Abstract
Gene expression profiling may be used to stratify patients by disease severity to test the hypothesis that variable disease outcome has a genetic component. In order to define unique expression signatures in African American rheumatoid arthritis (RA) patients with severe erosive disease, we undertook a gene expression study using samples of RNA from peripheral blood mononuclear cells (PBMCs). RNA from baseline PBMC samples of 96 African American RA patients with early RA (<2 years disease duration) was hybridized to cDNA probes of the Illumina Human HT-V3 expression array. Expression analyses were performed using the ca. 25,000 cDNA probes, and then expression levels were compared to the total number of erosions in radiographs of the hands and feet at baseline and 36 months. Using a false discovery rate cutoff of Q = 0.30, 1,138 genes at baseline and 680 genes at 36 months significantly correlated with total erosions. No evidence of a signal differentiating disease progression, or change in erosion scores between baseline and 36 months, was found. Further analyses demonstrated that the differential gene expression signature was localized to the patients with the most erosive disease (>10 erosions). Ingenuity Pathway Analysis demonstrated that genes with fold change greater than 1.5 implicated immune pathways such as CTLA signaling in cytotoxic T lymphocytes. These results demonstrate that CLEAR patients with early RA having the most severe erosive disease, as compared to more mild cases (<10 erosions), may be characterized by a set of differentially expressed genes that represent biological pathways with relevance to autoimmune disease.
Keywords: Genome-wide gene expression, Sharp/van der Heijde, Pathway analysis, CLEAR, ABCoN
Introduction
A huge translational medicine challenge is to stratify patients with early RA according to the risk of radiographic severity, which would help facilitate tailored, individualized therapies to limit disease activity, joint damage, and disability [1]. A first step toward this goal is to identify biomarkers (e.g., genetic polymorphisms, gene expression levels, and serum proteins) that are associated with disease severity. RA is heritable, and HLA DRB1 shared epitope alleles [2] represent the largest genetic risk factor, with many additional genetic variants of small to moderate risk also identified [3, 4, 5]. The HLA-DRB1 shared epitope has also been shown to influence severity of RA [6]. To use genetics reliably in a clinical context for stratifying patients with RA by clinical phenotype, there is the need to definitively characterize genetic variation in disease activity and severity. In theory, identification of a genetic signature of gene expression associated with RA radiographic severity could lead to delineation of novel or distinct pathways of disease progression and help to personalize therapies at the earliest stages of disease onset. Ideally, such gene expression patterns may be identified from synovial tissue, the target of inflammation in RA. However, obtaining synovium is logistically difficult and unlikely to have widespread clinical utility, so many investigators have focused on peripheral blood cells.
Many studies have demonstrated that genetic variants and gene expression signatures can be associated with RA, but the clinical utility of these findings is unclear [1]. There have been many fewer investigations of the relationship between genetic polymorphisms, gene expression, and the severity of erosive disease. Plant et al. [7] detected moderate associations between single-nucleotide polymorphisms (SNPs) from the TRAF1/C5 locus and erosive disease in RA among persons of European ancestry. Individuals with the MIC-1 D-allele variants have been shown to have increased bone erosions over alternate genotypes [8]. Edwards et al. [9] defined a signal including 330 mRNA transcripts that appeared to differentiate 9 patients from healthy controls using PBMCs and suggested that a subset of these were informative for predicting susceptibility. Genes related to apoptotic pathways, including p53, have been shown to be consistently under-expressed in individuals with one of several autoimmune diseases relative to healthy controls by using DNA microarrays to measure expression of peripheral blood monocytes (PBMCs) [10]. Olsen et al. [11] also derived a set of eight genes that appeared to be under-expressed in a sample of patients with established RA (N = 11) relative to patients with early RA (N = 7). In addition, microarray technology has been used to detect differential gene expression in a small number of patients with early and longstanding RA using synovial tissue [12]. In an attempt to find genes differentially expressed between a larger sample of patients and controls using PBMCs, Batliwalla et al. [13] defined a signature showing a distribution over representing CD14, which could be attributed to a proportionally larger number of circulating monocytes in the RA patients. Junta et al. [14] attempted to make inference on the set of genes uniquely defining each of four different subsets of RA patients using a sample of 23 patients differing according to combinations of the presence/absence of anti-cyclic citrullinated peptide, DAS-28, and HLA-DRB1 shared epitope alleles. Investigations using genome-wide scans of gene expression and severity of erosive disease have not previously been reported in rheumatoid arthritis.
These previous studies illustrate the feasibility of using large-scale cDNA libraries to perform expression profiling and identify a gene expression signature that explains variation in RA susceptibility. However, the studies are small and their practicality is questionable due to sample size and phenotype measured. Additional studies in RA and related autoimmune disorders are needed to find unique genetic profiles related to RA pathogenesis. Furthermore, in order to insure broad applicability, investigations from samples in multiple populations and racial or ethnic groups are needed. Many applications have used cDNA arrays of synovial tissue, for example [15], or peripheral blood monocytes [16] to investigate molecular genetic signatures of response to anti-TNF therapies. From a clinical perspective, more research defining genetic signatures of eventual mild or aggressive RA would be highly useful. Longitudinal studies that recruit large numbers of patients early in the disease course with numerous follow-up visits are most appropriate. Here, we present our findings of gene expression patterns in PBMCs with radiographic severity in African American patients with early RA disease co-enrolled in the Consortium for the Longitudinal Evaluation of African Americans with Early Rheumatoid Arthritis (CLEAR) Registry who were co-enrolled in the Autoimmune Biomarkers Collaborative Network (ABCoN).
In order to test the hypothesis that differential gene expression in peripheral blood cells is associated with RA disease severity, we sought to (1) correlate gene expression at baseline with radiographic severity at baseline (<2 years disease duration) and three years of disease duration in a sample of African Americans with early RA and (2) demonstrate that there are variations in expression among particular genes in pathways with relevance for RA pathogenesis.
