Abstract
Information concerning the effects of genetic variation between different background strains on hemodynamic, morphometric, and gene expression response to hypoxia would be useful. Three strains of mice were kept in hypoxia and phenotyped followed by gene profiling analysis. Among the variables examined, hematocrit, right heart muscularization, and right ventricular systolic pressure showed a strain-specific effect. Increased gene expression of inflammatory, muscle, and angiogenesis genes were seen in all strains, though the specific genes changed varied among groups. These results suggest that different strains use different gene expression mechanisms to adapt to the challenge of chronic hypoxia, resulting in modified phenotypic changes.
Keywords: gene expression, hypoxia pulmonary circulation, strain difference
Chronic hypoxia as a contributing factor to pulmonary arterial hypertension (PAH) is associated with a variety of human diseases. Chronic obstructive pulmonary disease [1], cystic fibrosis [2], interstitial fibrosis [3], upper airway obstruction, and bronchopulmonary dysplasia [4], among others, are all associated with exposure to chronic hypoxia. Understanding the effects of and potential treatments for chronic hypoxia is thus relevant to many human disorders [5].
Hypoxic pulmonary hypertension is often studied in mouse models [6, 7]. These studies, examining molecular mechanisms or interventions, use many of the same phenotypic markers of PAH. Standard measurements include right ventricular systolic pressure (RVSP), cardiac output, right ventricular hypertrophy, muscularization of small pulmonary vessels, counts of alveoli, hematocrit, and blood gas measurements. Together, these are used to both gauge extent of PAH, as well as physiologic response to it.
Three of the most widely used experimental mouse strains are FVB/N, C57BL/6, and SV129. The goal of our current study was to determine whether these 3 strains had a differential response to chronic hypoxia according to the metrics listed above, and if so, whether this could be correlated with a differential response by gene expression. This would be useful to the mouse modeling community in understanding the differences among strains in previous studies of hypoxic pulmonary hypertension and in planning future studies. Moreover, strain-specific differences in gene expression may suggest modifier genes if correlated to a less severe phenotype.
We measured 16 different phenotypic variables in each of the 3 strains, under either normoxia or 4 weeks’ hypobaric hypoxia. Although most variables had significant changes with hypoxia, and some had significant differences at baseline, only 3 had significant differences in hypoxic response among strains. SV129 had greater increase in RVSP with hypoxia, FVB/N had less right ventricular hypertrophy, and C57BL/6 had less of an increase in hematocrit, although from a higher baseline.
By gene array, we found that all of the strains had changes in similar pathways, including muscle, angiogenesis, growth, adhesion, G-protein, and differentiation pathways, and that all strains had changes in some common genes within these pathways, but that each strain also had a large number of changed genes within these pathways that were strain specific.
METHODS
Animals
FVB/NJ, C57BL/6J, or 129X1/SvJ mice, 6 to 8 weeks old, were ordered from The Jackson Laboratory (Bar Harbor, MA) and allowed to acclimatize for 1 week to Denver altitude. Fifteen mice from each strain were kept at either Denver altitude (~85 kPa) or at 50 kPa in hypobaric hypoxia chambers. After 4 weeks, animals were phenotyped by echocardiography, followed by RVSP measurement and blood and tissue collection. All animal studies were preapproved by the University of Colorado Health Sciences Center (UCHSC) Institutional Animal Care and Use Committee (IACUC).
Echocardiographic Measurements
Transthoracic echocardiography was performed using 10- and 13-MHz ultrasound probes with a Vivid Five System (General Electrics Vingmed Ultrasound, Horton, Norway). Echocardiographic data were analyzed with EchoPac 6.3.6 software (General Electrics Vingmed Ultrasound). Left heart dimensions were obtained in short-axis view. The diameters of the aorta and pulmonary artery as well as flow in the pulmonary artery were obtained in parasternal longitudinal axis. The flow in the aorta was measured using a suprasternal longitudinal view. Cardiac output and pulmonary vascular resistance was calculated in standardized fashion [8–10]. All echocardiographic measurements were performed in triplicate by one investigator and averaged.
