Abstract
Label free imaging of the chemical environment of biological specimens would readily bridge the supramolecular and the cellular scales, if a chemical fingerprint technique such as Raman scattering can be coupled with super resolution imaging. We demonstrate the possibility of label-free super-resolution Raman imaging, by applying stochastic reconstruction to temporal fluctuations of the surface enhanced Raman scattering (SERS) signal which originate from biomolecular layers on large-area plasmonic surfaces with a high and uniform hot-spot density (>1011/cm2, 20 to 35 nm spacing). A resolution of 20 nm is demonstrated in reconstructed images of self-assembled peptide network and fibrilated lamellipodia of cardiomyocytes. Blink rate density is observed to be proportional to the excitation intensity and at high excitation densities (>10 kW/cm2) blinking is accompanied by molecular breakdown. However, at low powers, simultaneous Raman measurements show that SERS can provide sufficient blink rates required for image reconstruction without completely damaging the chemical structure.
Plasmonic structures made the acquisition of Raman spectra possible from sub-monolayer coverage molecular films, a technique known as Surface-Enhanced Raman Spectroscopy (SERS)1,2,3. Using SERS, single molecule level sensitivity can be achieved4,5,6,7, attributed earlier to extremely high enhancement factors of 109 to 1012, although it was later claimed that much lower enhancement factors (106 to 108) may be sufficient for such observations8. SERS signal typically shows a rapid blinking behavior, which can be captured with a camera or spectrometer. This non-stationary nature of the SERS signal and the rich spectral information contained in acquired spectra, in principle, provides an opportunity to exploit the time dependent SERS signal for chemically-resolved, label-free super-resolution stochastic imaging9,10,11,12,13,14. Stochastic image reconstruction methods are versatile and fluorescence from multiple labels have been used to simultaneously image various components of cellular structures15. Raman spectra are conductive to label-free multicolor imaging using similar mathematical methods. However, for SERS to be a viable way of reconstructing the nanoscale structure of a biomolecular layer rather than the field distribution of the underlying hot-spot itself, a large number of closely packed hot-spots is required within a focal spot with an area defined by the diffraction limit. Previously, meta-surfaces with periodicities larger than the diffraction limited spot size have been shown to produce high and uniform enhancement16. Recently, we used an engineered meta-surface with sub-wavelength periodicity to demonstrate high uniformity and high spatial resolution confocal SERS imaging17. In this article, we present two variations of an easy-to-fabricate SERS substrate that exhibit a high density of hot-spots while allowing single-molecule level Raman spectroscopy. Traditionally, Coherent Anti-Stokes and Stimulated Raman scattering have been used for label-free confocal Raman imaging of biological samples with diffraction limited resolution18,19,20. In contrast, the temporal fluctuations of the SERS signal and high enhancement uniformity featured by the SERS substrates presented here, allow super-resolution stochastic Raman imaging of nanostructured biological samples, a technique fundamentally different from confocal imaging.
Results
Plasmonic surfaces utilized in this study are prepared through the use of spontaneous self-organization of silver into nano-islands by de-wetting upon controlled vacuum evaporation (see Methods). Although semi-continous metal films have been previously used for SERS21, to distinguish the layer structure and geometry presented here we will refer to the surfaces as Spontaneously Organized Metasurfaces (SOMs). The SOM surface exhibits features about an order of magnitude smaller than the free-space wavelength of 532 nm. Such a surface can be approximated by an effective medium theory and behaves like a surface with uniform optical properties22,23,24. We therefore refer to the plasmonic surface as a meta-surface. A schematic cross-section of the surface is shown in Fig. 1a along with a scanning electron microscope (SEM) image (Fig. 1b), from which a particle diameter distribution (mean of 32 nm and width of ± 8 nm) can be deduced. Atomic force microscopy revealed an island height of 10 nm, separated from a continuous Ag film by a 10–20 nm thick dielectric layer of Al2O3. The island formation and dielectric thickness has been optimized by varying process parameters and matching the Plasmon resonance wavelength close to the working laser wavelength of 532 nm (See Fig. S1 and S2). The surface can be regarded as a scaled down version of the meta-surface described in our previous work17. Computational study of a periodic structure similar to that shown in Fig. 1a, reveals that field enhancement is greatest in the gap between the nanoislands upon excitation near the resonance (Fig. 1c). A non-uniformity of the size distribution of silver nano-islands and gaps between the nanoislands results in the observed microscopic non-uniformity of the optical properties, such as the field enhancement factor. The uniformity of optical properties within a 10 μm diameter area as observed in the surface reflectance is 4%. Confocal Raman map acquired on a SOM treated with methylene blue is used to calculate an intensity histogram, using the Raman range from 270 cm−1 to 2000 cm−1 (Fig. 1d), which shows 14% spatial uniformity of intensity. In order to compare the uniformity of the SOM with other popular techniques used in SERS, confocal Raman maps are collected on the SOM, a surface prepared by layer-by-layer deposition of Ag nanoparticles and metal-insulator-metal substrates prepared by nanosphere lithography (Supplementary Figures S3, S4 and S5). The superior uniformity of the SOM is evidenced by the narrow Gaussian distribution of the SERS signal which compares favorably to the wide log-normal distributions of Raman enhancement (>100% signal non-uniformity) and inherent spatial non-uniformity obtained by the other techniques.
Figure 1.
(a) Schematic description of Self-Organized Metasurface (SOM). (b) Scanning electron micrograph of the SOM is shown (Scale bar 200 nm). The Ag nanoislands are formed due to de-wetting and spontaneous segregation, with approximately 30 nm diameter and 10 nm thicknesses, separated from a continuous Ag film by a 10–20 nm thick dielectric layer (Al2O3). (c) Field profiles show confinement and relative enhancement of field along the surface cross-section (scale bar 10 nm). (d) Confocal Raman map on SOM treated with 1 μM Methylene Blue shows the high uniformity of enhancement (scale bar 2 μm, Raman intensity integrated between 500 cm−1 to 2000 cm−1). (e) Computational modeling allows calculation of the peak enhancement factor for the configuration in (c) (40 nm Ag islands with 10 nm spacing, 10 nm above Ag surface). Experimental data (green curve) is shown for a surface with 20 nm dielectric thickness. (f) The resonance wavelength can be tuned by altering the dielectric thickness during fabrication. As the dielectric thickness increases, a broad resonance around 600 nm remains. A large dielectric thickness approximates the limit case of free standing Ag nanoisland film supported by a dielectric sample or substrate.