Materials and methods
Study subjects
Gene expression profiling was performed on 96 African American patients with early RA from the CLEAR Registry. The CLEAR Registry was established in 2000 with the goal of enrolling African Americans with early RA, defined as disease duration of less than 2 years, and following them longitudinally. The subjects met the 1987 revised American Rheumatism Association (now the American College of Rheumatology [ACR]) criteria and were self-defined African Americans of 19 years of age and older. The participants were recruited at tertiary referral centers in Alabama (University of Alabama at Birmingham—coordinating center), Georgia (Emory University), North Carolina (University of North Carolina at Chapel Hill), Missouri (Washington University), and South Carolina (The Medical University of South Carolina) and were followed longitudinally for 5 years.
A total of 359 patients with early RA (<2 year disease duration) were enrolled starting in 2000 and followed longitudinally until 5 years disease duration. Comprehensive demographic, clinical, and radiographic data were obtained by interviews and examinations during the baseline visit and at 36 and 60 months from disease onset. Of the 359 subjects, 153 (43%) were co-enrolled in the Autoimmune Biomarkers Collaborative Network (ABCoN) ref [17]. Blood was obtained for serum/plasma, and RNA from these CLEAR subjects at three additional time points: (1) two weeks after the baseline CLEAR visit; (2) one year after the initial ABCoN blood draw; (3) two years after the initial ABCoN blood draw. This analysis used baseline RNA from peripheral blood cells from 96 CLEAR patients who were co-enrolled in ABCoN.
At the initial CLEAR visit, peripheral blood was collected for the measurement of serum rheumatoid factor (RF) and anti-cyclic citrullinated peptide (anti-CCP) antibodies. At each CLEAR study visit, questionnaires were used to document current and previous drug treatments with disease-modifying anti-rheumatic drugs (DMARDs) and glucocorticoids. Radiographs of hands and feet were obtained at baseline, 36 months disease duration, and 60 months disease duration. The method of Sharp, as modified by van der Heijde, was used to score the radiographs, as previously described [18]. For detailed information on data and materials available for research on these subjects, please refer to: http://medicine.uab.edu/rheum/70918/.
The CLEAR Registry was approved by the Institutional Review Boards of the participating institutions.
Gene expression microarrays
RNA from whole blood was stabilized using PAXgene Blood RNA Tubes (Qiagen). There were baseline RNA samples from a total of 106 CLEAR patients who also had baseline and 36-month disease duration radiograph scores. Total RNA from these 106 baseline samples was isolated using PAXgene Blood RNA Kit (Qiagen) as per manufacturer’s instructions. The quality of RNA was assessed using Agilent RNA 6000 Nano Kit. Of these 106 samples, 10 had either poor-quality RNA or did not amplify well enough to allow hybridization, leaving 96 samples for final analysis. Gene expression profiling was performed using the Illumina HumanHT-12 v3 Expression BeadChip platform containing 48,000 probes covering RefSeq and Unigene annotated genes (Illumina Inc.). And 200 ng total RNA was labeled using a Illumina® Total Prep ™ RNA Amplification Kit (Applied Biosystems). Briefly, the protocol features a first- and second-strand reverse transcription step, followed by a single in vitro transcription (IVT) amplification that incorporates biotin-labeled nucleotides to generate biotinylated, antisense RNA (cRNA or aRNA). The purified aRNA is then quantified and the fragment size ascertained on Bioanalyzer (Agilent) using Agilent RNA 6000 Nano Kit. Then, 750 ng of labeled biotinylated aRNA probe was hybridized overnight to HumanHT-12 beadchips (Illumina). The hybridization, washing, and scanning were performed according to the manufacturer’s instructions. The chips were scanned using a iScan Reader (Illumina). The microarray images were registered and extracted automatically during the scan using the manufacturer’s default settings.
Statistical analysis
Data preprocessing
Raw data from the Illumina gene expression bead array were first normalized using quantile normalization [19]. Data were filtered to exclude probes with less than six samples showing detection P values less than 0.05.
Statistical testing
To identify genes that are correlated with the total erosion scores at baseline, 36 months disease duration, and the change in score between baseline and 36 months disease duration, we fitted three separate linear models (one for each response variable) to the gene expression of the 96 samples using a gene-by-gene approach, with array and RNA batch as covariates. The relationship between the preprocessed microarray data (gene expression) and the erosion score (baseline, 36 months, or the change) was estimated using the following model
with yijk representing preprocessed microarray data, μ as the overall mean, E as the erosion score (continuous variable), Ri as the RNA extraction batch i (i = 1,…,5), Cj as the chip j (j = 1,…,8), and ε ijk as the residual. A variance shrinkage F test [20] was used to test the effect of E. P values were obtained based on permutation [21], and false discovery rate (FDR) [22] was used for multiple testing adjustments. For obtaining the expression fold change between the individuals with high and low erosion scores, we separated the samples into two groups, erosion score >10 and the remaining. The two groups were compared using the same linear model described as above except that E is categorical with two levels. Hierarchical clustering was used to cluster gene expression fold change among the genes meeting the FDR criterion for both baseline and 36 months. To further visualize the cluster membership, the gene expression fold change data according to the FDR criterion were analyzed with principal components (PC), and the first two PC scores were plotted.
All the statistical analyses of the expression data were conducted using MAANOVA package [23] in the Bio-conductor (http://www.bioconductor.org/). Finally, to test the null hypothesis that the distribution of erosions at baseline and 36 months was the same, we used generalized mixed model regression to fit a model with total erosion score as the response (assumed Poisson) and two predictor variables: (1) time of sampling, dummy coded as 0 (baseline) or 1 (36 months) and (2) a random effect to account for the correlated observations of number of erosions on the same patients. The random effect insures that the observations are not treated as independent and a proper hypothesis test of the fixed effect variable, time.
