Abstract
The genetic diversity of siderophore-producing bacteria of tobacco rhizosphere was studied by amplified ribosomal DNA restriction analysis (ARDRA), 16S rRNA sequence homology and phylogenetics analysis methods. Studies demonstrated that 85% of the total 354 isolates produced siderophores in iron limited liquid medium. A total of 28 ARDRA patterns were identified among the 299 siderophore-producing bacterial isolates. The 28 ARDRA patterns represented bacteria of 14 different genera belonging to six bacterial divisions, namely β-, γ-, α-Proteobacteria, Sphingobacteria, Bacilli, and Actinobacteria. Especially, γ-Proteobacteria consisting of Pseudomonas, Enterobacter, Serratia, Pantoea, Erwinia and Stenotrophomonas genus encountered 18 different ARDRA groups. Results also showed a greater siderophore-producing bacterial diversity than previous researches. For example, Sphingobacterium (isolates G-2-21-1 and G-2-27-2), Pseudomonas poae (isolate G-2-1-1), Enterobacter endosymbiont (isolates G-2-10-2 and N-5-10), Delftia acidovorans (isolate G-1-15), and Achromobacter xylosoxidans (isolates N-46-11HH and N-5-20) were reported to be able to produce siderophores under low-iron conditions for the first time. Gram-negative isolates were more frequently encountered, with more than 95% total frequency. For Gram-positive bacteria, the Bacillus and Rhodococcus were the only two genera, with 1.7% total frequency. Furthermore, the Pseudomonas and Enterobacter were dominant in this environment, with 44.5% and 24.7% total frequency, respectively. It was also found that 75 percent of the isolates that had the high percentages of siderophore units (% between 40 and 60) belonged to Pseudomonas. Pseudomonas sp. G-229-21 screened out in this study may have potential to apply to low-iron soil to prevent plant soil-borne fungal pathogen diseases.
Keywords: genetic diversity, siderophore, tobacco rhizosphere, ARDRA, rRNA, sequencing
INTRODUCTION
Tobacco is an important economic crop in China. However, soil-borne bacterial and fungal pathogen diseases cause severe losses annually because of succession planting. So effective plant growth-promoting rhizobacteria (PGPR) including siderophore-producing bacteria were used to prevent infectious diseases of plant roots (6,14).
Siderophores are low molecular weight (generally<10, 000Da), iron-coordinating, organic compounds produced by most aerobic and facultative anaerobic microorganisms to combat low-iron stress (11). Data showed that the excretion of siderophores by rhizosphere bacteria may stimulate plant growth by improving Fe nutrition of the plants (13) or by inhibiting the establishment of plant pathogens or other harmful microorganisms (8,19).
Production of siderophore and antifungal activity were simultaneously exhibited by free-living rhizospheric isolates of Azotobacter (16.22%), fluorescent Pseudomonas (11.11%) and Bacillus (10%) (1). The siderophore-producing Pseudomonas SF4c can increase the shoot and dry biomass of wheat by 23% and 45% under greenhouse conditions, respectively (5).
The pioneer works on tobacco rhizosphere microorganisms were concentrated on the isolation and identification of various rhizosphere microbes mainly using traditional physiological and biochemical methods (10,18). Recently, amplified ribosomal DNA restriction analysis (ARDRA) technique has been successfully used for bacterial diversity analysis in a great variety of environments, including soil and tobacco rhizosphere (4).
The aim of this work was to analyze the diversity of cultivable siderophore-producing bacteria of tobacco rhizosphere by ARDRA analysis and rDNA sequencing. Better knowledge of cultivable siderophore-producing bacteria composition in tobacco rhizosphere will undoubtedly contribute to screening for PGPR to prevention of tobacco soil-borne fungal pathogen diseases.
MATERIALS AND METHODS
Rhizosphere soil
Samples from tobacco plants were collected from two different locations (designated as locations 1 and 2) that were less than 0.1 km apart of Science Park of Zunyi Region (27º252 N, 106º332 E) in August 2006. Zunyi Region (Guizhou Province) is an important tobacco production base of the People’s Republic of China, but soil-borne bacterial and fungal pathogen diseases cause severe losses annually in this base. The tobacco plants (NC-82) were grown in yellow soil (pH value is 6.36).
Bacterial isolation
Ten grams of each sample was dissolved in 90 ml of aseptic water. After appropriate dilutions, samples were spreaded to beef extract peptone agar medium (beef extract 0.3%, peptone 1%, NaCl 0.5%, agar 2%, w/v, pH 7.4~7.6). Plates were incubated at 30ºC for 2 to 3 days. Bacterial isolates were obtained from agar plates presenting between 30 and 150 colonies. Stock cultures were prepared in aseptic water supplemented with glycerol and kept frozen at -80ºC prior to use.