Although the fabrication of the SOM is quite simple, it must be noted that a suitable combination of geometry and materials is critical for the observed SERS performance. A similar surface was previously investigated for the presence of propagating and localized plasmons; however, no optimization for SERS or molecular imaging was attempted25. Thicker Ag films are seen not to produce significant SERS enhancement, as island formation is suppressed and continuous films are produced. In addition, evaporation of correct mass thickness of the Ag film results in required height, average diameter and separation of islands, all critical for obtaining optimal resonance wavelength and field enhancement. Computational results indicate that field enhancement factors as large as 4.5 × 103 (corresponding to a SERS enhancement factor of 2 × 107) can be achieved with the right combination of geometrical parameters (Fig. 1e). The dominant plasmonic resonance is observed in the extinction spectrum of the surface and reflectance is found to display an extinction peak around 550 nm for the given set of parameters. The resonance can be adjusted by altering the dielectric thickness (Fig. 1f). The resonance wavelength is matched to the laser excitation wavelength of 532 nm used in our experiments. For the limit case of infinite dielectric thickness, the resonance wavelength converges to around 600 nm for an island width of 40 nm, island thickness of 10 nm, and island spacing of 10 nm. Such a case still possesses a SERS enhancement factor of about 106.
Within a first order semi-classical approximation, the SERS photon generation areal density rate φ(x, y, ωs, t) as observed on a plasmonic substrate is a function of a large number of variables, such as spatial orientation of fields, molecular configurations and hot spots. Also, the SERS signal depends on Raman excitation and scattering frequencies, ωexc and ωs, as well as time t. The time dependence is observed to be strongly dependent on other parameters, such as excitation intensity, local field enhancement factor and chemical composition of imaged structures. At a given time t, the infinitesimal intensity dI = φ(x, y, ωs, t)dA generated at an area dA at a location (x,y) from a molecular component i is given as3
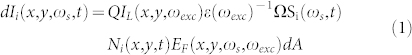 |
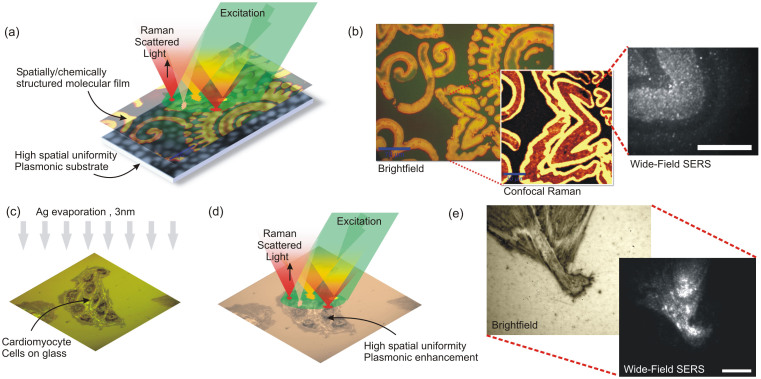
where 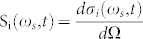 is the differential scattering cross-section of molecular component i, characterizing its Raman spectrum. Here, Q is the overall optical collection and detection efficiency, IL(x,y,ωexc) is the excitation power density, ε(ωexc) is the photon energy at excitation frequency, Ω is the solid-angle collected by the objective, Ni(x,y) is the areal density distribution of molecular component i, and EF(x,y,ωs,ωexc) is the function describing the spatial distribution of SERS enhancement factor (see Fig. 2a for a schematic description). Assuming spatially uniform illumination, IL(x,y,ωexc) = IL0, a wide-field image can be formed using the SERS signal from a spatial pattern contained in the molecular layer Ni(x,y) provided that the SERS enhancement distribution EF(x,y,ωs,ωexc) is spatially constant or highly uniform. In our experimental setup (Fig. S14a) the excitation laser can be focused to illuminate a diffraction limited spot or defocused to illuminate a large diameter spot to perform widefield imaging. A mirror in the optical path can be switched to perform confocal Raman spectroscopy and imaging, or the optical signal can be routed to a camera to perform widefield imaging. The optical configuration can be modified by a beam splitter to record both spectra and widefield images. Illumination power density is calculated by dividing the total excitation power with the average spot diameter as observed by the camera. Although interference effects and speckle pattern formed during wide-field illumination can be observed to be present, uniform illumination assumption is seen to be reasonably satisfied. Under wide field illumination, even with excitation power densities below 1 kW/cm2, a large number of uniformly distributed blinking spots can be observed through wide-field SERS imaging. We confirm that a spatially structured molecular layer is observed through wide-field SERS imaging of Amyloid Inspired Peptides (AIPs) (see Methods, Supplementary Fig. S6) on the SOM (Fig. 2b). The synthesis and characterization of the peptides were given in the literature in details26. Video sequences captured on the surface display a large number of blinking SERS spots, correlated with the presence of the peptides (see Supplementary Video 1). In our system, the minimum time required to acquire a spectrum is 11 msec. Individual blinking events take place spanning a timeframe of up to few seconds. Within each blinking event, the intensity and spectrum of the SERS signal shows fluctuations with tens of millisecond characteristic times as measured by the spectrometer. When measured by a photomultiplier, the blinking events are observed to occur rapidly and display intensity fluctuations even at time scales below 1 μs, with rates and areal densities dependent on excitation power (Supplementary Fig. S7). In addition, SERS images are recorded on a SOM surface uniformly covered with peptides using wide-field illumination and resulting video sequence is processed using stochastic optical reconstruction (STORM)27. The resulting SERS-STORM image (Supplementary Fig. S8) exhibits a spatially uniform distribution of spot locations with no identifiable sharp features, as expected from a uniformly distributed, featureless molecular layer. The nanoscale uniformity within a diffraction limited spot is also measured, and found to display blink events uniformly distributed within the illumination area (250 nm in diameter, 300 μW power, Fig. S9). If the plasmonic modes responsible for Raman enhancement can be regarded as uncoupled or weakly coupled, hot spots can be thought of as operating independently. In such a case, regardless of the details of fluctuation mechanisms, the fluctuations originate from local effects which are uncorrelated. Such an optical signal, i.e. one with uncorrelated nanoscale spatial and temporal structure at the source, can be used for super-resolved stochastic imaging. In an alternate method (Fig. 2c), the biological sample can be prepared on a glass substrate followed by evaporation of a thin Ag layer (3 nm mass thickness, see Methods). Membranes or other cellular structure can be regarded as thin dielectric media, on which Ag segregates into closely packed islands. Therefore, the resulting configuration can be regarded as the limit case of the SOM, where the bottom Ag layer is completely absent. Under wide-field illumination (Fig. 2d), such a sample is observed to exhibit blinking SERS spots correlated with the presence of biological material observed in bright-field images (Fig. 2e). In the absence of Ag overlayer, no blinking spots are observed (see Supplementary Video 2). In addition, without Ag evaporation, a broad auto-fluorescence can be observed to obscure the Raman bands, which cannot be clearly identified during confocal Raman imaging.
