Abstract
A Gram-negative bacterium, designated as strain KB2, was isolated from activated sludge and was found to utilize different aromatic substrates as sole carbon and energy source. On the basis of morphological and physiochemical characteristics and 16S rRNA gene sequence analysis, the isolated strain KB2 was identified as Stenotrophomonas maltophilia. Strain KB2 is from among different Stenotrophomonas maltophilia strains the first one described as exhibiting the activities of three types of dioxygenases depending on the structure of the inducer. The cells grown on benzoate and catechol showed mainly catechol 1,2-dioxygenase activity. The activity of 2,3-dioxygenase was detected after phenol induction. Protocatechuate 3,4-dioxygenase was found in crude cell extracts of this strain after incubation with 4-hydroxybenzoic acid, protocatechuic acid and vanillic acid. Because of broad spectrum of dioxygenases’ types that Stenotrophomonas maltophilia KB2 can exhibit, this strain appears to be very powerful and useful tool in the biotreatment of wastewaters and in soil decontamination.
Keywords: biodegradation, Stenotrophomonas, aromatic compounds, dioxygenases
INTRODUCTION
Aromatic compounds are environmental pollutants due to their widespread use and potential toxicity to higher organisms. The catabolic versatility exhibited by microorganisms plays an important role in the carbon cycle and the bioremediation of polluted environments. Pathways for aerobic biodegradation of phenolic compounds are diverse, but they usually involve the formation of catechols as common intermediates. Catechol 1,2-dioxygenase [EC 1.13.11.1] catalyzes an intradiol cleavage reaction of catechol at 1,2-(ortho) position to form cis,cismuconate. The catechol 2,3-dioxygenase [EC 1.13.11.2], an extradiol-ring fission enzyme, catalyzes the conversion of catechol to 2-hydroxymuconic semialdehyde by breaking the C-C bond at 2,3-(metha) position (6, 19, 26). Protocatechuate is one of the key intermediates for microbial catabolism of benzoic molecules. These compounds are degraded through one of two (ortho or metha) aromatic ring cleavage pathways, too. Protocatechuate may be cleaved by protocatechuate 3,4-dioxygenase [EC 1.13.11.3], which catalyzes the intradiol addition of molecular oxygen and forms 3-carboxy-cis,cis-muconate or by protocatechuate 4,5-dioxygenase [1.13.11.8]. This last enzyme catalyzes extradiol addition of molecular oxygen, which results in the aromatic ring cleaving and 4-carboxy-2-hydroxy-cis,cismuconic semialdehyde forming (6,20,26).
In our lab we isolated a bacterial strain, which was analyzed for the type of ring-cleavage dioxygenase physiologically induced during growth in the presence of six monocyclic aromatic compounds (phenol, catechol, benzoic acid, 4-hydroxybenzoic acid, protocatechuic acid and vanillic acid), which served as carbon and energy sources for strain KB2. Although Stenotrophomonas species capable of degrading aromatic compounds have been reported earlier (4,17) little has been investigated on the possibility of occurrence of the heterogeneous metabolic pathways in one strain of this genus (28). So far Stenotrophomonas species were mainly described as polycyclic hydrocarbon degraders or opportunistic human pathogens (2,3,4,8,10).
In this study, for the first time the isolation and characterization of a Gram-negative Stenotrophomonas maltophilia strain, designated as KB2, which exhibits activities of three types of dioxygenases while growing in the presence of monocyclic hydrocarbons, has been reported.
MATERIALS AND METHODS
Isolation of an aromatic-degrading bacterium
An aromatic compound degrading strain was isolated from the activated sludge of a sewage treatment plant in Bytom Miechowice in Poland using the classical enrichment technique with phenol as a selection factor. The mixed microbial population from the activated sludge was adapted to grow on phenol at a concentration of 10 mM. The adaptation process was carried out at 20ºC in an aeration chamber. On the 10th day of adaptation, a 2 ml sample was taken from the culturing chamber and 100 μl of 10-1 to 10-7 were spread on agar plates containing mineral salts medium (Na2HPO4 · 12H2O 3.78 g; KH2PO4 0.5 g; NH4Cl 5 g; MgSO4 · 7H2O; per litre distilled water) with 3 mM phenol to obtain pure cultures. Agar plates were incubated at 30ºC for 24h and single colonies were isolated and transferred to nutrient agar plates to test their purity. Strain was kept on nutrient agar slopes at 4ºC and systematically transferred to the new ones every month.