Gene set analyses
Of the number of probes tested (N = 25,868), a fraction of these were over- or under-expressed with fold change > 1.5 at baseline (N = 410) and 36 months (N = 289), when comparing individuals with zero erosions and > 10 erosions. To determine whether these genes were overrepresented among relevant biological functions, pathways, or networks, the data were analyzed using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com). The four combinations of over- and under-expressed genes at baseline and 36 months were separately analyzed. Evidence for enrichment comes from testing the overrepresentation of defined sets of genes with predefined biological functions or pathways compared to a null distribution, generated by permuting the phenotypes, using a right-tailed Fisher’s exact test to calculate P values. For pathway analysis, the overrepresentation of canonical pathways (e.g., Toll-like receptor signaling, CTLA4 signaling in cytotoxic T lymphocytes) among a specified set of target genes is assessed, with significance determined by computing a Fisher’s exact test with Benjamini–Hochberg correction for multiple testing.
Results
Microarray analyses
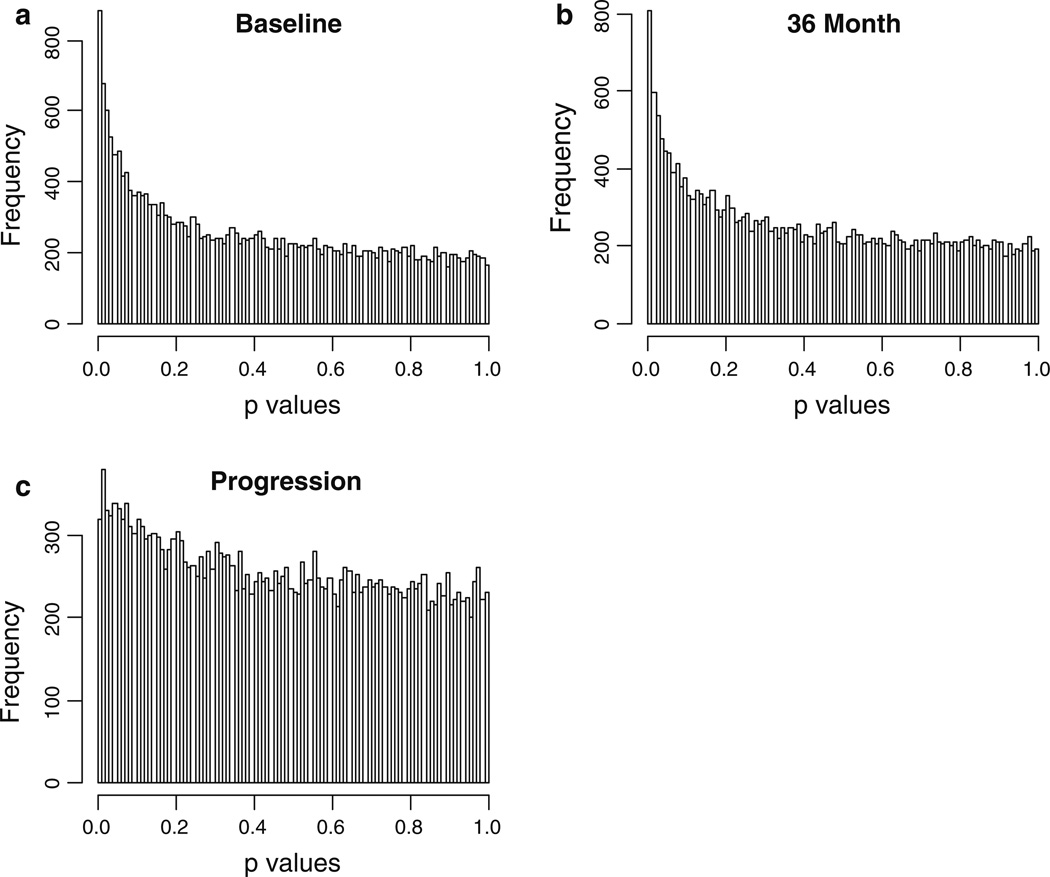
Total erosion score was significantly correlated with gene expression for a substantial proportion of genes at baseline and 36 months (Fig. 1a, b). Using an FDR cutoff of Q = 0.30, we identified 1,138 genes at baseline (and 680 genes at 36 months disease duration) that are above detection level and significantly correlated with the erosion score. To assess the consistency by which genes are differentially expressed over time, we compared the proportion of the same genes that were differentially expressed at both time points; eighty percent of the genes with significant differential expression at 36 months were also differentially expressed at baseline. However, when considering all ca. 25,000 probes in the analysis, there was substantial evidence for asymmetry between baseline and 36 months among quintiles of P values representing the tests of gene expression and number of erosions (symmetry statistic = 110, df = 10, P < 0.0001). Thus, results for both baseline and 36 months are presented. A very modest set of genes appeared to be differentially expressed when the outcome measure was disease progression, or the change in number of erosions between 36 months and baseline (Fig. 1c). The frequency distribution of P values will be uniform if the hypothesis of no linear differential signal across the range of erosion scores or change in erosion scores is true. Because the P values were obtained from permutation tests, we assuredly maintain the type 1 error rate at 5% [21]. Evidently, the distribution is much closer to uniform for the progression outcome variable.
Fig 1.