is the differential scattering cross-section of molecular component i, characterizing its Raman spectrum. Here, Q is the overall optical collection and detection efficiency, IL(x,y,ωexc) is the excitation power density, ε(ωexc) is the photon energy at excitation frequency, Ω is the solid-angle collected by the objective, Ni(x,y) is the areal density distribution of molecular component i, and EF(x,y,ωs,ωexc) is the function describing the spatial distribution of SERS enhancement factor (see Fig. 2a for a schematic description). Assuming spatially uniform illumination, IL(x,y,ωexc) = IL0, a wide-field image can be formed using the SERS signal from a spatial pattern contained in the molecular layer Ni(x,y) provided that the SERS enhancement distribution EF(x,y,ωs,ωexc) is spatially constant or highly uniform. In our experimental setup (Fig. S14a) the excitation laser can be focused to illuminate a diffraction limited spot or defocused to illuminate a large diameter spot to perform widefield imaging. A mirror in the optical path can be switched to perform confocal Raman spectroscopy and imaging, or the optical signal can be routed to a camera to perform widefield imaging. The optical configuration can be modified by a beam splitter to record both spectra and widefield images. Illumination power density is calculated by dividing the total excitation power with the average spot diameter as observed by the camera. Although interference effects and speckle pattern formed during wide-field illumination can be observed to be present, uniform illumination assumption is seen to be reasonably satisfied. Under wide field illumination, even with excitation power densities below 1 kW/cm2, a large number of uniformly distributed blinking spots can be observed through wide-field SERS imaging. We confirm that a spatially structured molecular layer is observed through wide-field SERS imaging of Amyloid Inspired Peptides (AIPs) (see Methods, Supplementary Fig. S6) on the SOM (Fig. 2b). The synthesis and characterization of the peptides were given in the literature in details26. Video sequences captured on the surface display a large number of blinking SERS spots, correlated with the presence of the peptides (see Supplementary Video 1). In our system, the minimum time required to acquire a spectrum is 11 msec. Individual blinking events take place spanning a timeframe of up to few seconds. Within each blinking event, the intensity and spectrum of the SERS signal shows fluctuations with tens of millisecond characteristic times as measured by the spectrometer. When measured by a photomultiplier, the blinking events are observed to occur rapidly and display intensity fluctuations even at time scales below 1 μs, with rates and areal densities dependent on excitation power (Supplementary Fig. S7). In addition, SERS images are recorded on a SOM surface uniformly covered with peptides using wide-field illumination and resulting video sequence is processed using stochastic optical reconstruction (STORM)27. The resulting SERS-STORM image (Supplementary Fig. S8) exhibits a spatially uniform distribution of spot locations with no identifiable sharp features, as expected from a uniformly distributed, featureless molecular layer. The nanoscale uniformity within a diffraction limited spot is also measured, and found to display blink events uniformly distributed within the illumination area (250 nm in diameter, 300 μW power, Fig. S9). If the plasmonic modes responsible for Raman enhancement can be regarded as uncoupled or weakly coupled, hot spots can be thought of as operating independently. In such a case, regardless of the details of fluctuation mechanisms, the fluctuations originate from local effects which are uncorrelated. Such an optical signal, i.e. one with uncorrelated nanoscale spatial and temporal structure at the source, can be used for super-resolved stochastic imaging. In an alternate method (Fig. 2c), the biological sample can be prepared on a glass substrate followed by evaporation of a thin Ag layer (3 nm mass thickness, see Methods). Membranes or other cellular structure can be regarded as thin dielectric media, on which Ag segregates into closely packed islands. Therefore, the resulting configuration can be regarded as the limit case of the SOM, where the bottom Ag layer is completely absent. Under wide-field illumination (Fig. 2d), such a sample is observed to exhibit blinking SERS spots correlated with the presence of biological material observed in bright-field images (Fig. 2e). In the absence of Ag overlayer, no blinking spots are observed (see Supplementary Video 2). In addition, without Ag evaporation, a broad auto-fluorescence can be observed to obscure the Raman bands, which cannot be clearly identified during confocal Raman imaging.
Figure 2.
(a) A highly uniform plasmonic substrate can be used for confocal or wide-field Surface Enhanced Raman Spectroscopic (SERS) imaging of biological architectures. (b) AIPs imprinted on the substrate can be observed in bright-field images, due to the contrast generated by a shift of the plasmonic absorption band. Uniformity of the surface allows confocal Raman mapping of the peptide overlayer. Under wide-field excitation (scale bar 10 μm), blinking spots correlated with the presence of peptides can be resolved (see Supplementary Video 1). (c) Cardiomyocytes are covered with 3 nm Ag, which segregate into nanoislands with 20 nm average diameter. (d) Thin Ag overlayer provides high-spatial uniformity plasmonic enhancement. (e) Brightfield and wide-field SERS images (scale bar 10 μm) show correlation of blinking spots with the presence of organic material from the cells (see Supplementary Video 3).