Screening of siderophore-producing bacterial isolates
Modified CAS-liquid medium (MM9, Tris buffer, casamino acids 0.3%, sucrose 0.2%, thiamine·HCl 0.0002%, and succinate 0.2%, w/v) (2) was used to screen siderophore-producing bacteria. The detection experiment was repeated three times. Siderophore production of the isolates were detected by CAS-liquid medium and CAS agar plates with a little modified (200 ml of blue agar with 20 ml resultant dark blue liquid, 150 ml double distilled-deionized water, 20 ml 10× MM9 salts, 3 g agar, 2.4 g of a 50% (w/w) NaOH solution, 6 ml casamino acids (10%, w/v), 2 ml glucose (10%, w/v), 0.2 ml thiamine·HCl (0.2%, w/v), and 0.6 ml L-tryptophan (1%, w/v)) (20), respectively. The siderophore production was evaluated as siderophore units (%) in liquid Bactéria of tobacco rhizosphere medium (12). Double distilled-deionized water was used to prepare all media and reagents. Glasswares were soaked for three days in 6 M HCl and rinsed with distilled-deionized water several times to remove traces of iron.
Total DNA extraction and PCR amplification
The DNA of bacterial isolates was prepared according to the procedures of Murray and Thompson (15)with the exception that for Gram-positive bacteria, no Lysozyme was used. The amplified 16S rRNA gene was obtained from each isolate by PCR with the universal primer P1 (5’-AGAGTTTGATCCT GGTCAGAACGCT-3’) and P6 (5’-TACGGCTACCTTG TTACGACTTCACCCC-3’), which were targeted to universally conserved regions and permitted the amplification of an approximate 1, 500-bp fragment. PCR was carried out in a Biometra TGRADIENT thermocycler (Whatman Biometra, Göttingen, Germany). 50 μl reaction volume contained about 25 ng of DNA extract, 2.5 U of TaqTM DNA polymerase (TaKaRa Biotechnology (Dalian) Co., Ltd.), 1× buffer (10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2, 500 mM KCl), 0.2 mM deoxynucleoside triphosphate (each), and 0.4 μM of each primer. Initial DNA denaturation and enzyme activation steps were performed at 95ºC for 5 min, followed by 35 cycles of denaturation at 94ºC for 1 min, annealing at 56ºC for 30 s and extension at 72ºC for 1.5 min, and a final extension at 72ºC for 10 min. The presence and yield of specific PCR product (16S rRNA gene) was monitored by 1% agarose (w/v) gel electrophoresis at 80 V for 1 h in 1× Tris-Boric Acid-EDTA (TBE) buffer and made visible by ethidium bromide staining and UV transillumination. The images of the agarose gels were documented using a UVP GelDoc-It Imaging System (UVP Inc. USA) under an ultraviolet lamp.
ARDRA and rarefaction analysis of siderophore-producing bacterial isolates
PCR products were purified using the 3S Spin PCR Product Purification Kit (Shenergy Biocolor Bioscience and Technology Company, P. R. China) following the manufacturer’s instructions. Purified PCR products were digested with the four chosen enzymes Alu I, Hae III, Hinf I, and Msp I (TaKaRa Biotechnology (Dalian) Co., Ltd.) in separated reactions. The selection of the four restriction enzymes was based on the studies of Laguerre et al. (9). The digestions were performed for 4 h at 37ºC in 10 μl reaction volume containing 5 μl of purified PCR products, 1 μl of the commercially supplied incubation 10× buffers, 3.5 μl of water, and 0.5 μl (10 U/μl) of the restriction enzyme. Reaction products (10 μl) were run on a 3% (w/v) agarose gel in 1× TBE buffer for 4 h at 100 V under refrigeration. Agarose gels were stained, visualized and digitalized as described above. The visible bands greater than 100 bp were used to construct the dendrogram. For each restriction enzyme’s band, a binary data matrix was constructed on the basis of the presence or absence of each band (coded as 1 or 0, respectively). The band patterns obtained with each enzyme were combined to obtain a single pattern for each isolate. The patterns were used to construct a dendrogram by using the unweighted pair group method using arithmetic averages (UPGMA), clustering algorithm using the Sorensen’s Coefficient along with the fine optimization option with MultiVariate Statistical Package (MVSP) version 3.13h (GeoMem, Blairgowrice, United Kingdom). Diversity of siderophore-producing bacterial isolates was further investigated by rarefaction analysis. Rarefaction curves were produced by using the analytical approximation algorithm of Hurlbert (7). Calculations were performed on a personal computer with the freeware program aRarefactWin (http://www.uga.edu/strata/software/).
16S rRNA gene sequencing and sequence analysis
Purified PCR products obtained from isolates of each ARDRA group were sequenced. The Nucleotide-nucleotide BLAST (blastn) of the National Center for Biotechnology Information database, accessible on the internet (http://www.ncbi.nlm.nih.gov/BLAST/) was used to find nearly identical sequences for the 16S rRNA gene sequences determined. The higher similarity sequences and the 16S rRNA gene sequences of type strains were retrieved from the Ribosomal Database Project a! (http://rdp.cme.msu.edu/). Sequences were aligned using the CLUSTAL X software version 1.8 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX/). The evolutionary distances were calculated using the software package TREECON version 1.3b (http://biocwww.uia.ac.be/u/yvdp/treeconw.html). The construction of neighbor-joining tree and bootstrap analysis of 1000 resamplings were performed using software package TREECON too.