Histogram of the P values from testing the relationship between erosion score and gene expression. The predictor variable is the continuous variable erosion score at baseline (a), 36 months (b) and the change in erosions from baseline to 36 months (c)
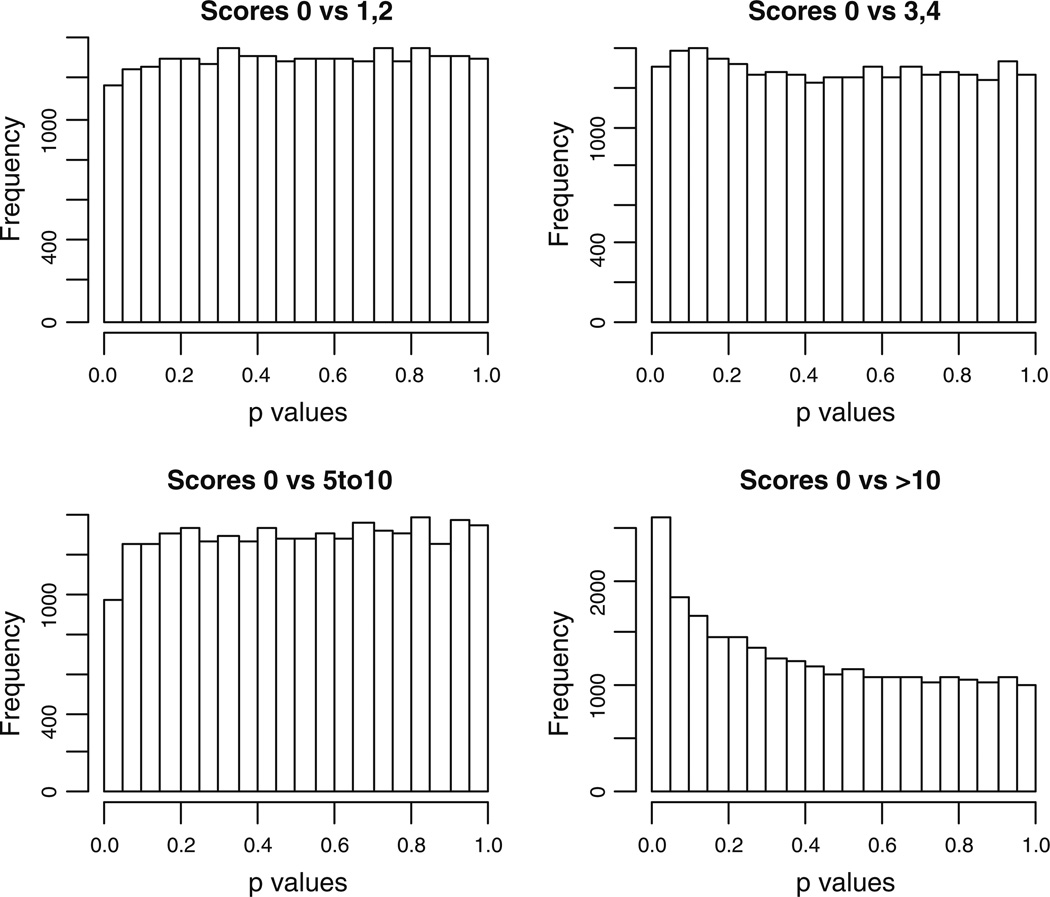
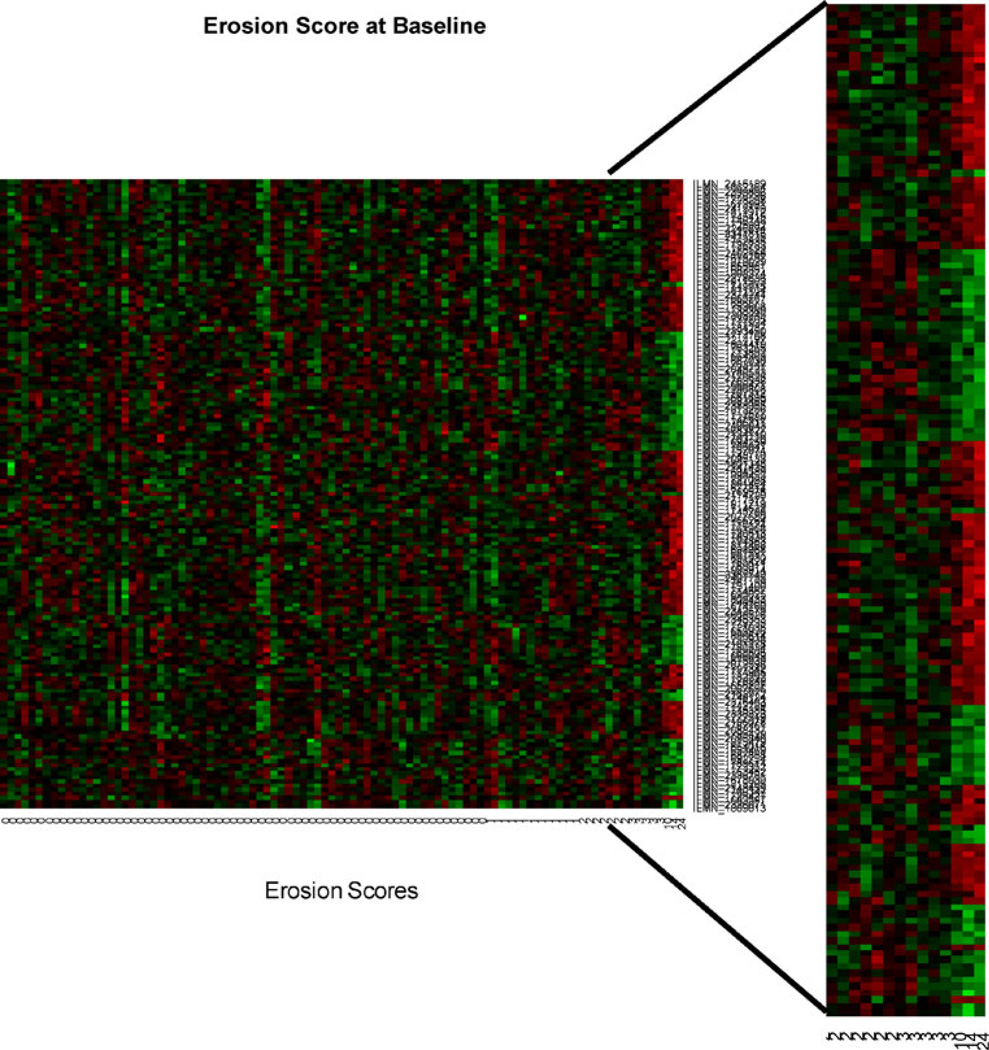
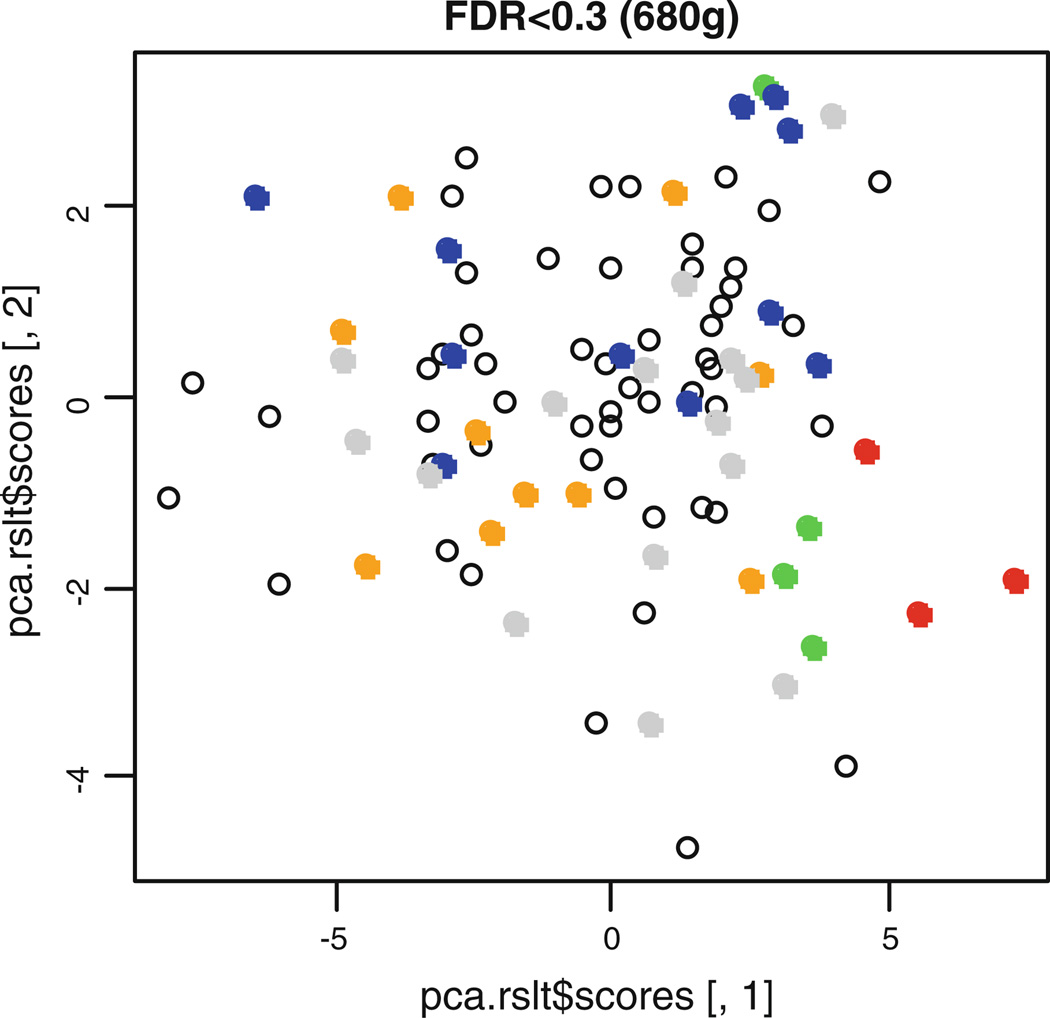
In order to investigate the main source of the relationship between gene expression and erosive damage, we further analyzed the relative differences in expression among groups of individuals according to severity of erosive disease. The four groups according to erosion scores were {0 vs. 1–2}, {0 vs. 3–4}, {0 vs. 5–10}, and {0 vs. > 10}. A similar pattern of significant P values as obtained in Fig. 1 was found when testing the difference in gene expression between individuals with the largest erosion scores and the individuals with zero erosions (Fig. 2). The heat map illustrates the subset of differentially expressed genes, as determined by testing the fold change compared between patients with zero and > 10 erosions at baseline and 36 months (FDR = 0.3), in the three individuals with highest disease severity (Fig. 3). Principal component analysis of the gene expression data also shows that the three patients with greater than 10 erosions clustered similarly in principal component space. This further illustrates that these patients demonstrate a similar molecular signature of gene expression compared to patients with less than 10 erosions (Fig. 4).
Fig 2.
The histogram of P values from the comparison between each group of samples with the samples with erosion score of 0. Differentially expressed genes are only observed in the comparison between the comparison between the group with erosion score of 0 and that above 10
Fig 3.
Heat map for genes significant at FDR 0.30 for testing linear relationship with erosion score and having a fold change above 1.5 between the three largest scores and the remaining individuals
Fig 4.
The first two principal component scores of the gene expression data with FDR < 0.30. Points representing patients with > 10 erosions are colored red
Gene set analysis
The differentially expressed genes with fold change greater than 1.5 from the test of the relationship with erosion score were used to examine the associated biological pathways using the software Ingenuity. The up- and down-regulated genes (only genes with FC > 1.5) at baseline and 36 months were examined separately for gene set analysis. The list of up-regulated genes at baseline and 36 months yielded significantly enriched gene sets of pathways representing CTLA4 signaling in cytotoxic T lymphocytes (Fig. 5), B cell development, calcium-induced T lymphocyte apoptosis and interferon signaling, among others (Table 1). The list of down-regulated genes at baseline and 36 months also high-lighted pathways of immune function including Toll-like receptor signaling (Table 1).