Morphological, physiological and biochemical characterization of the isolated strain
The isolated strain was phenotypically and biochemically characterized using standard techniques (Gram staining, colony shape, size and colour on nutrient agar plate, catalase and oxidase test, etc.), according to Bergey’s Manual of Determinative Bacteriology (12). Additional biochemical and physiological characteristics were determined using the API 20NE and API 20E system (BioMerieux, Lyon, France).
Isolation of fatty acids was performed according to Sasser (23). Analysis of FAMEs was performed using an HP 5890 gas chromatograph (Hewlett Packard, Rolling Meadows, IL, US) equipped with an HP 25 m x 0.2 mm cross-linked methyl-silicone capillary column. The initial oven temperature was 170ºC, increased 5ºC min -1 to 260ºC, the increased 40ºC min-1 and held constant at 320ºC for 1.5 min (9). Helium was used as the carrier gas. FAMEs were identified with Sherlock software (TSBA library, version 3.9, Microbial ID, Newark, NJ, USA), based on the actual calibration retention times run prior to sample analysis.
Analytical methods
The aromatic substrates were determined by a Merck HITACHI HPLC with a LiChromospher® RP-18 column (4 × 250 mm) and a DAD detector (Merck HITACHI). The wavelength of detection, composition of elution solvent and the flow rate were developed separately for each aromatic compound.
Culture conditions
In order to verify, which aromatic substrate can serve as the sole source of carbon and energy, adaptations to the increasing concentrations of various aromatic substrates were carried out. Cells were proliferated in mineral medium with 3 mM of phenol (at 30ºC on a rotary shaker at 130 rpm), harvested by centrifugation (5,000 × g at 4ºC for 15 min) and washed with fresh sterile medium. Such prepared cells were used as inoculum for the experiments with adaptation. Cultures in 250-ml flask containing 100 ml of sterile mineral salts medium supplemented with 1 mM of the tested aromatic compound, were inoculated with previously prepared cells to the final optical density about 0.1 in absorbance scale at λ = 600 nm, and incubated shaking at 130 rpm at 30ºC for 24 hours. Chromatographic analyses of the culture fluid and measurements of the cultures growth (spectrophotometrically at 600 nm) were carried out every 24 hours. If growth of the cultures and complete degradation of the aromatic substrate was observed, the successive dose (2 mM and higher) of the aromatic substrate was introduced and the cultures were left for incubation for the next 24 hours. Lack of aromatic substrate degradation and absence of growth for three following days finished the adaptation process.
The aromatic substrates, which could serve as the source of carbon and energy, were used as the inducers of enzymes activities. Induction experiments were carried out in 1-litre flasks, containing 500 ml of mineral salts medium and aromatic substrate in concentration of 3 mM. Inducer was added every 24 hours for three days in order to obtain final A600 of 0.9-1.0.
These induction’s were preceded by preparation of the cells suspensions as follows: colonies were picked from nutrient agar and transferred to a 50-ml conical flask containing 20 ml of mineral salt medium with 3 mM of the appropriate aromatic substrate. After 24 hours of incubation the suspension of the cells was transferred to a 250-ml flask containing 80 ml of mineral medium with 3 mM of inducer. The cells were allowed to grow the next 24 hours and this inoculum was used to prepare induction experiments. No-induced enzyme levels were determined in the cells coming from the cultures carried out in mineral salts medium supplemented with 1 g/L glucose.
Preparation of cell extracts
Cells were harvested in the late exponential phase of growth by centrifugation at 4,500 g for 15 min at 4ºC. The cells were washed with 50 mM phosphate buffer, pH 7.0, and resuspended in the same buffer. Cell-free extracts were prepared by sonication 6 times for 15 s and centrifugation at 9,000 g for 30 min at 4ºC. The clear supernatant was used as crude extract for enzyme assays.