RVSP Measurements
Animals were anesthetized with intraperitoneal injections of ketamine 200 mg/kg and xylazine 10 mg/kg. If further anesthesia was necessary, repeat doses of ketamine 100 mg/kg and xylazine 5 mg/kg were administered. Studies were conducted with mice positioned supine on a heated operating table while spontaneously breathing room air. Right ventricular (RV) pressure was directly measured with a 1.4-French Pressure Volume Conductance System SPR-839 (Millar Instruments, Houston, TX) inserted into the right ventricle via the surgically exposed right jugular vein. Hemodynamics were continuously recorded with a Millar MPVS-300 unit coupled to a Powerlab 8-SP A/D converter, acquired at 1000 Hz, and captured to a Macintosh G4 computer utilizing Chart5.3 software. Blood was drawn for gas measurements by cardiac puncture, and immediately transferred to a Radiometer Medical (Copenhagan, Denmark) ABL5 blood gas analyzer. After lethal injection of pentobarbital, the heart and lungs were removed. Lungs were divided and processed for immunohistochemistry or molecular studies.
Morphometry
Tissue blocks were made from the left lungs. The embedded tissue was cut at 4 μm on an AO 820 microtome (American Optical, Southbridge, MA) and placed onto Superfrost Plus slides (Fisher Scientific, Pittsburgh, PA). Slides were baked for 1 hour at 60°C and then deparaffinized through Citrisolv (Fisher Scientific, Pittsburgh, PA) and a graded alcohol series. Antigen retrieval was then performed by heating the slides 4 × 3 minutes at 30% power in a 1650-watt microwave in a solution containing 16 mM sodium citrate–trisodium and 4 mM citric acid, pH 5.6. Slides were then rehydrated in phosphate-buffered saline (PBS) and incubated in a blocking solution of PBS/4% serum/0.1% Triton X-100 for 30 minutes and then in primary antibody overnight at 4°C in PBS/4% serum/0.1% Triton X-100 (PBS). Polyclonal anti-smooth muscle actin was purchased from Abcam (Cambridge, MA) and used at a dilution of 1:1000.
Images from 12 random 20× fields per slide were taken using a Zeiss Axioscop II microscope, and medial thickness, fully muscularized (FM) vessels of <100 μm diameter, partially muscularized (PM vessels) of <100 μm diameter, and alveoli per field were counted in a blinded fashion using tools included in the Axioscop software.
Affymetrix Arrays
Samples were prepared for Affymetrix arrays using 2.5 μg of total RNA. First- and second-strand complimentary DNA was synthesized using standard techniques. Biotin-labeled antisense complimentary RNA was produced by an in vitro transcription reaction. Mouse Genome 430 2.0 microarrays (Affymetrix, Foster City, CA) were hybridized with 20 μg cRNA. Target hybridization, washing, staining, and scanning probe arrays were done following an Affymetrix Gene Chip Expression Analysis Manual. All array results have been submitted to the National Center for Bioinformatics (NCBI) gene expression and hybridization array data repository (GEO; http://www.ncbi.nlm.nih.gov/geo/), as series GSE7823.
Array Analysis
Affymetrix Cel files were loaded into dChip 2005 array analysis software. The dChip algorithm is capable of detecting significant differences at signal strengths lower than those usable in Microarray Suite(11) (Affymetrix, Santa Clara, CA). Overall signal strength from arrays was normalized to the median array, and expression levels determined using the perfect match/mismatch (PM/MM) algorithm. Gene ontology was determined using the Classify Genes tool within dChip(12), with gene ontology files down loaded from the Gene Ontology Consortium (www.geneontology.org)(13). In order to avoid problems with either false negatives, or with determining an arbitrary fold-change cutoff, we set very loose definitions for changed genes (1.4×, with a minimum change of 200) and then determined statistically overrepresented gene ontology groups at a high stringency (p < .001) within the genes called as differentially regulated. By this method, the number of gene ontology groups produced by chance should be close to zero. Other specific details of analytic methods are included in Results. Statistics for array analysis were handled by algorithms internal to dChip.
