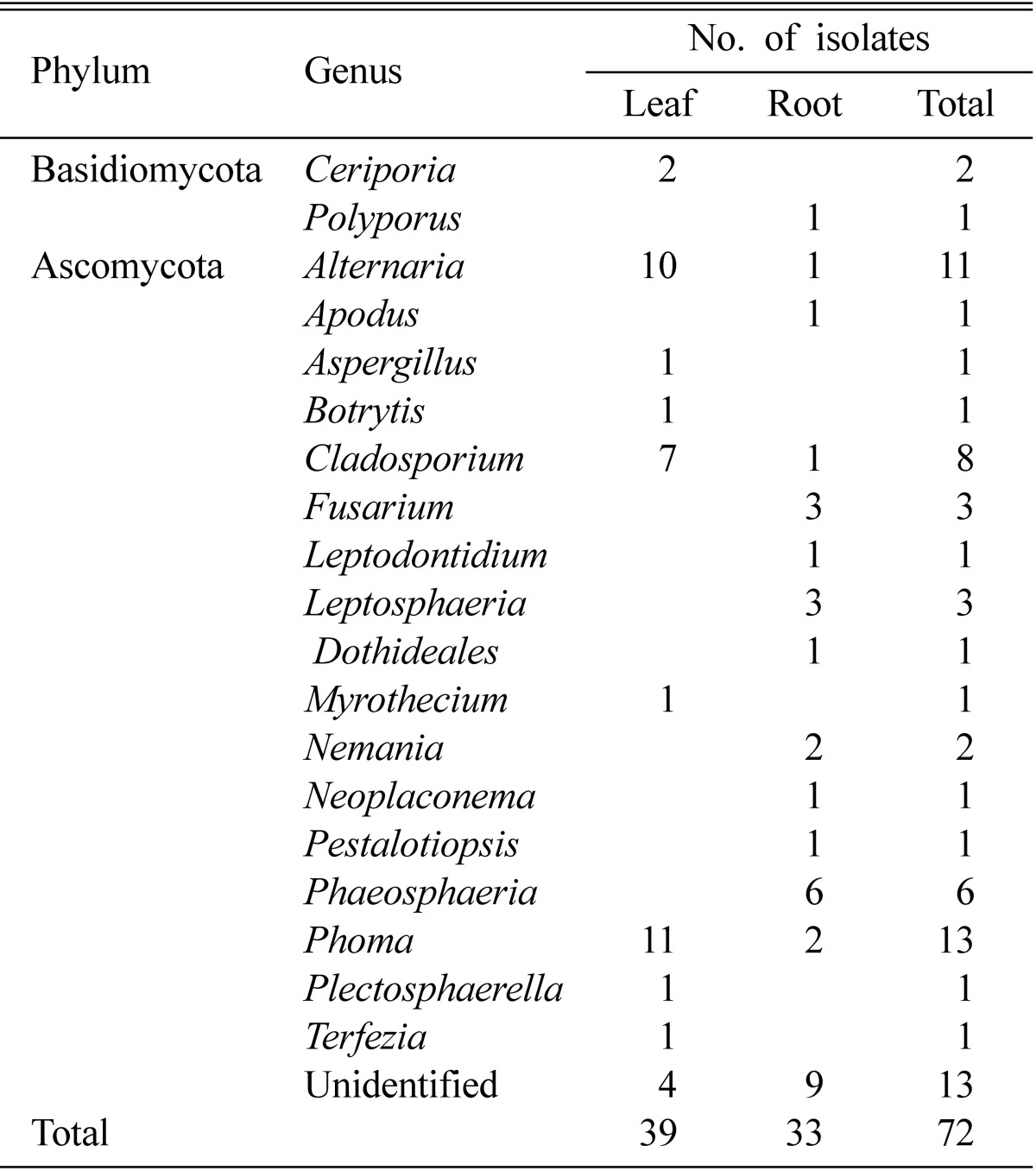
A total of 72 fungal isolates was recovered from the leaf and root samples of white dandelion plant. There was little difference in the isolation frequencies from leaf and root samples from white dandelion. Thirty nine (54%) isolates were originated from leaf samples and 33 (46%) were from roots (Table 1).
Table 1.