Enzyme assays
Activity of catechol 1,2-dioxygenase [EC 1.13.11.1] was measured spectrophotometrically by formation of cis,cismuconic acid at 260 nm (ε260 = 16,800 M-1 cm-1). The reaction mixture contained 20 μl of catechol (50 mM), 67 μl Na2EDTA (20 mM), 893 μl of phosphoric buffer pH 7.4 (50 mM) and 20 μl of crude extracts in a total volume of 1 ml. When activity of catechol 2,3-dioxygenase was detected, crude cell extract was incubated with 5% H2O2 prior to determination of catechol 1,2-dioxygenase. In order to determine catechol 2,3-dioxygenase [EC 1.13.11.2] activity, the formation of 2-hydroxymuconic semialdehyde was measured at 375 nm (ε375 = 36,000 M-1 cm-1) in a reaction mixture containing 20 μl of catechol (50 mM), 960 μl of phosphoric buffer pH 7.4 (50 mM) and 20 μl of crude extract in a total volume of 1 ml (11). Activity of protocatechuate 3,4-dioxygenase [EC 1.13.11.3] was measured by protocatechuate depletion at 290 nm in a reaction mixture containing 20 μl of protocatechuate (50 mM), 960 μl of Tris-HCl buffer pH 7.4 (50 mM) and 20 μl of crude extract in a total volume of 1 ml. A molar extinction coefficient of 2,300 M-1 cm-1 was used, which is the difference between ε290 of protocatechuate (3,890 M-1 cm-1) and ε290 of the product 3-carboxy-cis cismuconate (1,590 M-1cm-1). In order to determine protocatechuate 4,5-dioxygenase [EC 1.13.11.8] activity, the formation of 2-hydroxy-4-carboxymuconic semialdehyde was measured at 410 nm (ε410 = 9,700 M-1cm-1). The reaction mixture contained 20 μl of protocatechuate (50 mM), 960 μl of Tris-HCl buffer pH 7.4 (50 mM) and 20 μl of crude extract in a total volume of 1 ml (25). One unit of enzyme activity was defined as the amount of enzyme required to generate 1 μmol of product per minute. Protein concentrations of the crude extract from different inducer-cultured bacteria were determined by the Bradford method using lysozyme as a standard (5).
Phylogenetic analysis
Bacterial DNA was isolated from pure culture using the DNA Mini Prep Kit (Qiagen). For 16S rRNA gene amplification the Bacteria-specific primers 8F 5’AGTTTGATCATCGCTCAG 3’ and 1492R 5’GGTTACCTTGTTACGACTT3’ were used (18). Amplification was carried out through a program consisting of initial denaturation at 94ºC for 300 s, 3 cycles at 94ºC for 45 s, 57ºC for 30 s, 72ºC for 120 s; 3 cycles at 94ºC for 45 s, 56ºC for 30 s, 72ºC for 120 s; 3 cycles at 94ºC for 45 s, 55ºC for 30 s, 72ºC for 120 s; 26 cycles at 94ºC for 45 s, 53ºC for 30 s, 72ºC for 120 s; and a final elongation cycle at 72ºC for 300 s. The nucleotide sequencing of the gene was done by using Big DyeR Terminator Cycle Sequencing Kit (Applied Biosystem) and AbiPrism®3100 Genetic Analyzer. The MegaBLAST program was used for homology searches with the standard program default. Multiple sequence alignments were performed and the neighbour-joining phylogenetic tree was constructed using CLC Free Workbench 4.5.1 program. The 16S rRNA gene sequence determined in Monocyclic hydrocarbon degradation by S. maltophilia this study has been deposited in the GeneBank database of NCBI under the accession number DQ230920.
Chemicals
Phenol, monochlorophenols, cresols, 2,3-, 2,4-, 2,6-dichlorophenols, catechol, hydroquinone, cinnamic and gallic acid were obtained from E. Merck AG, Darmstadt, Germany. 2,5- and 3,4-dichlorophenol, vannilic, syryngic, 4-hydroxybenzoic and protocatechuic acid were obtained from FLUKA, Fluka AG, Buchs, Switzerland. Resorcinol, caffeic, sinapic and p-coumaric acid were obtained from Sigma Chemical Co., St. Louis, Mo. Ferulic acid and 2,5-dihydroxybenzoic acid were were obtained from Aldrich Chemical Co., Inc., Milwaukee, Wis. Benzoic and salicylic acid, and other chemicals were of analytical quality and were obtained from local suppliers.