RESULTS
Murine Phenotypic Response to Chronic Hypoxia Is Strain Specific
Fifteen animals per group of each of FVB/N, SV129, or C57BL/6 were subjected to either hypobaric hypoxia (50 kPa) or normoxia (85 kPa). After four weeks, mice were weighed, and examined by echocardiography. Echocardiography was used to determine the diameters of the aorta and pulmonary artery (PA), measure cardiac output (CO), and calculate pulmonary vascular resistance (PVR). Next, closed-chested catheterization by pressure transducer was used to determine right ventricular systolic pressure (RVSP) and heart rate under anesthesia. Blood was drawn and immediately analyzed for hematocrit, pH, pCO2, and PO2. Hearts were dissected and weighed to determine right ventricular muscularization (RV/LV+S). Finally, lung sections were fixed and analyzed by immunohistochemistry for numbers of alveoli and vessel muscularization. Total numbers of mice used for each measurement varied between 6 and 15, with average and median of ~11. Results are summarized in Table 1 and Figure 1.
TABLE 1.
Measurements ± SEM
| Pressure | Echocardiogram
|
Blood gas
|
||||||
|---|---|---|---|---|---|---|---|---|
| Aorta (mm) | PA diameter | CO (mL/mm) | PVR (calculated) | pH | pCO2 | pO2 | ||
| C57BL/6 | 85 kPa | 1.03 ± 0.03§ | 1.08 ± 0.05 | 6.8 ± 1.0‡ | 0.89 ± 0.23 | 7.38 ± 0.05 | 36 ± 3 | 70 ± 2§ |
| 50 kPa | 0.96 ± 0.03§* | 1.01 ± 0.02* | 6.8 ± 0.5 | 1.47 ± 0.16 | 7.37 ± 0.08 | 46 ± 3* | 52 ± 3* | |
| FVB/N | 85 kPa | 0.94 ± 0.02 | 1.09 ± 0.04 | 8.8 ± 0.8 | 0.91 ± 0.08 | 7.27 ± 0.05 | 39+2 | 76 ± 2 |
| 50 kPa | 0.90 ± 0.02* | 1.01 ± 0.03* | 5.7 ± 0.6‡ | 2.02 ± 0.19* | 7.25 ± 0.09 | 41 ± 2 | 59 ± 2* | |
| SV129 | 85 kPa | 0.96 ± 0.02 | 1.04 ± 0.04 | 8.9 ± 0.9 | 0.79 ± 0.08 | 7.26 ± 0.05 | 38 ± 4 | 77 ± 2 |
| 50 kPa | 0.92 ± 0.02* | 1.00 ± 0.03* | 8.1 ± 0.7 | 1.73 ± 0.31 | 7.22 ± 0.08 | 43 ± 3 | 63 ± 3* | |
| Presssure | Body mass | Heart rate | Morphometry
|
||||
|---|---|---|---|---|---|---|---|
| FM/field | PM/field | Alveoli/field | Medial thickness | ||||
| C57BL/6 | 85 kPa | 23.9 ± 0.3 | 184 ± 22 | 1.0 ± 0.2 | 5.5 ± 0.5 | 159 ± 9 | 9.3 ± 1.5 |
| 50 kPa | 22.0 ± 0.3* | 196 ± 9 | 3.1 ± 0.3* | 6.5 ± 0.6* | 176 ± 8 | 9.6 ± 1.0 | |
| FVB/N | 85 kPa | 24.6 ± 0.3 | 217 ± 11 | 1.4 ± 0.4 | 3.9 ± 0.5 | 156 ± 5 | 7.6 ± 1.3 |
| 50 kPa | 23.4 ± 0.5* | 174 ± 12 | 2.8 ± 0.6* | 6.1 ± 0.6* | 176 ± 6 | 9.2 ± 0.8 | |
| SV129 | 85 kPa | 24.0 ± 0.4 | 195 ± 14 | 1.2 ± 0.2 | 5.4 ± 0.6 | 185 ± 10 | 8.7 ± 1.3 |
| 50 kPa | 21.3 ± 0.4* | 189 ± 9 | 2.2 ± 0.3* | 5.1 ± 0.5 | 169 ± 8 | 7.9 ± 0.5 | |
All p values are for 2-way ANOVA with Fisher’s poet hoc for differences caused by strain or hypoxia.
p < .01 for difference compared across strains.
p < .01 for differences caused by hypoxia, except pCO2 (p = .029), PM (p = .047), PA diameter (p = .029).