Phylum and genus diversity of endophytic fungi in white dandelion plant
Molecular methods for fungal identification and classification were applied. PCR-fragments from the ITS were amplified for DNA sequence analysis in order to discriminate isolates at or close to the species level (Table 2). BLAST database searches were performed with full-length ITS-fragments as queries to reveal relationships to published sequences. Sequenced isolates and the BLAST searches are listed in Table 1. When the white dandelion fungal endophytes sequences and the respective matches from the BLAST searches were aligned pairwise similarity scores were obtained that was mostly > 90% (Table 2).
Table 2.
Endophytic fungi isolated from white dandelion in Korea
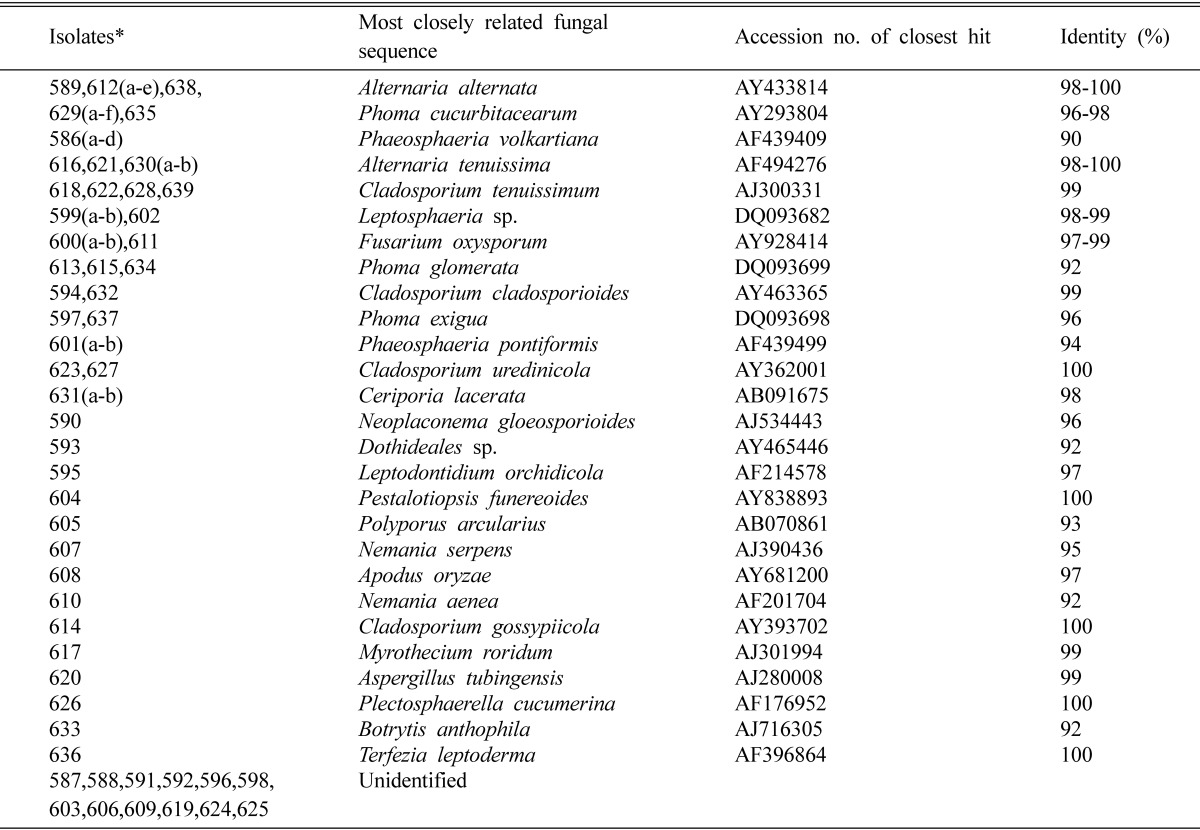
*All isolate number started with 050.