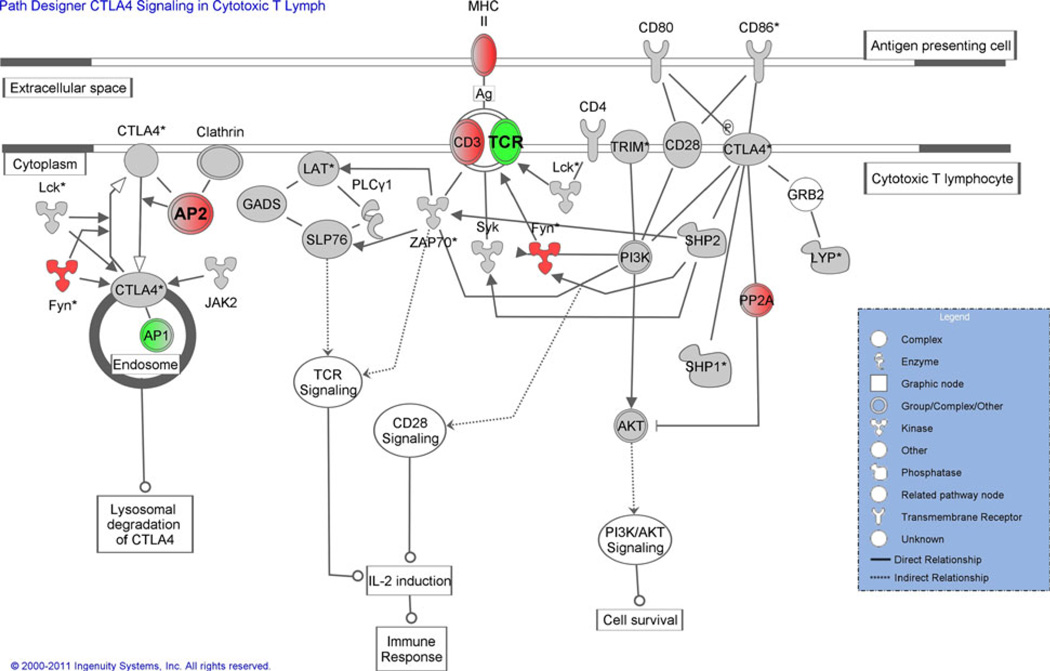
Fig 5.
Pathway diagram of the CTLA4 signaling in cytotoxic T lymphocytes from the IPA. Genes that have a positive fold change (up-regulated) are indicated in red, and those with a negative fold change (down-regulated) in green. Genes in gray were present in the analysis, but did not meet the 1.5-fold change cutoff used in this analysis. The intensity of the color is indicative of the degree of regulation. Genes in white did not have expression data available. For interpretability, both up- and down-regulated genes at baseline are shown in the same diagram, although the pathway was significant for only up-regulated at baseline and 36 months and not for either time point for the down-regulation. TCR and AP2 (indicated in bold) met the 1.5-fold change threshold at baseline, but not at 3 years, all other values remained the same
Table 1.
Top 5 canonical pathway results using Ingenuity Pathways Analysis (Ingenuity® Systems, www.ingenuity.com)
| Baseline-up | Baseline-down | 36 month-up | 36 month-down |
|---|---|---|---|
| B cell development (7.66 × 10E-04) | Oxidative phosphorylation (3.22 × 10E-09) | Interferon signaling (2.72 × 10E-07) | Mitochondrial dysfunction (5.54 × 10E-06) |
| Phospholipase C signaling (8.16 × 10E-04) | Mitochondrial dysfunction (9.18 × 10E-08) | Calcium-induced T lymphocyte apoptosis (1.28 × 10E-04) | Oxidative phosphorylation (1.02 × 10E-05) |
| Calcium-induced T lymphocyte apoptosis (3.4 × 10E-03) | Ubiquinone biosynthesis (8.85 × 10E-05) | Activation of IRF by cytosolic pattern recognition receptors (2.2 × 10E-04) | Toll-like receptor signaling (6.60 × 10E-04) |
| Aminoacyl tRNA biosynthesis (5.65 × 10E-03) | NRF2-mediated oxidative stress response (1.15 × 10E-03) | Nur77 signaling in T lymphocytes (5.89 × 10E-04) | Cdc42 signaling (8.31 × 10E-03) |
| CTLA4 signaling in cytotoxic T lymphocytes (5.77 × 10E-03) | Protein ubiquitination pathway (5.24 × 10E-03) | Role of pattern recognition receptors in recognition of bacteria and viruses (6.07 × 10E-04) | Ubiquinone biosynthesis (1.14 × 10E-02) |
Enrichment P values are in parentheses
Discussion
Using peripheral blood mononuclear cell RNA samples from the largest cohort of African Americans with early RA available to date, and the Illumina HT-12 V3 human expression bead chip, we have identified a gene set including 1,138 (680) differentially expressed genes, which were correlated with the number of baseline (three year) erosions. These results are encouraging but must be viewed cautiously as the majority of these effects were evident from the analysis of the three patients with the most severe erosive disease (Fig. 2). We used gene set analysis to discover relevant biological pathways and functions that overrepresented the set of genes with moderate to high fold change (when comparing gene expression for individuals with zero erosions to those with greater than 10). The results from the Ingenuity Pathway Analysis tool successfully illuminated biological pathways with relevance to autoimmune disease. Although the evidence is preliminary, the results indicate we can detect a meaningful gene expression signature at baseline that is associated with disease severity at baseline and three years.
The gene expression signature that appears to correlate with the number of erosions at baseline and 36 months is a measure of transcriptional activity associated with RA severity, but not necessarily disease progression. We also correlated gene expression intensity with the change in the erosion score between baseline and three years disease duration. The signal that was readily evident when examining the correlation of gene expression and erosions at baseline and 36 months was greatly attenuated when measuring the change in erosions. The weak signal between gene expression and disease progression is likely due to a limitation inherent to this particular cohort of patients with early RA. Although the change in the mean of the distribution of erosions at 36 months was significantly higher than baseline (Table 2, Z = 3.7, P = 0.0002), still the majority of patient radiographs (55%) showed no erosive disease at 36 months. It is possible that given a study of longer duration, we could observe a gene expression signature indicative of the progression of erosive disease.
Table 2.