0.94 for strain-specific and p = .057 for pressure specific differences in CO.
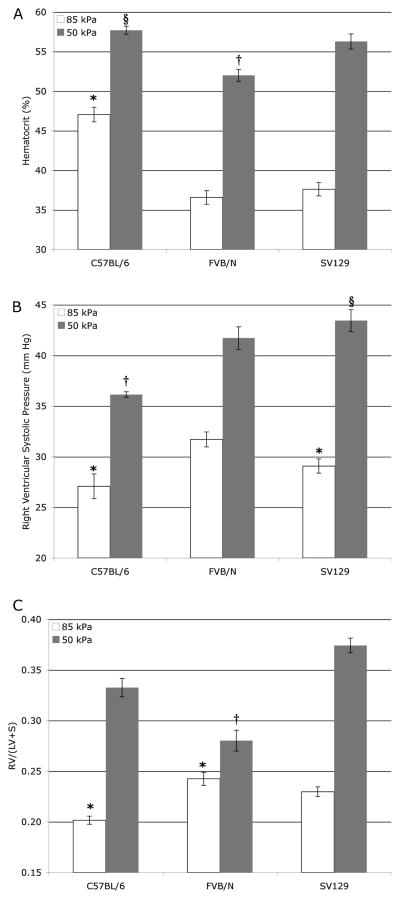
FIGURE 1.
Hematocrit (A), RVSP (B), and RV/LV+S (C) all have significant differences dependent on pressure, strain, and pressure-dependent on strain by 2-way ANOVA. Differences in individual values were determined using Tukey’s post hoc test. All values at 50 kPa are significantly different (p < .05) than the within-strain 85 kPa value. (A) Hematocrit in C57BL/6 is higher at 85 kPa than other strains (*) but increases less with 50 kPa hypoxia (§). Hematocrit in FVB/N at 50 kPa is less than in other strains (†). (B) RVSP at 85 kPA in C57BL/6 is significantly lower than in SV129 (*), but neither are different than FVB/N. RVSP at 85 kPa in C57BL/6 is lower than other strains (†), and increase in RVSP between 85 and 50 kPa is greater in SV129 than in other strains (§). (C) Right heart muscularization is lower at 85 kPa in C57BL/6 than FVB/N mice but neither is different than SV129. RV/LV+S is different at 50 kPa in all strains, and increase in RV/LV+S between pressures is lower in FVB/N than in other strains (†).
We found that 5 of these 16 metrics were different between strains in baseline animals at P < .05 by 2-way analysis of variance (ANOVA), 11 out of 16 were changed by hypoxia at P < .05, but only 3, hematocrit, RVSP, and RV/LV+S, showed a differential response to hypoxia by strain. Hematocrit was less increased in C57BL/6 mice, although starting from a higher baseline (Figure 1A); RVSP was more increased in SV129 mice (Figure 1B); right heart muscularization was less pronounced in FVB/N mice (Figure 1C).
Murine Transcriptional Response to Chronic Hypoxia Is Strain Specific
In order to examine transcriptional response to chronic hypoxia, Affymetrix arrays were probed with RNA from whole lung from hypoxic or normoxic FVB/N, SV129, or C57BL/6 mice. Array data were analyzed by dChip software and sorted into gene ontology groups. We found that gene expression changes in all strains fell into the same categories, and many of the same genes were differentially regulated in the same direction in all strains. However, some transcriptional changes were specific to a particular strain, or 2 of the 3 strains.
All strains had changes in muscle, angiogenesis, growth, adhesion, G-protein, differentiation, apoptosis, and stress-response genes. For most of these, there were many genes in common across strains (Table 2). However, in the case of stress response and apoptosis, each strain activates different sets of genes (Table 3).