RESULTS AND DISCUSSION
Isolation and identification of strain degrading aromatic compounds
The present work was focused on isolation and characterization of bacterial strain capable of degrading high concentrations of aromatic compounds. Although phenol was supplied as the carbon and energy source during the enrichment process, this strain could also utilize different kind of aromatic compounds such as catechol, benzoic acid, 4-hydroxybenzoic acid (4-HB), protocatechuic acid (3,4-DHB), vanillic acid. All substrates that were tested for their ability to support growth of KB2 strain, are summarized in Table 1.
Table 1.
Utilization of aromatic compounds as the sole source of carbon by the strain KB2 (in brackets the highest concentration degraded by strain within 24 hours is given).
| Aromatic | Results | Aromatic | Results |
|---|---|---|---|
| compounds | compounds | ||
| phenol | + (12 mM) | catechol | + (6 mM) |
| 2-chlorophenol | _ | resorcinol | |
| 3-chlorophenol | _ | hydroquinone | + (4 mM) |
| 4-chlorophenol | _ | vanillic acid | + (6 mM) |
| o-cresol | + (6 mM) | syringic acid | _ |
| m-cresol | + (6 mM) | protocatechuic acid | + (13 mM) |
| p-cresol | + (5 mM) | benzoic acid | + (10 mM) |
| 2-nitrophenol | _ | 4-hydroxybenzoic acid | + (9 mM) |
| 3-nitrophenol | _ | gentisic acid | −_ |
| 4-nitrophenol | _ | cinnamic acid | _ |
| 2,3-dichlorophenol | _ | salicylic acid | _ |
| 2,4-dichlorophenol | _ | gallic acid | _ |
| 2,5-dichlorophenol | _ | caffeic acid | _ |
| 2,6-dichlorophenol | _ | ferulic acid | _ |
| 3,4-dichlorophenol | _ | p-coumaric acid | _ |
| 3,5-dichlorophenol | _ | sinapic acid | _ |
Morphological, physiological and biochemical characterisation and 16S rRNA gene sequencing were used to identify strain KB2. Cells of strain KB2 were Gram-negative, rod shaped, motile and nonspore forming. Colonies of strain KB2 were found to be circular, smooth, convex and with the entire margin during growing on agar plates for 48 hours. The biochemical and physiological characteristic of strain KB2 are summarized in Table 2. The pattern generated with API 20NE test was a good match with Xanthomonas maltophilia. Similar results were obtained by Radianingtyas and co-workers (22). These authors identified the particular microorganisms of consortium degrading 4-chloroaniline. Strain SB5 which was able to grow on 4-chlorocatechol was identified using API 20NE test as Xanthomonas maltophilia and by using 16S rRNA method as Stenotrophomonas maltophilia (22). In Table 3 fatty acids from strain KB2 are listed. The cellular fatty acid content gave little information what species the isolated strain KB2 could belong to. Glucksman and co-workers (9) during analysis of fatty acid content of Stenotrophomonas maltophilia ATCC 13637 observed the largest contribution of some typical for Pseudomonas genus fatty acids: 16:0 and 16:1 ω7c (9). These cellular fatty acids were also observed in fatty acid profile of the strain KB2. Because there is no clear data what kinds of cellular fatty acids are the most typical for genus Stenotrophomonas, in order to confirm phylogenetic relationship of strain KB2 with Stenotrophomonas genus, genomic DNA was isolated and the gene coding for 16S rRNA was amplified. The nearly complete 1445-bp-long 16S rRNA gene sequence of strain KB2 was found to be 99% identical to that of Stenotrophomonas maltophilia Fatty % of total Fatty % of total strain D1 (EU340025.1). A high percentage identity was also found with Stenotrophomonas sp. strain Sea-8, a phenol degrader (28) (Fig. 1). In accordance with these data, the isolate was included in the genus Stenotrophomonas and named as Stenotrophomonas maltophilia KB2.
Table 2.
Differential phenotypic characteristics of strain KB2.
| Characteristic | Results | Characteristic | Results |
|---|---|---|---|
| Growth in the absence of NaCl | + | Assimilation of: | |
| Growth in the presence of 1.5% (w/v) NaCl | + | Glucose | + |
| Growth in the presence of 3% (w/v) NaCl | + | Arabinose | + |
| Growth at 4°C | − | Mannose | + |
| Growth at 42°C | + | Mannitol | + |
| Oxidase | + | Inositol | + |
| Catalase | + | Sorbitol | + |
| Hydrolysis of esculin | + | Rhamnose | + |
| Hydrolysis of gelatin | + | Sucrose | + |
| Fermentation of glucose | − | Melobiose | − |
| Arginine dihydrolase | − | Maltose | + |
| Urease | − | Gluconate | − |
| H2S production | − | Caprate | + |
| Indol production | − | Adipate | − |
| Nitrate reduction | − | Malate | + |
| Acetoin production | − | Citrate | + |
| Tryptophane desaminase | − | Phenylacetate | − |
| Ornithine decarboxylase | − | N-acetyl- | |
| glucosamine | + | ||
| Beta-galactosidase | − | Amygdalin | + |
+, positive reaction; −, negative reaction.