Demographic data for the 96 patients co-enrolled in CLEAR and ABCoN
| Variable | Percent or mean (SD) |
|---|---|
| Gender, % female | 85 |
| Age at diagnosis (years) | 49 (12) |
| Disease duration (months) | 15 (7.3) |
| Smoking (ever/never + ever) | 54 |
| CCP | 71 |
| RF | 79 |
| Erosions at baseline | 0.98 (3.4) |
| Erosions at 36 months | 1.6 (3.8) |
One of the goals of gene expression profiling in rheumatoid arthritis is to differentiate patients according to the severity of disease [13], and to identify corresponding genes and pathways underlying the signal. Olsen et al.’s work with gene expression differences between 11 patients with early RA and eight patients with long-standing disease implicated the up-regulated gene, TGFBR2 among others, which is related to cell growth and proliferation processes [11]. Similarly, another transforming growth factor TGFBR3 was among the top 10 genes up-regulated in the patients with the most severe disease at baseline and 36 months in our study. What role these cell growth and proliferative genes have in RA disease pathogenesis is unknown, but perhaps genes of this biological process or pathways warrant further study.
One of the limitations of these large-scale genome-wide expression studies is the sheer volume of genes and information that results. Clearly, we have been able to establish a signal (Figs. 1, 2, 3), but much work remains to define a conclusive signature for RA severity and progression in the cohort of African American patients representing the longitudinal arm of CLEAR. Gene set analysis is one approach that has proven valuable for taking large sets of genes and quantitatively assigning biological pathways representing the gene set in a nonrandom way [24, 25]. Given the few samples that defined the expression signature, we should treat the gene set analysis with a degree of reservation. Nevertheless, the differentially expressed genes identified using the statistical analysis were indicative of immune-related pathways. For example, Ingenuity identified the CTLA4 signaling pathway, based not on the actual significant fold change of the CTLA gene itself, but on significantly up- and down-regulated genes in this particular pathway (Fig. 5). Clearly, CTLA4 has been a focus of interest in RA genetics [26, 27] and therapy, for example [28], and this gene and its role in RA pathobiology remains an active area of research. Lequerre et al. [12] in a study of differential expression of 12,000 cDNA probes tested from RNA of synovial tissue sampled from early and long-standing RA also performed gene set analysis using the PANTHER database. Lequerre et al. [12] noted that T cell activation was implicated as a significant pathway using the set of genes overexpressed in early RA. By contrast, in our study, T cell apoptotic pathways were implicated by the set of genes up-regulated in patients with more severe disease. Because our study design was very different, for example, only patients with less than two years disease duration were enrolled and our use of PBMCs, our studies are difficult to compare. At the very least, it appears that gene expression profiling of both PBMCs and RA synovium can implicate pathways consistent with T cell-mediated pathogenesis.
While the study results are encouraging, there are additional limitations to consider aside from the ones previously mentioned, for example small sample size of patients with more aggressive disease progression. Using a population of PBMCs also has its disadvantages in that changing cell populations (e.g., percentage of CD4 + T cells, CD20 + B cells) with age or disease severity are possible but uncontrolled. Furthermore, sampling of cells from the peripheral blood may complicate the interpretation of organ-specific disease [13]. In particular, it could be argued that the gene expression signal derived in our study from PBMCs may bear no relationship to the synovial cells directly involved in RA pathogenesis. Furthermore, if the differential gene expression signal that we observed in this particular study is not due to disease severity per se but instead to an unobserved but correlated clinical or biological feature, then we may find it difficult to validate our findings in another independent sample of patients. Finally, successful validation of the gene expression signature may not be possible in late stage RA if, as Lequerre et al. [12] have shown, heterogeneous patterns of expression exist between early and long-standing RA. Nonetheless, this exploratory study forms the basis of ongoing investigations of gene expression in African American patients with early RA in an attempt to validate specific genes showing significant correlation between expression levels and erosion score. We are encouraged that future expression studies that robustly address the limitations will yield important and novel genetic features for predicting RA disease progression.
Acknowledgments
The authors kindly thank the CLEAR investigators: Moreland, Conn, Smith, Callahan, Jonas, Brasington, Howard. This research was supported by NIH 2P60 AR048095-06 (RP Kimberly, P.I.) Multidisciplinary Clinical Research Center Project 3: Predictors of Rheumatoid Arthritis Severity in African Americans; and NIH N01-AR-6-2278 (SLB, PI) Continuation of the Consortium for the Longitudinal Evaluation of African Americans with Early Rheumatoid Arthritis (CLEAR) Registry. Support also provided by the UAB Center for Clinical and Translational Science through the NIH National Center for Research Resources as part of its Clinical and Translational Science Award Program (5UL1RR025777-03, 5KL2RR025776-03, 5TL1RR025775-03). RJR was supported in part by NIH K01- AR060848 and LKV by K01-DK080188. The authors gratefully acknowledge the kind willingness of the CLEAR study participants whose PBMCs were used for this study.
Contributor Information
Richard J. Reynolds, Email: rreynolds@uab.edu, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA.
Xiangqin Cui, Section on Statistical Genetics, Department of Biostatistics, University of Alabama at Birmingham, Birmingham, AL, USA.
Laura K. Vaughan, Section on Statistical Genetics, Department of Biostatistics, University of Alabama at Birmingham, Birmingham, AL, USA
David T. Redden, Department of Biostatistics, University of Alabama at Birmingham, Birmingham, AL, USA
Zenoria Causey, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA.
Elizabeth Perkins, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA.
Tishi Shah, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA.
Laura B. Hughes, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA
Aarti Damle, Robert S. Boas Center for Genomics and Human Genetics, Manhasset, NY, USA.
Marlena Kern, Robert S. Boas Center for Genomics and Human Genetics, Manhasset, NY, USA.
Peter K. Gregersen, Robert S. Boas Center for Genomics and Human Genetics, Manhasset, NY, USA
Martin R. Johnson, Department of Pharmacology and Toxicology, University of Alabama at Birmingham, Birmingham, AL, USA
S. Louis Bridges, Jr, Email: lbridges@uab.edu, Division of Clinical Immunology and Rheumatology, Department of Medicine, University of Alabama at Birmingham, 1530 3rd Avenue South, SHEL 178, Birmingham, AL 35294-2182, USA.