TABLE 2.
Genes Changed In All Strains (Examples)
| BL6 | FVBN | SV129 | |
|---|---|---|---|
| Muscle | |||
| caldesmon 1 | 1.7 | 2.3 | 2.1 |
| myocyte enhancer factor 2C | 1.7 | 2.6 | 2.6 |
| myosin X | 1.7 | 2.4 | 1.7 |
| restin | 1.6 | 3.9 | 2.4 |
| tropomodulin 2 | 1.6 | 2.4 | 2.4 |
| tropomyosin 1, alpha | 2.1 | 3.4 | 2 |
| Angiogenesis | |||
| angiopoietin 1 | 1.6 | 2.5 | 1.6 |
| quaking | 2 | 2.6 | 1.4 |
| semaphorin 5A | 2 | 2.1 | 1.7 |
| transforming growth factor, beta 2 | 1.4 | 1.5 | 1.5 |
| vascular endothelial growth factor A | 1.5 | 1.5 | 1.4 |
| vascular endothelial growth factor C | 1.4 | 2.3 | 1.4 |
| Growth | |||
| cell division cycle associated 5 | − 1.8 | −3.2 | −1.7 |
| ephrin B1 | 1.8 | 2.7 | 1.6 |
| growth associated protein 43 | 2.4 | 2.2 | 1.8 |
| large tumor suppressor 2 | 2.2 | 2.2 | 2.4 |
| RAD21 homolog | −3.3 | −2.2 | −2.3 |
| tumor necrosis factor receptor 5 | 1.7 | 3.1 | 1.6 |
| Adhesion | |||
| integrin alpha 8 | 1.4 | 2.5 | 1.4 |
| integrin beta 6 | 3.2 | 1.8 | 2.4 |
| integrin, beta-like 1 | 1.3 | 2.3 | 1.8 |
| LIM and senescent cell 1 | 1.9 | 2.7 | 2.2 |
| nephronectin | 1.7 | 2.3 | 2.4 |
| nidogen 1 | 1.4 | 2.8 | 1.5 |
| G-Protein | |||
| A kinase anchor protein 9 | 1.7 | 2.8 | 2.9 |
| calmodulin 1 | −1.7 | −1.4 | −1.4 |
| guanine nucleotide bp alpha 13 | 1.8 | 1.8 | 1.4 |
| guanine nucleotide bp alpha inh. 3 | −3.9 | −2.8 | −2.9 |
| phosphodiesterase 7A | 1.6 | 1.8 | 1.4 |
| protein kinase, cAMP dep, catalytic, b | 1.8 | 2.2 | 1.4 |
| Development | |||
| GLI-Kruppel family member GLI2 | −1.5 | −1.5 | −1.2 |
| jagged 1 | −3.1 | −2.9 | −1.9 |
| MAP3K10 | −1.5 | −1.9 | −1.4 |
| muscleblind-like 3 | 1.5 | 1.9 | 1.6 |
| Notch gene homolog 3 | 1.4 | 2 | 1.9 |
| transducer of ErbB-2.1 | −1.8 | −3 | −1.4 |
Note. Number is fold change from control.
TABLE 3.