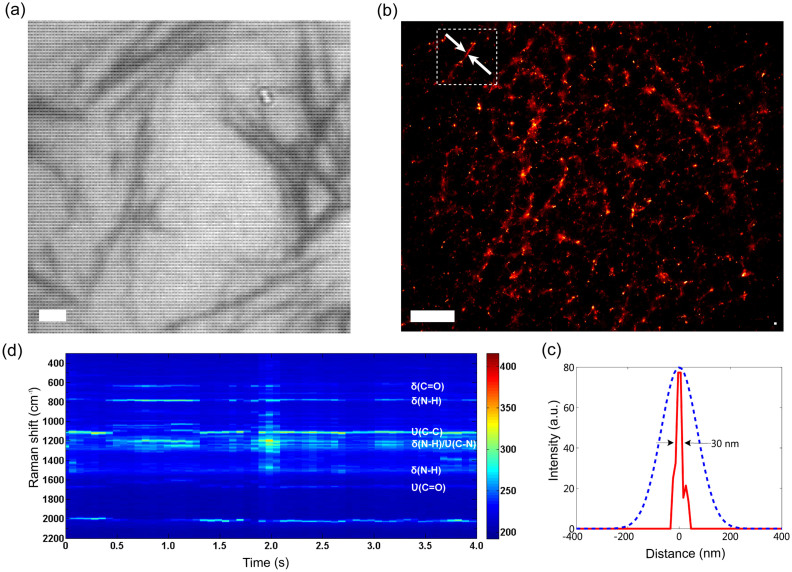
SERS imaging is demonstrated on several complex biological architectures such as supramolecular self-assembled peptide networks, microalgae membranes and eukaryotic cells. We use oppositely charged short amyloid-inspired peptide molecules: Ac-EFFAAE-Am (AIP-1) and Ac-KFFAAK-Am (AIP-2; Supplementary Fig. S6) that self-assemble into amyloid like nanofibers upon mixing at pH 7 in water26. The formed peptide network was diluted to 0.8 μM concentrated AIP solution at pH 7, and dropped on the SOM. During air-drying, AIPs formed peptide bundles on the SOM (bright-field image shown in Fig. 3a) and SERS video sequences were recorded and processed to obtain a reconstructed image of the network (Fig. 3b). Due to the single molecule sensitivity of the SOM, presence of AIP nanofibers and residual peptide coverage on the surface, which are not visible in bright-field images, can be observed in the reconstruction. The resolution obtained in the reconstructed image is about 30 nm, as seen in a line profile across nanofibers (Fig. 3c). In a separate experiment, the resolution of the microscope was observed to be about 300 nm, as measured in the confocal Raman map of a thin layer on a bare glass substrate (Fig. S9). The SERS and Raman spectra of peptide nanofiber networks were recorded for comparison and it is observed that, although main Amide bands (Amide I and III) are preserved for both Raman and SERS spectra, peak positions and intensities of the Raman bands differ significantly than SERS (Fig. S10). On the other hand, SERS spectra themselves are repeatable for low excitation powers (Fig. S10b). During imaging, high excitation densities (e.g. > 10 kW/cm2) were observed to result in chemical and structural damage due to the organic decomposition of biological samples, even in the absence of plasmonic enhancement (Supplementary Fig. S10, S11, S12, S13). However, sample damage can be avoided during wide-field SERS imaging and chemical information (i.e. distinct Raman bands corresponding to chemical bonds) remains present in the optical signal if low excitation powers are used. The presence of chemical information and dominance of Raman bands (not auto-fluorescence) in the optical signal is confirmed by simultaneously recording video images and spectra from a central spot using a beam-splitter and fiber optic cable (Supplementary Fig. S14). At low excitation densities (<1 kW/cm2), Raman bands were observed to be stable during blink events over durations long enough to record sufficient data to produce stochastic reconstructions. SERS spectra of AIP networks revealed several amide bands associated with main chain conformation and side chain composition of self-assembled AIP network (Fig. 3d); amide VI band (C = O in plane bending, 630–750 cm−1)28, amide V band (N-H out-of plane bending, 700–750 cm−1)29, amide III band (N-H bending and C-N stretching, 1230–1300 cm−1)30,31, amide II band (N-H bending, 1550 cm−1)32, and amide I band (C = O stretching, 1600–1700 cm−1)33 can be identified in the spectra.
Figure 3.
(a) Bright field optical micrograph of AIPs (Amyloid Inspired Peptides) network, directly self-assembled on the SOM substrate by air-drying a peptide solution. (b) A wide-field SERS video sequence is recorded at the same location and resulting images are used to reconstruct an image of the self-assembled AIP network. Due to high SERS sensitivity, residual peptide presence on the surface complicates resolution of individual peptide nanofibers. A well-separated, thin peptide nano-bundle (highlighted by arrows) is used to plot the line profile. (see Supplementary Video 4). (c) The peptide bundle is observed to be approximately 30 nm in diameter (solid line), a width smaller than the diffraction limited resolution (represented by dashed line). (d) During wide-field SERS video imaging of AIPs, SERS spectra were simultaneously recorded from a spot located at the center. Amide VI (630–750 cm−1), Amide V (700–750 cm−1), Amide III (1230–1300 cm−1), Amide II (~1550 cm−1) and Amide I (1600–1700 cm−1) bands associated with peptide backbone structure and side chain composition can be identified in the spectra. δ, in plane bending vibration; ν, stretching vibrations. Scale bars are 1 μm wide.
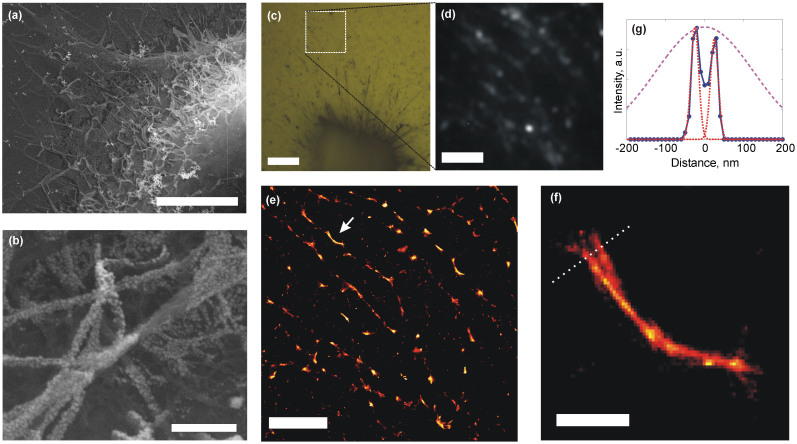
The over-layer version of the SOM, prepared by evaporation of a thin Ag overlayer on a biological sample, was also used for stochastic SERS imaging. Imaging of fixed cardiomyocyte samples are performed using a 3 nm thick Ag over-layer as shown in Fig. 4a and 4b, where closely packed 20 nm diameter Ag islands are seen to cover the sample. Wide-field illumination is used at 532 nm and with up to 4 kW/cm2 excitation power density. Fibrilated lamellipodia of the cardiomyocytes provide good contrast and feature well-defined spatial structures that can be resolved using SERS-STORM (Fig. 4c and 4d). Reconstructed images show (Fig. 4e and 4f) fibrillar structure, and individual fibers separated by 50 nm are observed to be 20 nm in diameter (Fig. 4g). Chemical information (i.e. distinct Raman bands) are observed to be present in the SERS signal (as observed in complementary confocal Raman measurements recorded on the samples using 100 μW excitation, Supplementary Fig. S15). However, unambiguous identification of spectral features is complicated by chemicals used during the fixation of cells.