RESULTS
Bacterial isolation
Bacteria were isolated from tobacco rhizosphere soil at densities ranging from 6.73 to 7.86 log CFU/g (dry weight). A total of 354 isolates were obtained from the soil samples of tobacco rhizosphere (166 isolates from location 1, 188 isolates from location 2).
Screening of siderophore-producing bacterial isolates
Two hundred and ninty nine isolates (143 isolates from location 1, 156 isolates from location 2) produced siderophores in iron limited liquid medium. Ninety-nine percent of the isolates produced big or small size of orange halos on CAS agar plates, except for the isolates PC-1012, PC-1011, P-6-3h, and G-2-17-1 which couldn’t grow on CAS agar plates (Fig. 1). The siderophores production was evaluated as siderophore units (%). Result showed that more than 75% total frequency of the siderophore-producing strains had the siderophore units (%) between 20 and 40. All of the other strains had the siderophore units (%) between 40 and 60 with less than 25% total frequency.
Figure 1.
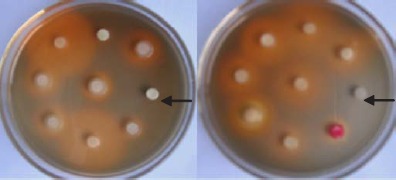
Screening of siderophore-producing bacterial isolates using CAS agar plates. The arrows indicated the isolates PC-1012 (the left one), and G-2-17-1 (the right one) which couldn’t grow on CAS agar plates. The circles in the center of the orange halos were filter papers soaked up with bacterial supernatant.
ARDRA analysis of siderophore-producing bacterial isolates
Total 299 siderophore-producing isolates were subjected to ARDRA analysis by digestion of the amplified 16S rRNA gene with the four restriction enzymes. Similar banding patterns obtained after combination of the four independent digestions (Fig. 2) were grouped to finally obtain a total of 28 groups (location 1 and location 2 contained 28 groups, respectively) (Table 2). As shown in Fig. 2, each group presented a specific banding pattern, and groups of different species were clearly differentiated, with a similarity lower than 95%.
Figure 2.
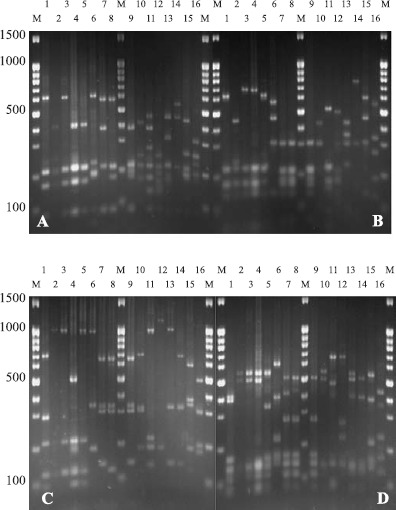
16S-ARDRA patterns obtained from restriction digestion gel photos with Alu I, Hae III, Hinf I, and Msp I. Lanes (1-16) represent the ARDRA groups 1, 2, 4, 7, 10, 11, 12, 13, 14, 17, 18, 21, 24, 25, 26, and 27. Lanes M are the marker 100 bp + 1.5 kb DNA ladder; A, B, C, and D represent restriction digestion gel photos with Alu I, Hae III, Hinf I, and Msp I.
Table 2.
Distribution of siderophore-producing bacterial isolates of tobacco rhizosphere according to ARDRA groups and restriction patterns.
| Group No. | ARDRA banding pattern according to restriction enzyme used | Numbers of isolates | Group total frequency (%) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alu I | Hae III | Hinf I | Msp I | L1 | L2 | L1/2 | L1 | L2 | L1/2 | |
| 1 | A | A | A | A | 11 | 12 | 23 | 3.7 | 3.9 | 7.6 |
| 2 | B | B | B | B | 7 | 9 | 16 | 2.3 | 3.1 | 5.4 |
| 3 | C | C | B | B | 4 | 4 | 8 | 1.3 | 1.4 | 2.7 |
| 4 | D | C | B | B | 7 | 9 | 16 | 2.2 | 3.2 | 5.4 |
| 5 | C | C | C | B | 5 | 3 | 8 | 1.6 | 1.1 | 2.7 |
| 6 | C | C | C | C | 3 | 2 | 5 | 1.0 | 0.7 | 1.7 |
| 7 | C | D | C | D | 10 | 11 | 21 | 3.3 | 3.7 | 7.0 |
| 8 | C | E | B | E | 2 | 3 | 5 | 0.7 | 1.0 | 1.7 |
| 9 | C | F | B | F | 6 | 7 | 13 | 2.0 | 2.3 | 4.3 |
| 10 | C | F | B | E | 8 | 10 | 18 | 2.7 | 3.3 | 6.0 |