Genes Changed Differentially Between Strains
| BL6 | FVBN | SV129 | |
|---|---|---|---|
| Development | |||
| BMP2 | 1.4 | 2 | — |
| BMPR2 | −1.4 | 2 | — |
| forkhead box O3a | −1.6 | — | −1.9 |
| Kruppel-like factor 9 | −2.3 | — | — |
| SMAD1 | −1.5 | — | −1.4 |
| spondin 2 | 5.3 | 1.8 | — |
| Muscle | |||
| adrenergic receptor, alpha 1a | 2.4 | — | 1.6 |
| chloride channel 3 | — | −2.8 | — |
| chloride channel 5 | — | −4.2 | — |
| myosin IB | — | 2.1 | — |
| myosin IC | — | 2 | — |
| phospholipase C, beta 4 | −2.2 | — | −1.5 |
| Stress | |||
| chemokine CXCL12 | 2.1 | — | 1.6 |
| coagulation factor III | −2.4 | — | −1.4 |
| Fibronectin 1 | — | 1.4 | 1.5 |
| heat shock protein 1B | 3.3 | 1.4 | −1.7 |
| S100 calcium binding A8 | −1.6 | — | −2.9 |
| sialophorin | — | — | −1.7 |
| Growth | |||
| c-fos induced growth factor | 2.3 | — | 2.7 |
| cyclin D2 | — | 2.8 | 1.4 |
| cyclin T2 | — | 1.8 | — |
| cysteine rich protein 61 | — | −3.2 | — |
| retinoblastoma-like 2 | −1.9 | — | −1.4 |
| Zinc finger and BTB domain 16 | −1.8 | −1.7 | −2.4 |
| Apoptosis | |||
| caspase recruitment domain 4 | — | 1.6 | — |
| cytochrome c, somatic | — | −4.7 | — |
| deoxyribonuclease II alpha | −1.9 | −6.5 | −1.7 |
| phosphatidylinositol 3-kinase, a | −2 | — | −2.3 |
| programmed cell death 8 | — | −3.6 | – |
| TSC22 domain family 3 | −2.2 | — | −1.9 |
DISCUSSION
The goal of this study was to determine mouse strain-specific differences in phenotype and gene expression in response to chronic hypoxia. As a secondary benefit, we believe we have produced a more complete characterization of murine response to hypoxia than was previously available. Most of the changes to hypoxia we saw have been found in many previous studies: decreased body mass [14], increased hematocrit [14], increased RVSP [15], right heart and pulmonary vessel muscularization [15], and pulmonary vascular resistance [15]. Decreased aortic and pulmonary artery diameters are similar to results found in humans in the Operation Everest III study [16]. Because the mice were at room air for approximately half an hour prior to phenotyping, the increased pCO2 and decreased pO2 in chronically hypoxic animals likely resulted from ventilation-perfusion mismatch.
There were interesting baseline differences between strains, as well. We found that C57BL/6 mice had higher baseline hematocrit but lower baseline RVSP and right heart weight than the other strains, with a slightly larger aortic diameter. Because the study was performed at Denver ambient pressure (85 kPa), it is unclear whether any of these are an early response to mild hypoxia.
Only 3 of our metrics had a differential response to hypoxia by strain. Hematocrit had a smaller increase in C57BL/6 than the other strains, but to approximately the same final value. RVSP was increased to a greater extent in SV129 than the other strains, by ~14 mm Hg instead of 9 to 10 mm Hg in C57BL/6 and FVB/N mice. FVB/N mice showed less right heart muscularization than the other strains, with only a 20% increase in RV/LV+S, instead of the ~60% increase seen in C57BL/6 and SV129.
Gene expression changes also generally followed expectation, with increased expression of muscle, angiogenesis, and adhesion-related genes, and differential regulation of growth, differentiation, G-protein, stress, and apoptosis genes. All of these are pathways long associated with hypoxic response[17–19].
The most interesting finding of the array data was that, whereas the overall pathways were the same, the specific genes dysregulated were strain specific. This suggests that it is possible to accomplish the same phenotypic ends with different molecular means, and indeed that different strains use different genes to do so.
Although ours is the first study to reach this conclusion in comparisons among strains in the pulmonary vasculature of mice, similar results have been obtained in studies of the systemic vasculature and in comparisons between mice and rats.
In the systemic vasculature, Korshunov and Berk showed the magnitude of remodeling of the carotid artery after partial ligation is significantly different among the mouse strains using C3H/HeJ, SJL/J, DBA/2J, C57Bl/6J, and FVB/NJ strains. Their study suggested fundamental alterations in sensing or transducing hemodynamic signals among strains[20]. Similarly, Ward and colleagues showed that cerebral expression of the angiogenic factors, such as vascular endothelial growth factor (VEGF), angiopoietin (Ang)-1 and -2, are different in response to long-term hypoxia, and are variable among frequently used mouse strains, including CD1, C57Bl/6,129/SV, and Balb/c [21]. Both of these studies show that, in other organs, different mouse strains have similar but unique responses to hemodynamic stress.