Figure 4.
(a) Scanning electron micrograph of the edge of a cardiomyocyte (scale bar 4 μm). (b) Evaporation of 3 nm thick Ag results in spontaneous formation of Ag islands on the sample, with 20 nm average diameter (scale bar 300 nm). (c) Bright field optical micrograph of a collapsed cardiomyocyte covered with a 3 nm mass thickness Ag film (scale bar 7 μm). (d) Frame from a video sequence recorded under wide-field illumination shows blinking events on a fibrillar section of the collapsed cell (scale bar 2 μm, see Supplementary Video 5). (e) Stochastic reconstruction of the video sequence allows improved resolution imaging of the fibrillar structure (scale bar 2 μm). (f) Detail of the region denoted by the arrow in (e) shows that the resolution is significantly below the diffraction limit (scale bar 250 nm). (g) Two features separated by 50 nm are clearly resolved across the cross-section shown by dotted line in (f). Stochastically reconstructed images display fibrillar features whose apparent diameters can be fitted with Gaussian widths of 2σ = 20 nm, while diffraction limited spot would be represented by the broad dashed curve, with a Gaussian width of 2σ = 250 nm.
Discussion
Description of the SERS signal shown in Eq. 1, while convenient for the optimization of the effect for practical applications, fails to capture the complete picture of SERS. The mechanisms related to generation of the enhanced Raman signal are complex and considerable effort has been spent in understanding various features of the SERS effect. Other factors such as chemical enhancement are known to play a role in the SERS phenomena. A more detailed description of SERS is given through quantum mechanical calculation of the Raman polarizability tensor34,35,36. For a single scatterer, the intensity of a Raman transition is related to the polarizability tensor ασρ through the expression 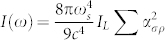 , where IL is the excitation laser intensity, c is the speed of light, and σ, ρ represent the components of the tensor, which is a function of molecular wavefunctions. Perturbation of the polarizability tensor is thought to be due to a very wide variety of microscopic phenomena37. For SERS-STORM to be a viable way of observing nanoscale structure, blinking must be due to non-diffusive and non-destructive phenomena. The experiments presented here show that mechanisms other than molecular diffusion produce blinking events with a rate sufficient to perform stochastic imaging while conserving the Raman bands. The role of thermally activated molecular jumps has been designated as a source for blinking in SERS43,44,45. In our experiments, nanoscale local heating and thermal activation is also a potential source of the observed blinking. The presence of local heating is evident in carbonization of the biomolecular structures at high illumination levels. Optical absorption of organic molecules are increased for wavelengths below 600 nm. Plasmonic structures also possess ohmic losses due to the imaginary part of the dielectric constant of the metal, which decreases towards the infrared for Ag. In our experiments, we are limited in the choice of excitation wavelength and use 532 nm as the laser source, where significant optical absorption is present both in the metal nanostructures and in the molecules. If a near infrared wavelength (such as 785 nm) can be used in the Plasmon enhanced imaging experiments, denaturatization of biomolecules can be reduced and the longevity of the Raman signatures can be improved. The presence of an Ag underlayer or overlayer is essential for SERS imaging. Without the use of an Ag overlayer, for example, no blinking spots can be observed in wide-field images and only a broad and strong auto-fluorescence can be observed at high excitation powers (10 mW spot illumination), accompanied with sample damage. The Raman effect has been designated as a photostable mechanism for chemically specific imaging of biological samples38. Our results show that under intense illumination photo-stability becomes a limiting issue in SERS imaging, however; chemical structure is preserved at low excitation levels, while blink rates sufficient for stochastic reconstruction are still present. We note that, while SERS imaging can be performed in solution using the SOM (data not shown), the Ag overlayer method requires dry samples. SERS-STORM imaging in solution can potentially be used in the study of self-assembly of peptidic nanostructures, or in visualization of membranes. It must be emphasized that, due to finite hot-spot density, or nanoscale nonuniformity of the SERS enhancement, imaging continous membranes will potentially suffer from a sampling effect, where closely separated parts of a membrane that are at high enhancement regions will contribute to an image. Absence of fluorescent labels that might interfere with biomolecular phenomena makes the imaging technique unique, in the sense that imaging of biomolecular structures can be done in their undisturbed native state. Fundamentally, localization precision of point-like objects imaged in two dimensions, is given by
, where IL is the excitation laser intensity, c is the speed of light, and σ, ρ represent the components of the tensor, which is a function of molecular wavefunctions. Perturbation of the polarizability tensor is thought to be due to a very wide variety of microscopic phenomena37. For SERS-STORM to be a viable way of observing nanoscale structure, blinking must be due to non-diffusive and non-destructive phenomena. The experiments presented here show that mechanisms other than molecular diffusion produce blinking events with a rate sufficient to perform stochastic imaging while conserving the Raman bands. The role of thermally activated molecular jumps has been designated as a source for blinking in SERS43,44,45. In our experiments, nanoscale local heating and thermal activation is also a potential source of the observed blinking. The presence of local heating is evident in carbonization of the biomolecular structures at high illumination levels. Optical absorption of organic molecules are increased for wavelengths below 600 nm. Plasmonic structures also possess ohmic losses due to the imaginary part of the dielectric constant of the metal, which decreases towards the infrared for Ag. In our experiments, we are limited in the choice of excitation wavelength and use 532 nm as the laser source, where significant optical absorption is present both in the metal nanostructures and in the molecules. If a near infrared wavelength (such as 785 nm) can be used in the Plasmon enhanced imaging experiments, denaturatization of biomolecules can be reduced and the longevity of the Raman signatures can be improved. The presence of an Ag underlayer or overlayer is essential for SERS imaging. Without the use of an Ag overlayer, for example, no blinking spots can be observed in wide-field images and only a broad and strong auto-fluorescence can be observed at high excitation powers (10 mW spot illumination), accompanied with sample damage. The Raman effect has been designated as a photostable mechanism for chemically specific imaging of biological samples38. Our results show that under intense illumination photo-stability becomes a limiting issue in SERS imaging, however; chemical structure is preserved at low excitation levels, while blink rates sufficient for stochastic