References
- 1.Scherer H, Dörner T, Burmester G. Patient-tailored therapy in rheumatoid arthritis: an editorial review. Curr Opin Rheumatol. 2010;22:237–245. doi: 10.1097/BOR.0b013e328337b832. [DOI] [PubMed] [Google Scholar]
- 2.Gregersen P, Silver J, Winchester R. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 3.Coenen M, Gregersen P. Rheumatoid arthritis: a view of the current genetic landscape. Genes Immun. 2009;10:101–111. doi: 10.1038/gene.2008.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding B, Padyukov L, Lundström E, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl EA, Raychaudhuri S, Remmers EF, et al. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat Genet. 2010;42:508–514. doi: 10.1038/ng.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weyand CM, Hicok KC, Conn DL, Goronzy JJ. The influence of HLA-DRB1 genes on disease severity in rheumatoid arthritis. Ann Intern Med. 1992;117:801–806. doi: 10.7326/0003-4819-117-10-801. [DOI] [PubMed] [Google Scholar]
- 7.Plant D, Thomson W, Lunt M, et al. The role of rheumatoid arthritis genetic susceptibility markers in the prediction of erosive disease in patients with early inflammatory polyarthritis: results from the Norfolk Arthritis Register. Rheumatology. 2011;50:78–84. doi: 10.1093/rheumatology/keq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown DA, Moore J, Johnen H, et al. Serum macrophage inhibitory cytokine 1 in rheumatoid arthritis: a potential marker of erosive joint destruction. Arthritis Rheum. 2007;56:753–764. doi: 10.1002/art.22410. [DOI] [PubMed] [Google Scholar]
- 9.Edwards CJ, Feldman JL, Beech J, et al. Molecular profile of peripheral blood mononuclear cells from patients with rheumatoid arthritis. Mol Med. 2007;13:40–58. doi: 10.2119/2006-000056.Edwards. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olsen NJ, Moore JH, Aune TM. Gene expression signatures for autoimmune disease in peripheral blood mononuclear cells. Arthritis Res Ther. 2004;6:120–128. doi: 10.1186/ar1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen N, Sokka T, Seehorn C, et al. A gene expression signature for recent onset rheumatoid arthritis in peripheral blood mononuclear cells. Ann Rheum Dis. 2004;63:1387–1392. doi: 10.1136/ard.2003.017194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lequerré T, Bansard C, Vittecoq O, et al. Early and long-standing rheumatoid arthritis: distinct molecular signatures identified by gene-expression profiling in synovia. Arthritis Res Ther. 2009;11:R99. doi: 10.1186/ar2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Batliwalla FM, Baechler EC, Xiao X, et al. Peripheral blood gene expression profiling in rheumatoid arthritis. Genes Immun. 2005;6:388–397. doi: 10.1038/sj.gene.6364209. [DOI] [PubMed] [Google Scholar]
- 14.Junta CM, Sandrin-Garcia P, Fachin-Saltoratto AL, et al. Differential gene expression of peripheral blood mononuclear cells from rheumatoid arthritis patients may discriminate immunogenetic, pathogenic and treatment features. Immunology. 2009;127:365–372. doi: 10.1111/j.1365-2567.2008.03005.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindberg J, Wijbrandts C, van Baarsen L, et al. The Gene Expression Profile in the Synovium as a Predictor of the Clinical Response to Infliximab Treatment in Rheumatoid Arthritis. Plos One. 2010;5 doi: 10.1371/journal.pone.0011310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lequerré T, Gauthier-Jauneau A, Bansard C, et al. Gene profiling in white blood cells predicts infliximab responsiveness in rheumatoid arthritis. Arthritis Res Ther. 2006;8:R105. doi: 10.1186/ar1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu C, Batliwalla F, Li W, et al. Genome-wide association scan identifies candidate polymorphisms associated with differential response to anti-TNF treatment in rheumatoid arthritis. Mol Med. 2008;14:575–581. doi: 10.2119/2008-00056.Liu. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bridges SL, Causey ZL, Burgos PI, et al. Radiographic severity of rheumatoid arthritis in African Americans: results from a multicenter observational study. Arthritis Care Res. 2010;62:624–631. doi: 10.1002/acr.20040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolstad B, Irizarry R, Astrand M, Speed T. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 20.Cui X, Hwang JT, Qiu J, Blades NJ, Churchill GA. Improved statistical tests for differential gene expression by shrinking variance components estimates. Biostatistics. 2005;6:59–75. doi: 10.1093/biostatistics/kxh018. [DOI] [PubMed] [Google Scholar]
- 21.Yang H, Churchill G. Estimating p-values in small microarray experiments. Bioinformatics. 2007;23:38–43. doi: 10.1093/bioinformatics/btl548. [DOI] [PubMed] [Google Scholar]
- 22.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Roy Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 23.Wu H, Kerr MK, Cui X, Churchill GA. MAANOVA, a software package for the analysis of spotted cDNA microarray experiments. In: Parmigiani G, Garret ES, Irizarry RA, Zeger SL, editors. The analysis of gene expressions data: an overview of methods and software. New York: Springer; 2003. [Google Scholar]
- 24.Nam D, Kim SY. Gene-set approach for expression pattern analysis. Brief Bioinform. 2008;9:189–197. doi: 10.1093/bib/bbn001. [DOI] [PubMed] [Google Scholar]
- 25.Huang dW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plenge RM, Padyukov L, Remmers EF, et al. Replication of putative candidate-gene associations with rheumatoid arthritis in >4,000 samples from North America and Sweden: association of susceptibility with PTPN22, CTLA4, and PADI4. Am J Hum Genet. 2005;77:1044–1060. doi: 10.1086/498651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hughes LB, Reynolds RJ, Brown EE, et al. Most common single-nucleotide polymorphisms associated with rheumatoid arthritis in persons of European ancestry confer risk of rheumatoid arthritis in African Americans. Arthritis Rheum. 2010;62:3547–3553. doi: 10.1002/art.27732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kremer JM, Westhovens R, Leon M, et al. Treatment of rheumatoid arthritis by selective inhibition of T-cell activation with fusion protein CTLA4Ig. N Engl J Med. 2003;349:1907–1915. doi: 10.1056/NEJMoa035075. [DOI] [PubMed] [Google Scholar]