Hoshikawa and colleagues compared some hemodynamic markers and gene expression between C57Bl6 mice and Sprague-Dawley rats [17]. They reported that mice showed less vascular remodeling than rat, though both demonstrated pulmonary hypertension. Although both had alterations in cell cycle and vasodilation-related genes, the precise changes were unique to each species.
Thus, the literature supports the hypothesis that although in broad terms the morphologic changes and molecular pathways involved in response to vascular stress are conserved, genotypic background determines the precise mechanisms used in this response. Because humans all have different genotypic background, human response to vascular stress is also likely to conserve pathways, but with different specific genes used depending on the individual genetic background, with implications for both modifier gene searches and the practice of molecular medicine.
Acknowledgments
This work was supported by NIH-HL071596, NIH-HL079315, NIH-HL072058, and Ministry of Education, Science, Culture and Sports of Japan grant 19590884.
Contributor Information
Yuji Tada, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA; and Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
Sven Laudi, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA; and Department of Anesthesiology and Intensive Care Medicine, University of Leipzig, Leipzig, Germany.
Julie Harral, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA.
Michelle Carr, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA.
Charles Ivester, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA.
Nobuhiro Tanabe, Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
Yuichi Takiguchi, Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
Koichiro Tatsumi, Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
Takayuki Kuriyama, Department of Respirology, Graduate School of Medicine, Chiba University, Chiba, Japan.
William C. Nichols, Division of Human Genetics, Cincinnati Children’s Hospital Medical Center, Cincinnati, Ohio, USA
James West, Division of Pulmonary Sciences and Critical Care Medicine, University of Colorado Health Sciences Center, Denver, Colorado, USA.
References
- 1.Naeije R. Pulmonary hypertension and right heart failure in chronic obstructive pulmonary disease. Proc Am Thorac Soc 2005. 2005;2:20–22. doi: 10.1513/pats.200407-037MS. [DOI] [PubMed] [Google Scholar]
- 2.Roy R, Couriel JM. Secondary pulmonary hypertension. Paediatr Respir Rev 2006. 2006;7:36–44. doi: 10.1016/j.prrv.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Strange C, Highland KB. Pulmonary hypertension in interstitial lung disease. Curr Opin Pulm Med 2005. 2005;11:452–455. doi: 10.1097/01.mcp.0000174250.38188.6d. [DOI] [PubMed] [Google Scholar]
- 4.Stenmark KR, Abman SH. Lung vascular development: implications for the pathogenesis of bronchopulmonary dysplasia. Annu Rev Physiol 2005. 2005;67:623–661. doi: 10.1146/annurev.physiol.67.040403.102229. [DOI] [PubMed] [Google Scholar]
- 5.Stenmark KR, Fagan KA, Frid MG. Hypoxia-induced pulmonary vascular remodeling: cellular and molecular mechanisms. Circ Res 2006. 2006;99:675–691. doi: 10.1161/01.RES.0000243584.45145.3f. [DOI] [PubMed] [Google Scholar]
- 6.Bull TM, Coldren CD, Geraci MW, Voelkel NF. Gene expression profiling in pulmonary hypertension. Proc Am Thorac Soc 2007. 2007;4:117–120. doi: 10.1513/pats.200605-128JG. [DOI] [PubMed] [Google Scholar]
- 7.Eddahibi S, Adnot S. Serotonin and pulmonary arterial hypertension. Rev Mal Respir 2006. 2006;23(Suppl 2):4S45–44S51. [PubMed] [Google Scholar]