reconstruction are still present. We note that, while SERS imaging can be performed in solution using the SOM (data not shown), the Ag overlayer method requires dry samples. SERS-STORM imaging in solution can potentially be used in the study of self-assembly of peptidic nanostructures, or in visualization of membranes. It must be emphasized that, due to finite hot-spot density, or nanoscale nonuniformity of the SERS enhancement, imaging continous membranes will potentially suffer from a sampling effect, where closely separated parts of a membrane that are at high enhancement regions will contribute to an image. Absence of fluorescent labels that might interfere with biomolecular phenomena makes the imaging technique unique, in the sense that imaging of biomolecular structures can be done in their undisturbed native state. Fundamentally, localization precision of point-like objects imaged in two dimensions, is given by 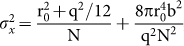 , where r0 is the standard deviation of the point spread function, N is the total number of photons collected, q is the size of an image pixel, and b is the background noise per pixel39. For single blink events resulting in ~ 105 SERS photons, the resolution is expected to be on the order of few nanometers. However, it is observed in our experiments that the resolution is limited by the Ag nanoisland separation, which is on the order of 20 to 30 nm. The plasmonic enhancement is restricted to the close vicinity (few nanometers) of the hot-spots, limiting the applicability of the technique to imaging of membranes or other biological nano-structures directly in contact with the plasmonic surface. Images are essentially produced by sampling of small volumes (few nm3) of biological structures within high enhancement regions of the plasmonic surface, spaced by 20 to 30 nm. Scaling of the metasurface suggests that, further improvement of hot-spot density may be possible by the use of smaller nanoisland diameters and a thinner dielectric layer17. However, even when the hot-spot density is improved, the technique will be limited in its application to imaging of samples that are directly in contact with the high enhancement regions of the surface.
, where r0 is the standard deviation of the point spread function, N is the total number of photons collected, q is the size of an image pixel, and b is the background noise per pixel39. For single blink events resulting in ~ 105 SERS photons, the resolution is expected to be on the order of few nanometers. However, it is observed in our experiments that the resolution is limited by the Ag nanoisland separation, which is on the order of 20 to 30 nm. The plasmonic enhancement is restricted to the close vicinity (few nanometers) of the hot-spots, limiting the applicability of the technique to imaging of membranes or other biological nano-structures directly in contact with the plasmonic surface. Images are essentially produced by sampling of small volumes (few nm3) of biological structures within high enhancement regions of the plasmonic surface, spaced by 20 to 30 nm. Scaling of the metasurface suggests that, further improvement of hot-spot density may be possible by the use of smaller nanoisland diameters and a thinner dielectric layer17. However, even when the hot-spot density is improved, the technique will be limited in its application to imaging of samples that are directly in contact with the high enhancement regions of the surface.
In summary, we demonstrated the possibility of using the fluctuating SERS signal in stochastic optical reconstruction microscopy of large area biological samples on uniform plasmonic substrates, providing significant resolution improvement beyond the diffraction limit. It has been observed that chemical structure of the samples can be retained at low excitation power densities for durations long enough to record data required for reconstruction. The demonstration can be regarded as an initial step towards more advanced SERS-STORM imaging; presentation of detailed chemical information (i.e. distinct Raman bands) directly in the reconstructed image would require data collection by filtering a subset of Raman bands and applying reconstruction to such narrow spectrum image sequences. This multispectral stochastic imaging approach requires high sensitivity cameras and sharp filters specifically chosen to fit the excitation wavelength and Raman spectra. We leave such advanced SERS-STORM imaging beyond the scope of this article.
Methods
Plasmonic surface preparation
Fabrication of Plasmonic Structures. Germanium (2–3 nm, 99.99% purity) is deposited on 10 cm2 diameter silicon substrates using e-beam evaporation system (Vaksis PVD Vapor 4S e-beam) as wetting and adhesion layer with a deposition rate 0.5 Å/s. Then 70 nm silver (99.99% Purity) is deposited with e-beam system with a deposition rate of 0.6–0.8 Å/s. For the dielectric spacer layer, 12–20 nm Al2O3 (99.99% Purity) is also deposited with the e-beam system with a deposition rate 0.3–0.4 Å/s. For the nanoisland layer, 3 nm silver (99.99% Purity) is deposited using the e-beam evaporation system with a deposition rate of 0.6–0.8 Å/s. See Supplementary Information for details of preparation of layer by layer and nanosphere lithography substrates.
Raman measurements and imaging
Reflection mapping is done with WITEC Alpha 300S system using white light illumination. For Raman enhancement uniformity measurements, samples are immersed in 1 μM methylene blue in deionized water solution for 1 h, rinsed with ethanol several times, and blow-dried with nitrogen. SERS measurements are performed using WITEC Alpha 300S Raman module. A diode-pumped solid-state 532 nm wavelength laser is used for excitation in the Raman measurements. Laser power is measured using a silicon photodiode at sample plane, power densities are calculated using apparent spot diameter of the illumination area. For Raman mapping measurements 20 ×, 50 ×, and 100 × objectives are used with integration times of 22–100 ms. For wide-field SERS measurements, a monochrome CMOS camera (Videology 24C1.3XUSB) is fitted on to the microscope system. Stochastic reconstruction is done using rapidStorm27 software freely available at the internet address: http://www.super-resolution.biozentrum.uni-wuerzburg.de/research_topics/rapidstorm/.
Electromagnetic simulations
Field profiles are simulated using a commercial computational tool (Lumerical). Dielectric function of silver is obtained from the literature. Germanium wetting layer is neglected in the simulations. The refractive index of electron-beam-evaporated Al2O3 is measured experimentally using a J.A.Woolam V-VASE ellipsometer, and a constant refractive index of nAl2O3 = 1.6 is used for the Al2O3 dielectric layer. The thickness of supporting Ag is 50–70 nm, and transmission to the silicon substrate is less than 1% of incident light over the visible spectrum; therefore, only reflection is calculated. Absorption is approximated as A = 1 - R. Perfectly matched layer (PML) boundary condition is used in the z-direction, and Bloch boundary condition is used in the x-direction (along the period) of structures. Germanium wetting layer is neglected in the simulations. The mesh size is used in the simulations are 0.25 nm by 0.25 nm. Field profiles are calculated by illuminating structures with monochromatic plane wave corresponding to resonance wavelengths.