Table 3.
Percentage of total fatty acid from Stenotrophomonas maltophilia KB2.
| Fatty acids | % of total fatty acids | Fatty acids | % of total fatty acids |
|---|---|---|---|
| Saturated | 18:0 | 0.94 | |
| 10:0 | 0.64 | 19:0 | 0.22 |
| 12:0 | 2.39 | 19:0 iso | 0.23 |
| 12:0 2OH | 4.81 | 19:0 cyclo m8c | 1.29 |
| 14:0 | 0.45 | Unsaturated | |
| 14:0 3OH | 0.08 | 16:1 m7c | 16.33 |
| 15:0 | 0.26 | 17:1 m8c | 0.18 |
| 15:0 iso 2OH | 3.31 | 18:1 m7c | 23.45 |
| 16:0 | 32.09 | 18:1 m9c | 0.3 |
| 17:0 | 0.25 | other | 0.24 |
| 17:0 cyclo | 12.46 | Sat./unsat. ratio | 1.48 |
| 17:0 iso | 0.08 | ||
w - methyl end of fatty acid;
c - cis configuration of the double bound;
cyclo-cyclopropane fatty acid;
-OH indicates the position of hydroxyl group from the acid end; iso-branched fatty acids.
Figure 1.
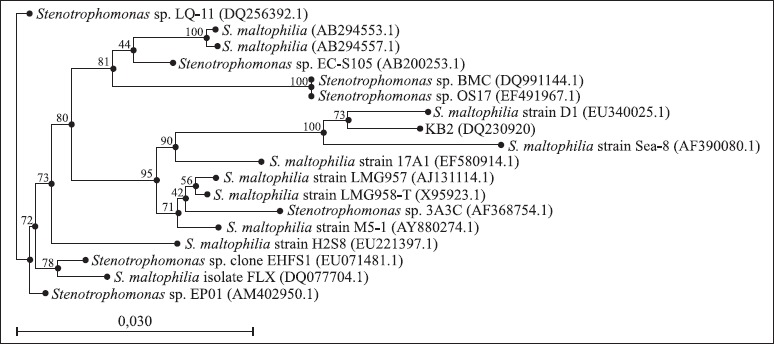
Neighbor-joining tree showing the phylogenetic position of the aromatic substances-degrading bacterium strain KB2 and related species of the genus Stenotrophomonas based on partial 16S rRNA gene sequences. The GenBank accession number for each microorganism used in the analysis is shown in parentheses after the species name. Bootstrap values (expressed as percentage of 100 replicons) are shown at the branch.