- 8.Kawahara Y, Tanonaka K, Daicho T, Nawa M, Oikawa R, Nasa Y, Takeo S. Preferable anesthetic conditions for echocardiographic determination of murine cardiac function. J Pharmacol Sci 2005. 2005;99:95–104. doi: 10.1254/jphs.fp0050343. [DOI] [PubMed] [Google Scholar]
- 9.Reffelmann T, Kloner RA. Transthoracic echocardiography in rats. Evalution of commonly used indices of left ventricular dimensions, contractile performance, and hypertrophy in a genetic model of hypertrophic heart failure (SHHF- Mcc-facp-Rats) in comparison with Wistar rats during aging. Basic Res Cardiol 2003. 2003;98:275–284. doi: 10.1007/s00395-003-0401-3. [DOI] [PubMed] [Google Scholar]
- 10.Slama M, Susic D, Varagic J, Ahn J, Frohlich ED. Echocardiographic measurement of cardiac output in rats. Am J Physiol Heart Circ Physiol 2003. 2003;284:H691–H697. doi: 10.1152/ajpheart.00653.2002. [DOI] [PubMed] [Google Scholar]
- 11.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A 2001. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong S, Li C, Wong WH. ChipInfo: software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res 2003. 2003;31:3483–3486. doi: 10.1093/nar/gkg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Gene Ontology (GO) project in 2006. Nucleic Acids Res. 2006;34:D322–D326. doi: 10.1093/nar/gkj021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunn CD, Smith LN, Leonard JI, Andrews RB, Lange RD. Animal and computer investigations into the murine erythroid response to chronic hypoxia. Exp Hematol 1980. 1980;8(Suppl 8):259–282. [PubMed] [Google Scholar]
- 15.Fagan KA, Fouty BW, Tyler RC, Morris KG, Jr, Hepler LK, Sato K, LeCras TD, Abman SH, Weinberger HD, Huang PL, McMurtry IF, Rodman DM. The pulmonary circulation of homozygous or heterozygous eNOS-null mice is hyperresponsive to mild hypoxia. J Clin Invest 1999. 1999;103:291–299. doi: 10.1172/JCI3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boussuges A, Molenat F, Burnet H, Cauchy E, Gardette B, Sainty JM, Jammes Y, Richalet JP. Operation Everest III (Comex ’97): modifications of cardiac function secondary to altitude-induced hypoxia. An echocardiographic and Doppler study. Am J Respir Crit Care Med 2000. 2000;161:264–270. doi: 10.1164/ajrccm.161.1.9902096. [DOI] [PubMed] [Google Scholar]
- 17.Hoshikawa Y, Nana-Sinkam P, Moore MD, Sotto-Santiago S, Phang T, Keith RL, Morris KG, Kondo T, Tuder RM, Voelkel NF, Geraci MW. Hypoxia induces different genes in the lungs of rats compared with mice. Physiol Genomics 2003. 2003;12:209–219. doi: 10.1152/physiolgenomics.00081.2001. [DOI] [PubMed] [Google Scholar]
- 18.McMurtry IF, Bauer NR, Fagan KA, Nagaoka T, Gebb SA, Oka M. Hypoxia and Rho/Rhokinase signaling. Lung development versus hypoxic pulmonary hypertension. Adv Exp Med Biol 2003. 2003;543:127–137. [PubMed] [Google Scholar]
- 19.Shreeniwas R, Koga S, Karakurum M, Pinsky D, Kaiser E, Brett J, Wolitzky BA, Norton C, Plocinski J, Benjamin W, et al. Hypoxia-mediated induction of endothelial cell interleukin-1 alpha. An autocrine mechanism promoting expression of leukocyte adhesion molecules on the vessel surface. J Clin Invest 1992. 1992;90:2333–2339. doi: 10.1172/JCI116122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korshunov VA, Berk BC. Strain-dependent vascular remodeling: the “Glagov phenomenon” is genetically determined. Circulation 2004. 2004;110:220–226. doi: 10.1161/01.CIR.0000134958.88379.2E. [DOI] [PubMed] [Google Scholar]
- 21.Ward NL, Moore E, Noon K, Spassil N, Keenan E, Ivanco TL, LaManna JC. Cerebral angiogenic factors, angiogenesis, and physiological response to chronic hypoxia differ among four commonly used mouse strains. J Appl Physiol 2007. 2007;102:1927–1935. doi: 10.1152/japplphysiol.00909.2006. [DOI] [PubMed] [Google Scholar]