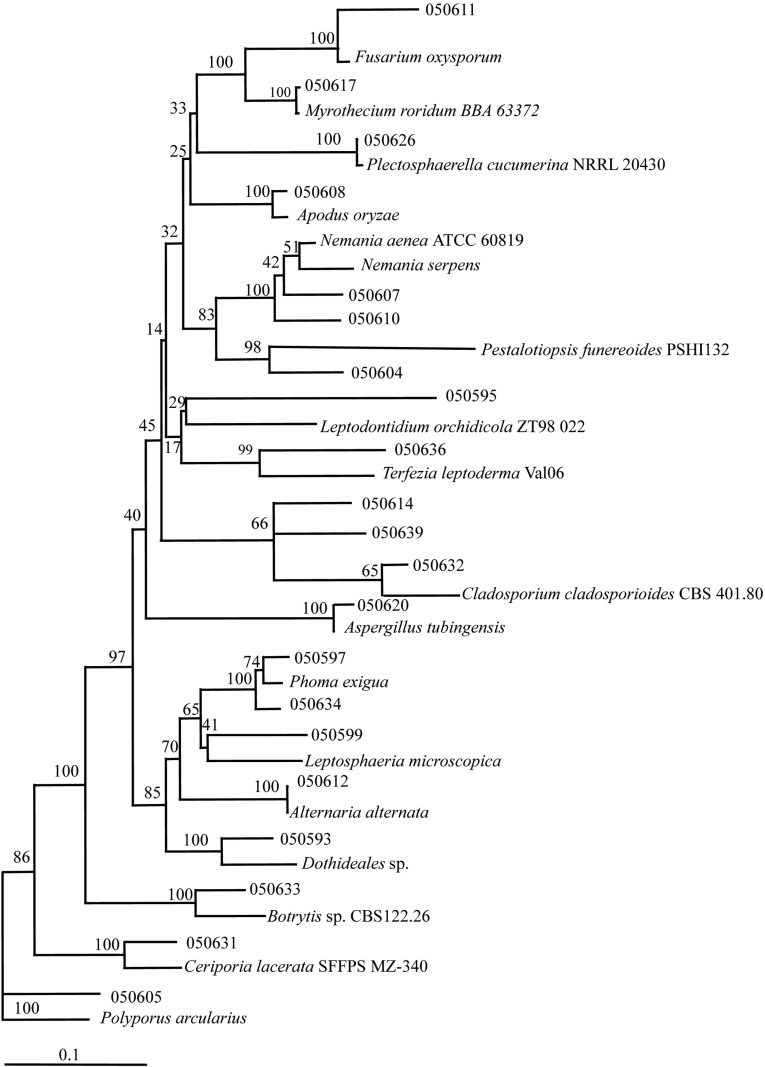
Nineteen different genera were discovered from the white dandelion, of which 17 were under the phylum Ascomycota and two (Ceriporia and Polyporus) were from the phylum Basidiomycota (Table 2). Among the fungi encountered, the following 9 genera were new to Korea (Annonymous, 2004); Apodus, Ceriporia, Dothideales, Leptodontidium, Nemania, Neoplaconema, Phaeosphaeria, Plectosphaerella and Terfezia. A phylogenetic tree was constructed to illustrate the overall molecular diversity of the endophytic fungi associated with white dandelion and to check sequence data within a phylogenetic context (Fig. 1).
Fig. 1.
Phylogenetic tree showing the relationship between white dandelion endophytic fungi and reference strains. The tree was constructed on the basis of rDNA sequence (ITS1 and ITS5) fragment by using neighbour-joining method. The boostrap analysis was performed with 1000 repetations. Table 1 showing the details of endophytic fungi.
A total of 27 different species were identified based on the ITS sequence analysis. The predominant fungi were Alternaria alternata (n = 7) and Phoma cucurbitacearum (n = 7) followed by Alternaria tenuissima (n = 4), Cladosporium tenuissimum (n = 4) and Phaeosphaeria volkartiana (n = 4). From the database of GenBank, 100% similar sequence were matched to isolates 050589, 050612 and 050638 with Alternaria alternata (AY433814), isolates 050616, 050621 and 050630 with Alternaria tenuissima (AF494276), isolates 050623, 050627 with Cladosporium uredinicola (AY362001), isolate 050604 with Pestalotiopsis funereoides (AY838893), isolate 050614 with Cladosporium gossypiicola (AY393702), isolate 050626 with Plectosphaerella cucumerina (AF176952) and isolate 050636 with Terfezia leptoderma (AF396864). By neighbour-joining algorithm, isolates 050587-88, 050591-92, 050596, 050598, 050603, 050606, 050609, 050619 and 050624-25 could not be identified.