| 11 | E | G | D | G | 7 | 6 | 13 | 2.3 | 2.0 | 4.3 |
| 12 | F | H | E | H | 19 | 21 | 40 | 6.2 | 7.1 | 13.3 |
| 13 | F | H | F | H | 7 | 9 | 16 | 2.3 | 3.1 | 5.4 |
| 14 | G | H | E | H | 3 | 2 | 5 | 1.0 | 0.7 | 1.7 |
| 15 | H | H | F | H | 4 | 6 | 10 | 1.3 | 2.0 | 3.3 |
| 16 | G | I | G | H | 2 | 1 | 3 | 0.7 | 0.3 | 1.0 |
| 17 | I | H | H | I | 1 | 2 | 3 | 0.3 | 0.7 | 1.0 |
| 18 | J | I | I | J | 5 | 3 | 8 | 1.7 | 1.0 | 2.7 |
| 19 | K | I | I | K | 5 | 5 | 10 | 1.6 | 1.7 | 3.3 |
| 20 | L | J | J | L | 2 | 1 | 3 | 0.7 | 0.3 | 1.0 |
| 21 | M | J | J | L | 4 | 4 | 8 | 1.3 | 1.4 | 2.7 |
| 22 | N | K | K | M | 1 | 2 | 3 | 0.3 | 0.7 | 1.0 |
| 23 | O | L | J | N | 4 | 6 | 10 | 1.3 | 2.0 | 3.3 |
| 24 | O | L | J | O | 4 | 4 | 8 | 1.3 | 1.4 | 2.7 |
| 25 | P | M | L | P | 2 | 3 | 5 | 0.7 | 1.0 | 1.7 |
| 26 | Q | N | M | R | 3 | 5 | 8 | 1.0 | 1.7 | 2.7 |
| 27 | R | O | N | S | 4 | 4 | 8 | 1.3 | 1.4 | 2.7 |
| 28 | S | P | O | T | 3 | 2 | 5 | 1.7 | 1.0 | 1.7 |
| Total | 19 | 16 | 15 | 20 | 143 | 156 | 299 | 47.8 | 52.2 | 100.0 |
L1: isolates of location 1; L2: isolates of location 2; L1/2: the total isolates of location 1 and 2.
Rarefaction analysis of the siderophore-producing bacterial isolates
Rarefaction analysis was used to estimate how percent of the biodiversity was sampled (Fig. 3). Results showed that by screening of 140 siderophore-producing bacterial isolates, the rarefaction curves reached saturation for both samples of the two locations and the variation between the two samples was minimal, that is to say, the repetition rate was well between the two samples (Table 2).
Figure 3.
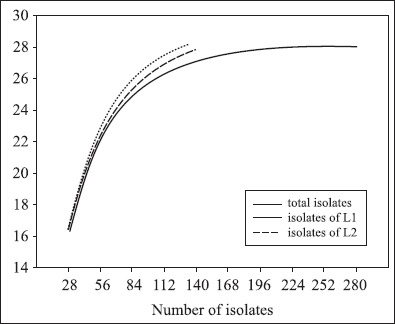
Rarefaction curves for the different ARDRA patterns of all the siderophore-producing bacterial isolates used in this study. The expected number of ARDRApatterns is plotted versus the number of isolates. Rarefaction curves were also calculated for the fraction of L1 and L2. L1: location 1; L2: location 2.
For the fact that there had no isolate in the two locations was unique, the rarefaction analysis suggests that 299 bacterial isolates is sufficient to circumscribe the siderophore-producing bacterial diversity of tobacco rhizosphere.
Sequencing and phylogeny of 16S rRNA gene of siderophoreproducing isolates
Representative isolates of each group were selected for partial 16S rRNA gene sequencing so as to retrieve sequence similarity and bacterial identity from NCBI (Table 1). After partial sequencing of the 16S rRNA gene of ARDRA group representatives, closest identities of isolates were retrieved from NCBI. Results showed a strong similarity with NCBI sequences, like those of group 10, 17, 26, and 28, were strongly related to NCBI sequences, with 100% similarity (Table 1). Relatedness between isolate and type strain sequences was demonstrated in Fig. 4 and Fig. 5. Isolates were therefore clustered in six different classes: β-, γ-, α-Proteobacteria, Sphingobacteria, Bacilli, andActinobacteria. γ-Proteobacteria which consisted of Pseudomonas, Enterobacter, Serratia, Pantoea, Erwinia and Stenotrophomonas genus encountered 18 different ARDRA groups.
Table 1.
List of siderophore-producing bacterial isolates obtained in this study representing each ARDRA group and their closest affiliation according to sequencing of 16S rRNA gene.