Imprinting of AIPs on SOM
Dow Corning Sylgard 184 silicone elastomer kit containing base (Part A) and curing agent (Part B) is used to prepare the patterned molds with different structures. Parts A and B are mixed in 10:1 mass ratio, degassed in rough vacuum, poured onto silicon wafers with 2.4 μm deep resist patterns, then degassed again and cured at 70°C for 4 h. Molds are immersed and waited a few minutes in AIPs solution at pH 7 in water, prepared by mixture of 1:1 volume ratio of 0.8 μM Ac-EFFAAE-Am (AIP-1) and Ac-KFFAAK-Am (AIP-2) solutions at pH 7 in water. The mold immersed into the peptide solution is air-dried and the peptide layer is formed on the mold. Then, peptide layer on the mold is transferred onto the plasmonic substrates by imprinting of the mold with no extra pressure.
Self-assembled AIPs network on SOM
26.6 mM AIP-1 and AIP-2 peptide solutions are mixed at pH 7 in water, and AIPs network formation is incubated for an hour for completion. The detailed characterization of the AIPs network is given in the literature26. The formed peptide network was diluted to 0.8 μM AIPs solution at pH 7, and dropped on the SOM. The self-assembled network on SOM is imaged using WITEC Alpha 300S Raman module (see Raman measurements and imaging section).
Microalgae preparation and analysis
Cells of Chlamydomonas reinhardtii CC124 grown to confluence in TAP medium were pipetted on an unpatterned plasmonic surface and left to dry for 30 min. Dried samples were directly imaged without staining. Three carotenoid peaks40,41 at 1008, 1160 and 1515 cm−1 were observed to be most prominent in the sample tested, in an experiment performed to identify Raman bands of non-synthetic, unstained biological samples.
Cardiomyocyte sample preparation
H9c2 cells derived from embryonic BD1X rat heart tissue, were cultured in Dulbecco's Modified Eagle Serum (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin42. Passaging of cells was performed when cells reached ~ 80–90% confluency. H9c2 cells were seeded onto 13 mm glass coverslips and cultured in standard cell culture conditions (37°C, humidified and 5% CO2 environment). After 48 h, media on cells was aspirated, and cells were washed with 1 × PBS twice. 2.5% glutaraldehyde fixation was followed by 1 wt% OsO4 fixation. After dehydration by gradual increases of ethanol, samples were dried by Tourismis Autosamdri-815B critical-point-drier. Dried samples were coated with Ag and imaged using WITEC Alpha 300S Raman module.
Author Contributions
S.A. designed, optimized, fabricated and characterized the surfaces, performed imaging and spectroscopy experiments, performed electromagnetic calculations, prepared figures. G.C. and M.O.G. prepared peptides, characterized peptides and self-assembled nanofibers, performed imprinting of peptides, analyzed Raman data, performed imaging experiments, prepared figures and wrote manuscript. Z.S. and A.B.T. prepared cardiomyocyte samples. A.D.O. and T.T. prepared Algae samples. O.O.E. and D.K. helped in SERS experiments and sample preparation. A.T. and H.Z. prepared nanosphere lithography samples. P.T. and I.T. prepared layer-by-layer samples. Y.K. performed noise reduction and data processing of video sequences. A.D. designed the experiments, performed imaging and spectroscopy experiments, analyzed data, supervised research and wrote the manuscript.
Supplementary Material
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6
Supplementary figures
Acknowledgments
This work was partially supported by TUBITAK under Grant 111M344, EU FP7:People-IAPP NanoBacterPhageSERS and the State Planning Organization of Republic of Turkey, project UNAM. AD acknowledges Hakan Ceylan and Hasan Guner's help in the clean room, during sample preparation.
References
- Jeanmaire D. L. & Van Duyne R. P. Surface raman spectroelectrochemistry: Part I. Heterocyclic, aromatic, and aliphatic amines adsorbed on the anodized silver electrode. J. Electroanal. Chem. 84, 1–20 (1977). [Google Scholar]
- Albrecht M. G. & Creighton J. A. Anomalously intense Raman spectra of pyridine at a silver electrode. J. Am. Chem. Soc. 99, 5215–5217 (1977). [Google Scholar]
- Stiles P. L., Dieringer J. A., Shah N. C. & Van Duyne R. P. Surface-Enhanced Raman Spectroscopy. Annu. Rev. Anal. Chem. 1, 601–626 (2008). [DOI] [PubMed] [Google Scholar]
- Nie S. & Emory S. R. Probing Single Molecules and Single Nanoparticles by Surface-Enhanced Raman Scattering. Science 275, 1102–1106 (1997). [DOI] [PubMed] [Google Scholar]
- Kneipp K. et al. Single Molecule Detection Using Surface-Enhanced Raman Scattering (SERS). Phys. Rev. Lett. 78, 1667–1670 (1997). [Google Scholar]
- Kneipp K., Wang Y., Dasari R. R. & Feld M. S. Approach to single molecule detection using surface-enhanced resonance Raman scattering (SERRS): A study using Rhodamine 6 G on colloidal silver. Appl. Spectrosc. 49, 780–784 (1995). [Google Scholar]
- Kneipp K. et al. Surface-enhanced Raman scattering (SERS)—a new tool for single molecule detection and identification. Bioimaging 6, 104–110 (1998). [Google Scholar]
- Le Ru E. C. & Etchegoin P. G. Single-Molecule Surface-Enhanced Raman Spectroscopy. Annu. Rev. Phys. Chem. 63, 65–87 (2012). [DOI] [PubMed] [Google Scholar]
- Rust M. J., Bates M. & Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–796 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stranahan S. M. & Willets K. A. Super-resolution Optical Imaging of Single-Molecule SERS Hot Spots. Nano Lett. 10, 3777–3784 (2010). [DOI] [PubMed] [Google Scholar]
- Cang H. et al. Probing the electromagnetic field of a 15-nanometre hotspot by single molecule imaging. Nature 469, 385–388 (2011). [DOI] [PubMed] [Google Scholar]
- Balzarotti F. & Stefani F. D. Plasmonics Meets Far-Field Optical Nanoscopy. ACS Nano 6, 4580–4584 (2012). [DOI] [PubMed] [Google Scholar]