Stefan et al. (2000) also identified 26 different fungal ITS sequences from common reed, 23 of them probably from Ascomycetes and 3 of them from Basidiomycetes. Carter et al. (1999) were recovered in a similar proportion of Ascomycetes and Basidiomycetes from the rhizospheres of wheat and oat. Diversity of endophytic fungal community of cacao was evaluated by Rubini et al. (2005) in Brazil. They isolated 150 endophytic fungi and characterized by rDNA sequences. They showed that the cultivable endophytic fungi associated with cacao cultivars were mainly to Ascomycetes group.
Estimates of the global diversity of endophytic fungi have run from minimal to a million of species (i.e., the maximal estimate based on four endophytes per each of 250,000 plant species) (Rebecca et al., 2004). Many factors affect the structure and species composition of the microbial communities that colonize roots, stems, branches and leaves. Endophytic communities vary spatially in the plant or may be dependent on the interaction with other endophytic or pathogenic microorganisms (Rubini et al., 2005).
From our experiment, isolated fungi were a little higher in the leaf samples and all the fungi were isolated from healthy tissues. Tejesvi et al. (2005) isolated endophytic fungi from healthy tissues of Terminalia arjuna, an important ethno pharmacological plant extensively used in ayurvedic medicines to treat heart ailments. Seena and Sridhar (2004) isolated endophytic fungi from cotyledons, seed coats, roots, stems and leaves of 2 sand dune wild legumes from the south east coast of India and endophytic fungi colonized over 90% of root, stem and leaf segments. Eight Cladosporium isolates were identified in our study. The genus Cladosporium, that exhibits interactions with plants ranging from facultative biotrophs, to endophytes and epiphytes, was frequently isolated from reed and established their taxonomic placement at the species level (Stefan et al., 2000).
Antifungal activity
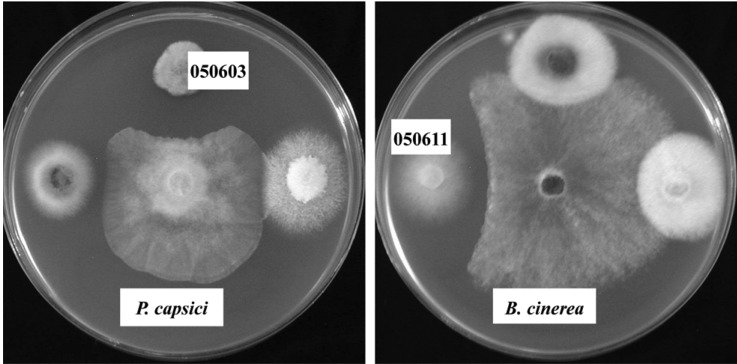
The antifungal activity of endophytic fungi isolated from the white dandelion was shown in Table 3. Majority of the fungi did not show any antifungal activity against six plant pathogenic fungi and all of the fungal endophytes were inactive against R. solani. Similar results were found by Taechowisan et al. (2003). They isolated 330 endophytic actinomycetes, out of them only three were strongly inhibited Colletotrichum musae, five were very active against F. oxysporum and two strongly inhibited growth of both the test fungi. In this experiment, the isolate 050603 had the widest antifungal spectra of activity (Table 3) and strongly inhibited the growth of P. capsici (Fig. 2), and two fungi such as isolate 050592 and 050611 showed moderate to weak antifungal activity against 3 plant pathogenic fungi. There were no endophytic fungi which were active against more than four pathogenic fungi. Aghighi et al. (2004) tested 110 isolates for antifungal activity and found 14 isolates which showed activity against at least one fungal isolate. Sardi et al. (1992) also observed same results. They tested 500 endophytic isolates for antimicrobial activity of 10 groups and found that all groups had antimicrobial activity against one or the other organisms, but not the both. Thus most of their isolates had narrow antimicrobial spectrum.
Table 3.