| Isolate | ARDRAgroup | Accession no. | Closest NCBI Strain and Accession no. | Sid | Similarity(%) |
|---|---|---|---|---|---|
| G-1-19-1 | 1 | EF102853 | Pseudomonas sp., AB076857 | ( + ) | 99.3 |
| G-1-21-2 | 1 | EF102855 | Pseudomonas sp., AB076857 | ( + ) | 99.9 |
| G-1-11-12 | 1 | EF102854 | Pseudomonas sp., AB076857 | ( + ) | 99.6 |
| G-5-14 | 1 | EF102857 | Pseudomonas sp., DQ118954 | ( + ) | 99.4 |
| G-229-21 | 2 | EF102850 | Pseudomonas sp., AF456229 | ( + ) | 99.6 |
| G-229-22 | 2 | EF102851 | Pseudomonas sp., AF456229 | ( + ) | 99.6 |
| G-229-23 | 2 | EF102852 | Pseudomonas sp., AF456229 | ( + ) | 99.8 |
| G-6-9-2 | 3 | EF102858 | Pseudomonas sp., DQ453823 | ( + ) | 99.8 |
| G-2-1-1 | 4 | EF102856 | Pseudomonas poae, DQ536513 | ( − ) | 99.3 |
| N-5-143XBH | 5 | EF102848 | Pseudomonas putida, DQ449024 | ( + ) | 99.4 |
| G-4-14-1 | 6 | EF102849 | Pseudomonas putida, DQ886481 | ( + ) | 99.9 |
| G-4-1-2 | 7 | EF102847 | Pseudomonas putida, DQ886481 | ( + ) | 99.3 |
| G-229-2 | 8 | EF102839 | Pseudomonas aeruginosa, CP000438 | ( + ) | 99.9 |
| NB-17-1 | 9 | EF102841 | Pseudomonas aeruginosa, CP000438 | ( + ) | 99.9 |
| P-5-3 | 9 | EF102842 | Pseudomonas aeruginosa, CP000438 | ( + ) | 99.2 |
| PB-5-2 | 9 | EF102843 | Pseudomonas aeruginosa, CP000438 | ( + ) | 100.0 |
| GA-15-1 | 9 | EF102837 | Pseudomonas aeruginosa, CP000438 | ( + ) | 99.0 |
| G-229-1 | 10 | EF102838 | Pseudomonas aeruginosa, CP000438 | ( + ) | 97.8 |
| GA-8-1 | 10 | EF102840 | Pseudomonas aeruginosa, CP000438 | ( + ) | 100.0 |
| PB-6-1 | 10 | EF102844 | Pseudomonas aeruginosa, CP000438 | ( + ) | 100.0 |
| PB-9-2 | 10 | EF102845 | Pseudomonas aeruginosa, CP000438 | ( + ) | 100.0 |
| G-1-7 | 11 | EF102835 | Pectobacterium carotovorum, AY207086 | ( + ) | 99.1 |
| G-1-5-1 | 11 | EF102836 | Pectobacterium carotovorum, AY207086 | ( + ) | 99.6 |
| G-4-13-2 | 12 | EF102834 | Pantoea agglomeran, AM184214 | ( + ) | 99.0 |
| G-226-1 | 12 | EF102825 | Enterobacter aerogenes, AB099402 | ( + ) | 99.6 |
| G-2-10-2 | 13 | EF102826 | Enterobacter endosymbiont, AY753172 | ( + ) | 99.2 |
| N-5-10 | 13 | EF102824 | Enterobacter endosymbiont, AY753172 | ( + ) | 99.1 |
| P-6-4R | 14 | EF102862 | Serratia marcescens, AY043386 | ( + ) | 99.8 |
| P-4-8R | 15 | EF102860 | Serratia marcescens, AY514435 | ( + ) | 99.5 |
| P-6-14RR | 15 | EF102863 | Serratia marcescens, AY514435 | ( + ) | 99.8 |
| P-4-8RR | 16 | EF102861 | Serratia marcescens, AY514432 | ( + ) | 99.1 |
| G-1-15 | 17 | EF102823 | Delftia acidovorans, AM180725 | ( − ) | 100.0 |
| G-5-5-1 | 18 | EF102868 | Stenotrophomonas maltophilia, AJ306833 | ( − ) | 99.7 |
| P-2-13 | 19 | EF102866 | Stenotrophomonas maltophilia, AY486381 | ( − ) | 99.7 |
| P-5-22 | 19 | EF102867 | Stenotrophomonas maltophilia, AY486381 | ( − ) | 98.3 |
| P-5-2 | 20 | EF102833 | Ochrobactrum sp., DQ486950 | ( + ) | 98.8 |
| G-1-22 | 21 | EF102818 | Agrobacterium tumefaciens, AB247617 | ( + ) | 98.8 |
| N-46-11HH | 22 | EF102816 | Achromobacter xylosoxidans, AY631060 | ( − ) | 99.4 |
| N-5-20 | 23 | EF102817 | Achromobacter xylosoxidans, AF411021 | ( − ) | 99.7 |
| G-4-3-1 | 24 | EF102820 | Alcaligenes faecalis, AJ509012 | ( + ) | 99.7 |
| G-1-6 | 25 | EF102819 | Alcaligenes sp., DQ211905 | ( + ) | 99.1 |
| PC-1012 | 26 | EF102822 | Bacillus subtilis, DQ012095 | ( + ) | 100.0 |
| PC-1011 | 26 | EF102821 | Bacillus subtilis, DQ012095 | ( + ) | 100.0 |
| G-2-21-1 | 27 | EF102864 | Sphingobacterium sp., AY556417 | ( − ) | 99.3 |
| G-2-27-2 | 27 | EF102865 | Sphingobacterium sp., AY556417 | ( − ) | 99.3 |
| G-2-17-1 | 28 | EF102859 | Rhodococcus erythropolis, DQ858961 | ( + ) | 100.0 |
Accession no.: GenBank accession number; Sid: siderophore; have been reported to produce siderophores ( + ); not have been reported to produce siderophores ( − )
Figure 4.