- Weber M. L. & Willets K. A. Correlated Super-Resolution Optical and Structural Studies of Surface-Enhanced Raman Scattering Hot Spots in Silver Colloid Aggregates. J. Phys. Chem. Lett. 2, 1766–1770 (2011). [Google Scholar]
- Titus E. J., Weber M. L., Stranahan S. M. & Willets K. A. Super-Resolution SERS Imaging beyond the Single-Molecule Limit: an Isotope-Edited Approach. Nano Lett. (2012). [DOI] [PubMed] [Google Scholar]
- Bates M., Huang B., Dempsey G. T. & Zhuang X. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science 317, 1749–1753 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perney N. M. B. et al. Tuning localized plasmons in nanostructured substrates for surface-enhanced Raman scattering. Opt. Express 14, 847–857 (2006). [DOI] [PubMed] [Google Scholar]
- Ayas S. et al. Raman Enhancement on a Broadband Meta-Surface. ACS Nano 6, 6852–6861 (2012). [DOI] [PubMed] [Google Scholar]
- Saar B. G. et al. Video-rate molecular imaging in vivo with stimulated Raman scattering. Science 330, 1368–1370 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freudiger C. W. et al. Highly specific label-free molecular imaging with spectrally tailored excitation-stimulated Raman scattering (STE-SRS) microscopy. Nature Photonics 5, 103–109 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian W. Freudiger et al. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science. 322(5909), 1857–1861 (2008) 10.1126/science.1165758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergman J. et al. Relationship between surface-enhanced Raman scattering and the dielectric properties of aggregated silver films. Opt. Lett. 6, 33–35 (1981). [DOI] [PubMed] [Google Scholar]
- Datta S., Chan C. T., Ho K. M. & Soukoulis C. M. Effective dielectric constant of periodic composite structures. Phys. Rev. B 48, 14936–14943 (1993). [DOI] [PubMed] [Google Scholar]
- Aspnes D. Local-field effects and effective-medium theory: A microscopic perspective. Am. J. Phys. 50 (1982). [Google Scholar]
- Ruppin R. Evaluation of extended Maxwell-Garnett theories. Opt. Commun. 182, 273–279 (2000). [Google Scholar]
- Maaroof A. & Sutherland D. S. Optimum plasmon hybridization at percolation threshold of silver films near metallic surfaces. J. Phys. D: Appl. Phys. 43, 405301 (2010). [Google Scholar]
- Cinar G. et al. Amyloid Inspired Self-Assembled Peptide Nanofibers. Biomacromolecules 13, 3377–3387 (2012). [DOI] [PubMed] [Google Scholar]
- Wolter S. et al. Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 237, 12–22 (2009). [DOI] [PubMed] [Google Scholar]
- Overman S. A. & Thomas Jr G. J. Amide modes of the α-helix: Raman spectroscopy of filamentous virus fd containing peptide 13C and 2H labels in coat protein subunits. Biochemistry 37, 5654–5665 (1998). [DOI] [PubMed] [Google Scholar]
- Song S., Asher S. A., Krimm S. & Bandekar J. Assignment of a new conformation-sensitive UV resonance Raman band in peptides and proteins. J. Am. Chem. Soc. 110, 8547–8548 (1988). [Google Scholar]
- Mirkin N. G. & Krimm S. Amide III Mode, ψ Dependence in Peptides: A Vibrational Frequency Map. J. Phys. Chem. A 106, 3391–3394 (2002). [Google Scholar]
- Tuma R. Raman spectroscopy of proteins: from peptides to large assemblies. J. Raman Spectrosc. 36, 307–319 (2005). [Google Scholar]
- Kobko N. & Dannenberg J. Cooperativity in amide hydrogen bonding chains. A comparison between vibrational coupling through hydrogen bonds and covalent bonds. Implications for peptide vibrational spectra. J. Phys. Chem. A 107, 6688–6697 (2003). [Google Scholar]
- Schweitzer-Stenner R. Advances in vibrational spectroscopy as a sensitive probe of peptide and protein structure: A critical review. Vib. Spectrosc. 42, 98–117 (2006). [Google Scholar]
- Albrecht A. C. On the Theory of Raman Intensities. J. Chem. Phys. 34, 1476–1484 (1961). [Google Scholar]
- Lombardi J. R. & Birke R. L. A unified approach to surface-enhanced Raman spectroscopy. J. Phys. Chem. C 112, 5605–5617 (2008). [Google Scholar]
- Lombardi J. R. & Birke R. L. The theory of surface-enhanced Raman scattering. J. Chem. Phys. 136, 144704 (2012). [DOI] [PubMed] [Google Scholar]
- Wang Y. & Irudayaraj J. Surface-enhanced Raman spectroscopy at single-molecule scale and its implications in biology. Phil. Trans. R. Soc. B 368 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J. W., Nguyen F. T., Lue N., Dasari R. R. & Heller D. A. Measuring Uptake Dynamics of Multiple Identifiable Carbon Nanotube Species via High-Speed Confocal Raman Imaging of Live Cells. Nano Lett. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess S. T., Girirajan T. P. K. & Mason M. D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. et al. In vivo lipidomics using single-cell Raman spectroscopy. Proc. Natl. Acad. Sci. USA 108, 3809–3814 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Beal C., Cai W., Ruoff R. & Terentjev E. Micro-Raman spectroscopy of algae: Composition analysis and fluorescence background behavior. Biotechnol. Bioeng. 105, 889–898 (2010). [DOI] [PubMed] [Google Scholar]
- Kimes B. & Brandt B. Properties of a clonal muscle cell line from rat heart. Exp. Cell. Res. 98, 367–381 (1976). [DOI] [PubMed] [Google Scholar]
- Maruyama Y., Ishikawa M. & Futamata M. Thermal Activation of Blinking in SERS Signal. J. Phys. Chem. B 108, 673–678 (2004). [Google Scholar]
- Emory R. Jensen A., Wenda, Hanb & Nie Re-examining the origins of spectral blinking in single-molecule and single-nanoparticle SERS. Faraday Discuss. 132, 249–259 (2006). [DOI] [PubMed] [Google Scholar]
- Otto A. Theory of First Layer and Single Molecule Surface Enhanced Raman Scattering (SERS). Phys. Stat. Sol. (a) 188(4), 1455–1470 (2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Video 1
Supplementary Video 2
Supplementary Video 3
Supplementary Video 4
Supplementary Video 5
Supplementary Video 6
Supplementary figures