Antifungal activity of ten selected endophytic fungi against seven plant pathogenic fungi
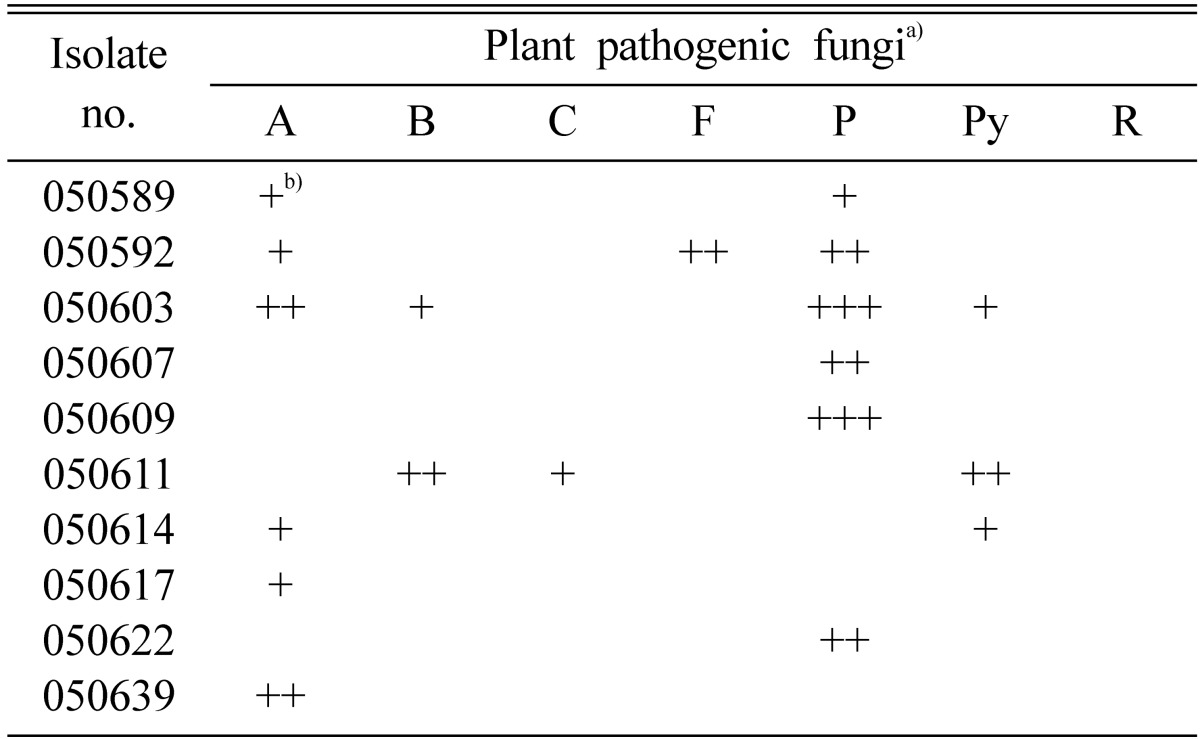
a)A, Alternaria panax; B, Botrytis cinerea; C, Colletotrichum gloeosporioides; F, Fusarium oxysporum; P, Phytophthora capsici; Py, Pythium ultimum; R, Rhizoctonia solani.
b)Inhibition zone. +, < 2 mm; ++, 2~8 mm; +++, > 8 mm.
Fig. 2.
Isolate 050603 (left) showing antifungal activity against Phytophthora capsici and isolate 050611 (right) showing antifungal activity against Botystis cinerea.
Acknowledgement
This study was supported by the BioGreen 21 Program of the Rural Development Administration, Korea.