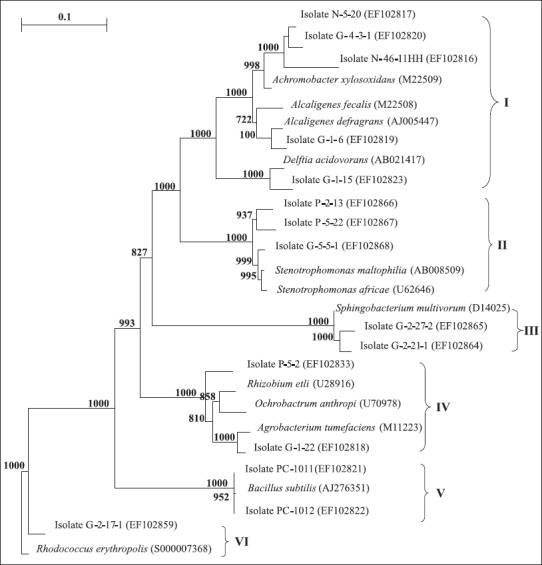
Aneighbor-joining phylogenetic tree based on 16S rRNA gene sequences of siderophore-producingrepresentative strains. The tree contains the closest type strain for each isolate which belong to I, II, III, IV, V, and VI class, respectively. Bootstrap analyses were made with 1000 cycles. Only bootstrap values of > 50% are shown at the branch points. The scale bar represents 0.1 substitutions per nucleotide. I, II, III, IV, V, and VI indicated β-Proteobacreria, γ-Proteobacteria (Xanthomonadacea), Spingobacteria, α-Proteobacteria, Bacilli, andActinobacteria class, respectively.
Figure 5.
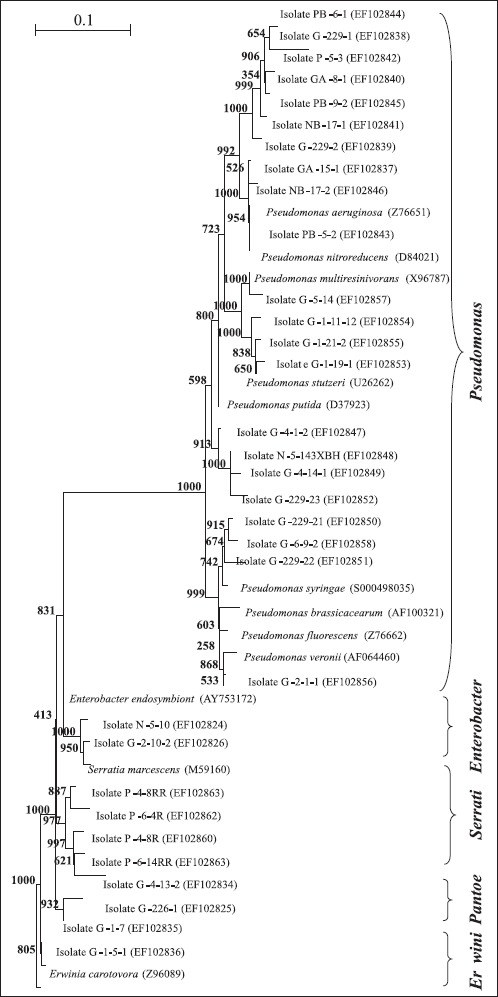
Figure 5. A neighbor-joining phylogenetic tree based on 16S rRNA gene sequences of siderophore-producing strains. The tree contains the closest type strain for each isolate which belong to Pseudomonas, Serratia, Enterobacter, Pantoea, and Erwinia genera, respectively. Bootstrap analyses were made with 1000 cycles. Only bootstrap values of > 50% are shown at the branch points. The scale bar represents 0.1 substitutions per nucleotide.
Distribution of siderophore-producing isolates
Table 1 demonstrated a wide variety of siderophoreproducing bacteria, with 14 different genera encountered. Gram-negative isolates were more frequently encountered, with 289 isolates, compared to 10 Gram-positive isolates. Distribution of siderophore-producing isolates according to ARDRA groups (Table 2) revealed that groups 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 12, 13, 14, 15, and 16 had the most numerous isolates and that these groups belonged to Gram-negative bacteria corresponding to the Pseudomonas and Enterobacter genera with 44.5% and 24.7% total frequency, respectively. That is to say, the siderophore-producing Pseudomonas isolated from tobacco rhizosphere soil ranged from 6.38 to 6.51 log CFU/g (dry weight). For Gram-positive bacteria, group 25 and 28 were the only two groups corresponding to the Bacillus and Rhodococcus genera, with 1.7% total frequency, respectively.
DISCUSSION
The diversity of cultivable siderophore-producing bacteria of tobacco rhizosphere was characterized in this study. Results have demonstrated that 85% of the total 354 isolates produced siderophores in iron limited liquid medium and a total of 28 ARDRA patterns were identified among the 299 siderophoreproducing bacterial isolates. The 28 ARDRA patterns represented bacteria of 14 different genera belonging to six bacterial divisions, namely β-, γ-, α-Proteobacteria, Sphingobacteria, Bacilli, andActinobacteria. Especially, γ-Proteobacteria which was consisted of Pseudomonas, Enterobacter, Serratia, Pantoea, Erwinia and Stenotrophomonas genus encountered 18 different ARDRA groups. Gram-negative isolates were more frequently encountered, with more than 95% total frequency. Furthermore, the Pseudomonas and Enterobacter were dominant in this environment, with 44.5% and 24.7% total frequency, respectively. For Gram-positive bacteria, the Bacillus and Rhodococcus were the only two genera, with 1.7% total frequency, respectively.