References
- 1.Aghighi S, Bonjar GHS, Rawashdeh R, Batayneh S, Saadoun I. First report of antifungal spectra of activity of Iranian actinomycetes strains against Alernaria solani, Alternaria alternata, Fusarium solani, Phytophthora megasperma, Verticillium dahliae and Saccharomyces cerevisiae. Asian J Plant Sci. 2004;3:463–471. [Google Scholar]
- 2.Anonymous. List of Plant Diseases in Korea. 4th edition. Korea: The Korean Society of Plant Pathology; 2004. [Google Scholar]
- 3.Bony S, Pichon N, Ravel C, Durix A, Balfourier F, Guillanumin JJ. The relationship between mycotoxin synthesis and isolate morphology in fungal endophytes of Lolium perenne. New Phytologist. 2001;152:125. doi: 10.1046/j.0028-646x.2001.00231.x. [DOI] [PubMed] [Google Scholar]
- 4.Carroll G. Fungal endophytes in stems and leaves: from latent pathogen to mutualistic symbiont. Ecology. 1998;69:2–9. [Google Scholar]
- 5.Carter JP, Spink J, Cannon PF, Daniels MJ, Osbourn AE. Isolation, characterization, and sensitivity of a diverse collection of Cereal-Root-Colonizing fungi. Appl Environ Microbiol. 1999;65:3364–3372. doi: 10.1128/aem.65.8.3364-3372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duran EL, Plopper LD, Ramallo JC, Grandi RAP, Giancoli ACH, Azevedo JL. The foliar fungal endophytes on citrus limon in Argentina. NRC Res. 2005;83:350–355. [Google Scholar]
- 7.Huang Y, Wang J, Li G, Zheng Z, Su W. Antitumor and antifungal activities in endophytic fungi isolated from pharmaceutical plants Taxus mairei, Cephalotaxus fortunei and Torreya grandis. FEMS Immunol Med Microbiol. 2001;31:163–167. doi: 10.1111/j.1574-695X.2001.tb00513.x. [DOI] [PubMed] [Google Scholar]
- 8.Koo HN, Hong SH, Kim CH, Yoo YH, Kim HM. Taraxacum officinale induces cytotoxicity through TNF-alpha and IL-alpha secretion in Hep G2 cells. Life Sci. 2004;74:1149–1157. doi: 10.1016/j.lfs.2003.07.030. [DOI] [PubMed] [Google Scholar]
- 9.Lee TB. Illustrated Flora of Korea. Seoul, Korea: Hyang Mun Sa; 1980. [Google Scholar]
- 10.Gamboa MA, Laureano S, Bayman P. Measuring diversity of endophytic fungi in leaf fragments: Does size matter? Mycopathologia. 2002;156:41–45. doi: 10.1023/a:1021362217723. [DOI] [PubMed] [Google Scholar]
- 11.Ganley RJ, Brunsfeld SJ, Newcombe G. A community of unknown, endophytic fungi in western white pine. Proc Natl Acad Sci U S A. 2004;101:10107–10112. doi: 10.1073/pnas.0401513101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubini MR, Silva-Ribeiro RT, Pomella AWV, Maki CS, Araujo WL, Santos DR, Azevedo JL. Diversity of endophytic fungal community of cacao (Theobroma cacao L.) and biological control of Crinipellis perniciosa, causal agent of witches broom disease. Int J Biol Sci. 2005;1:24–33. doi: 10.7150/ijbs.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos RMG, Rodrigues-Fo E, Rocha WC, Teixeira MFS. Endophytic fungi from Melia aedarach. World J Microbiol Biotechnol. 2003;19:767–770. [Google Scholar]
- 14.Sardi P, Saracchi M, Quaroni S, Petrolini B, Borgonovi GE, Merli S. Isolation of endophytic Streptomyces strains from surface sterilized roots. Appl Environ Microbiol. 1992;58:2691–2693. doi: 10.1128/aem.58.8.2691-2693.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seena S, Sridhar K.R. Endophytic fungal diversity of 2 sand dune wild legumes from the southwest coast of India. Can J Microbiol. 2004;50:1015–1021. doi: 10.1139/w04-094. [DOI] [PubMed] [Google Scholar]
- 16.Stefan GR, Leibinger W, Ernst M, Mendgen K. Genetic diversity of fungi closely associated with common reed. New Phytologist. 2001;149:589–598. doi: 10.1046/j.1469-8137.2001.00038.x. [DOI] [PubMed] [Google Scholar]
- 17.Strobel G, Daisy B. Bioprocessing for microbial endophytes and their natural products. Microbiol Mol Biol Rev. 2003;67:491–502. doi: 10.1128/MMBR.67.4.491-502.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taechowisan T, Peberdy JF, Lumyong S. Isolation of endophytic actinomycetes from selected plants and their antifungal activity. World J Microbiol Biotechnol. 2003;19:381–385. [Google Scholar]
- 19.Tejesvi M, Nahesh B, Nalini M, Prakash H, Kini K, Subbiah V, Shetty H. Endophytic fungal asemblanges from inner bark and twig of Terminalia arjuna W. & A. (Combretaceae) World J Michobiol Biotechnol. 2005;21:1535–1540. [Google Scholar]
- 20.Yang X, Strobel G, Stierle A, Hess WM, Lee J, Clardy J. A fungal endophyte-tree relationship: Phoma sp. in Taxus wallachiana. Plant Sci. 1994;102:1–9. [Google Scholar]