Seventy five percent of the isolates that had the siderophore units (%) between 40 and 60 belonged to Pseudomonas genus. This observation was also made by Alexander (2) who had found that most of the siderophoreproducing isolates coming from the roots of two grass species belonged to the genus Pseudomonas. The presence of the genus Pseudomonas was expected due to its importance in plant rhizosphere ecosystems, in relation notably with its siderophoremediated suppression and inducing local systemic host resistance activities (16, 17).
It was excited to find that Sphingobacterium sp. G-2-21-1 and G-2-27-2 had the siderophore units (%) between 40 and 60; in our knowledge, it was the first time to report that genus Sphingobacterium can produce siderophores under low-iron conditions. We also reported that P. poae (isolate G-2-1-1), Enterobacter endosymbiont (isolates G-2-10-2 and N-5-10), Delftia acidovorans (isolate G-1-15), and A. xylosoxidans (isolates N-46-11HH and N-5-20) can produce siderophores under low-iron conditions for the first time. These eight bacterial isolates are potential to be examined for their capability in enhancing plant growth. Furthermore, we also reported that Pseudomonas sp. G-229-21 could produce high-affinity carboxylate type siderophores under low iron conditions and its siderophores were capable of antagonising against Phytophthora parasitica var. nicotianae (Breda de Hann) Tucker strongly under low iron conditions (21).
In addition, the predominant presence of Gram-negative siderophore-producing bacteria isolated from tobacco rhizosphere may be due in part to the use of CAS agar plates. Because Gram-positive bacteria were especially sensitive to the HDTMA used in the CAS agar plates (20).
Although the samples were all collected from acerbic tobacco rhizosphere, we had detected a large number of siderophoreproducing bacteria under low-iron conditions. It indicated that the producing of siderophores was induced by low iron stress. At the same time, 36.4% of the total tobacco in China are planted in neutral and alkaline low-iron soil (3), so the siderophoreproducing bacteria screened out in this study have great potential to apply to low-iron soil to prevent plant soil-borne fungal pathogen diseases(concerned greenhouse experimentations are in progress).
Acknowledgments
This work was funded by the Research Award Fund for Outstanding Middle-aged and Young Scientist of Shangdong Province (People’s Republic of China) (2006BS06012). We thank Dr. Jie Liu for assistance with ARDRA procedure and phylogenetic analyses, Dr. Guang-yi Wang at the University of Hawaii for revising the manuscript and Mr. Xian-ming Zeng for his work on soil and root samples collection.
RESUMO
Diversidade genética de bactérias de rizosfera de tabaco produtoras de sideróforos
A diversidade genética de bactérias de rizosfera de tabaco produtoras de sideróforos foi estudada por meio da técnica de análise de restrição do DNA ribossomal amplificado (ARDRA), homologia de seqüência de 16s rRNA e métodos de análise filogenética. Observou-se que 85% do total de 354 isolados produziram sideróforos em meio liquido com restrição de ferro. Entre os 299 isolados produtores de sideróforos identificou-se 28 padrões ARDRA, que representaram 14 gêneros bacterianos diferentes, pertencentes a seis divisões bacterianas: β-, γ-, α-Proteobacteria, Sphingobacteria, Bacilli e Actinobacteria. γ-Proteobacteria, consistindo dePseudomonas, Enterobacter, Serratia, Pantoea, Erwinia eStenotrophomonas, pertenceram a 18 grupos ARDRA. Os resultados também mostraram uma diversidade maior de bactérias produtoras de sideróforos do que a relatada em outros estudos. Por exemplo, Sphingobacterium (isolados G-2-21-1 e G-2-27-2),Pseudomonas poae (isolado G-2-1-1), Enterobacter endosymbiont (isolados G-2-10-2 e N-5-10),Delftia acidovorans (isolado G-1-15) e Achromobacter xylosoxidans (isolados N-46-1HH e N-5-20), capazes de produzir sideróforos em condições de baixa disponibilidade de ferro, foram relatados pela primeira vez. Isolados Gram negativos foram encontrados com maior freqüência, correspondendo a mais de 95% da freqüência total. Entre as bactérias Gram positivas, foram encontrados apenas os gêneros Bacillus e Rhodococcus, com 1,7% da freqüência total. Além disso, neste ambiente houve predominância de Pseudomonas e Enterobacter, com 44,5% e 24,7% da freqüência total, respectivamente. Verificou-se também que 75% dos isolados com alta porcentagem de unidades de sideróforos (% entre 40 e 60) pertenceram a Pseudomonas. Pseudomonas sp. G-229-21, selecionado neste estudo, apresenta potencial de aplicação em solos com baixo teor de ferro para prevenção de doenças fúngicas em plantas.
Palavras-chave: diversidade genética, sideróforos, rizosfera de tabaco, ARDRA, sequenciamento de rRNA.
REFERENCES
- 1.Ahmad F., Ahmad L., Khan M.S. Screening of free-living rhizospheric bacteria for their multiple plant growth promoting activities. Microbiol. Res. 2008;163:173–181. doi: 10.1016/j.micres.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Alexander D.B., Zuberer D.A. Use of chrome azurol s regents to evaluate siderophore production by rhizosphere bacteria. Biol. Fertils. Soils. 1991;12:39–45. [Google Scholar]
- 3.Chen J.H., Li Z.H., Liu J.L., Wang G., Long H.Y., Lei Q.L., Zhang R.L., Zhang W.L. Evaluation of soil nutrients condition in major tobacco production region of China. Acta Tabacaria Sinica. 2004;10:14–18. (in Chinese with English abstract) [Google Scholar]
- 4.D’Angelo-Picard C., Faure D., Penot I., Dessaux Y. Diversity of N-acyl homoserine lactone-producing and-degrading bacteria in soil and tobacco rhizosphere. Environ. Microb. 2005;7:1796–1808. doi: 10.1111/j.1462-2920.2005.00886.x. [DOI] [PubMed] [Google Scholar]
- 5.Fischer S.E., Fischer S.I., Magris S., Mori G.B. Isolation and characterization of bacteria from the rhizosphere of wheat. World J. Microbiol. Biotechnol. 2006;23:895–903. [Google Scholar]
- 6.Haas D., Defago G. Biological control of soil-borne pathogens by fluorescent pseudomonas. Nat. Rev. Microbiol. 2005;3:307–319. doi: 10.1038/nrmicro1129. [DOI] [PubMed] [Google Scholar]
- 7.Hurlbert S.H. The nonconcept of species diversity: a critique and alternative parameters. Ecology. 1971;52:577–586. doi: 10.2307/1934145. [DOI] [PubMed] [Google Scholar]
- 8.Kloepper J.W., John L., Teintze M. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature. 1980;286:885–886. [Google Scholar]
- 9.Laguerre G., Allard M.R., Revoy F., Amarger N. Rapid identification of Rhizobia by restriction fragment length polymorphism analysis of PCR-amplified 16S rRNA genes. Appl. Environ. Microb. 1994;60:56–63. doi: 10.1128/aem.60.1.56-63.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X.L., Wang C., Wu F., Xue D.H., Chen K. Studies on tobacco rhizosphere microbes. Acta Ecologica Sinica. 2006;26:552–557. (in Chinese with English abstract) [Google Scholar]
- 11.Neilands J.B., Leong S.A. Siderophores in relation to plant growth and disease. Annu. Rev. Plant Physiol. 1986;37:187–208. [Google Scholar]
- 12.Machuca A., Milagres A.M.F. Use of CAS-agar plate modified to study the effect of different variables on siderophore production by Aspergillus. Lett. Appl. Microbiol. 2003;36:177–181. doi: 10.1046/j.1472-765x.2003.01290.x. [DOI] [PubMed] [Google Scholar]
- 13.Masalha J., Kosegarten H., Elmaci ö., Mengel K. The central role of microbial activity for iron acquisition in maize and sunflower. Biol. Fertils. Soils. 2000;30:433–439. [Google Scholar]
- 14.Motta A.S., Cladera-Olivera F., Brandelli A. Screening for antimicrobial activity among bacteria isolated from the Amazon Basin. Braz. J. Microbiol. 35:307–310. [Google Scholar]
- 15.Murray M.G., Thormpson W.F. Rapid isolation of high molecular weight plant DNA. Nucl. Acids Res. 1980;8:4321–4326. doi: 10.1093/nar/8.19.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Sullivan N.J., O’Gara F. Traits of fluorescent Pseudomonas spp. involved in suppression of plant root pathogens. Microbiol. Rev. 1992;56:662–676. doi: 10.1128/mr.56.4.662-676.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ran L.X., Liu C.Y., Wu G.J., van Loon L.C., Bakker P.A.H.M. Suppression of bacterial wilt in Eucalyptus urophylla by fluorescent Pseudomonas spp. in China. Biol. Control. 2005;32:111–120. [Google Scholar]
- 18.Reche M.H.L.R., Fiuza L.M. Bacterial diversity in Rice-field water in Rio Grande do Sul. Braz. J. Microbiol. 2005;36:253–257. [Google Scholar]
- 19.Schroth M.N., Hancock J.G. Disease-suppressive soil and root-clonizing bacteria. Science. 1982;216:1376–1381. doi: 10.1126/science.216.4553.1376. [DOI] [PubMed] [Google Scholar]
- 20.Schwyn B., Neilands J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 21.Tian F., Ding Y.Q., Zhu H., Yao L.T., Jin F.L., Du B.H. Screening, identification and antagonistic activity of a siderophore producing bacteria G-229-21T from rhizosphere of tobacco. Wei Sheng Wu Xue Bao. 2008;48(5):631–637. (in Chinese with English abstract) [PubMed] [Google Scholar]