Induction of the various types of dioxygenase in cells of strain KB2
To investigate which degradation pathway was used by the strain KB2 for growth on aromatic compounds, the presence of enzymes involved in the ortho- and metha- cleavage pathways for dissimilation of catechol or protocatechuate was determined using crude cell extracts after sonication. Depending on the structure of inductor three various dioxygenases were induced in Stenotrophomonas maltophilia KB2 strain cells (Table 4). These results suggest that strain KB2 has three main metabolic pathways to degrade aromatic compounds. The existence of two different metabolic routes for the aerobic breakdown of aromatic compounds is now well established in bacteria (26). Growth on benzoate mainly induced either catechol 1,2- and that Stenotrophomonas maltophilia KB2 degraded benzoate 2,3- dioxygenase (24) or 1,2-benzoate dioxygenases and catechol through the catechol pathway. Lack of monooxygenase activity 1,2-dioxygenase (1). Although activity of 1,2-benzoate after benzoate induction (data not shown) also confirmed that dioxygenase activity in the crude cell extract of strain KB2 was hypothesis. Among the Stenotrophomonas maltophilia species not determined, but the absence of 3,4-protocatechuate only one described as strain Sea-8 was found to express catechol dioxygenase activity after benzoic acid induction suggested dioxygenases activities after phenol induction, although the activity of catechol 2,3-dioxygenase in that case was rather small (28). Contrary to Sea-8 strain, Stenotrophomonas maltophilia strain KB2 growing on 3 mM phenol showed trace activity of intradiol catechol dioxygenase (Table 3). After growth on hydroxybenzoic acids, additional cleaving dioxygenase is usually induced (26). 4-Hydroxybenzoic acid as well as vanillic acid were found to induce mainly protocatechuate 3,4dioxygenase (7,13,14,16,21). Gentisate dioxygenase was also found in the cells grown on hydroxybenzoic acids (6,27). But to the authors’ knowledge, so far only one strain Pseudomonas sp. K82 was described as to be able to induce three different kinds of dioxygenases including protocatechuate 4,5dioxygenase for p-hydroxybenzoate degradation, catechol 1,2dioxygenase for benzoate decomposition and catechol 2,3dioxygenase for breakdown of aniline. By contrast in Stenotrophomonas maltophilia KB2 strain the protocatechuate 3,4-dioxygenase pathway was the only one induced by 4hydroxybenzoate, protocatechuate or vanillic acid exposure (Table 3). Presence of metabolic pathways for both catechol and protocatechuic acid breakdown increases the biodegradation potential of Stenotrophomonas maltophilia strain KB2 and its ability to perform a role of the universal tool to fight the increasing pollution of the environment. Cometabolic biodegradation of aromatics by strain KB2 and activity of dioxygenases in such conditions are under investigations.
Table 4.
Dioxygenase activities in cell-free extracts from Stenotrophomonas maltophilia KB2 grown on 3 mM aromatic compounds. Catechol was used as substrate in the reaction mixture for detection of catechol dioxygenase activities and protocatechuate - for protocatechuate dioxygenases. The results are an average of at least three independent measurements.
| Inductor | Specific activity µmol min.-1 mg-1 of protein | |||
|---|---|---|---|---|
| Catechol 1,2-dioxygenase | Catechol 2,3-dioxygenase | Protocatechuate 3,4-dioxygenase | Protocatechuate 4,5-dioxygenase | |
| Phenol | 0.06 ± 0.02 | 4.92 ± 0.34 | NDa | ND |
| Catechol | 1.03 ± 0.01 | 0.37 ± 0.00 | ND | ND |
| 4-HB | ND | ND | 6.37 ± 0.72 | ND |
| 3,4-DHB | ND | 0.01 ± 0.00 | 2.98 ± 0.72 | ND |
| Vanillic acid | 0.12 ± 0.03 | 0.04 ± 0.02 | 8.70 ± 1.36 | ND |
| Benzoic acid | 11.97 ± 3.04 | 0.02 ± 0.01 | ND | ND |
| Glucose | ND | ND | ND | ND |
ND - not detected; 4-HB - 4-hydroxybenzoic acid; 3,4-DHB - 3,4-dihydroxybenzoic acid.
Acknowledgments
Authors would like to thank Agnieszka Materna for excellent technical assistance.
RESUMO
Isolamento e caracterização de uma nova cepa de Stenotrophomonas maltophilia com várias dioxigenases para degradação de hidrocarbonetos monocíclicos
Uma bactéria Gram-negativa, denominada KB2, foi isolada de lodo ativado, verificando-se ser capaz de utilizar substratos aromáticos com única fonte de carbono e energia. Com base nas características morfológicas e físico-químicas, e na análise da sequencia do gene 16SrRNA, esta bactéria foi identificada como Stenotrophomonas maltophilia. Entre as diversas cepas de S. maltophilia já descritas, essa cepa é a primeira com atividade de três tipos de dioxigenases, dependendo da estrutura do indutor. As células cultivadas em benzoato e catecol apresentaram atividade de catecol 1,2-dioxigenase principalmente. A atividade de 2,3-dioxigenase foi detectada após indução com fenol. Após incubação com ácidos 4-hidrobenzoico, ácido protocatecuico e vanílico, encontrou-se protocatecuato 3,4-dioxigenase no extrato celular. Devido ao amplo espectro de atividade das diferentes dioxigenases de S. maltophilia KB2, esta cepa parece ser uma ferramenta poderosa e útil para o biotratamento de efluentes e descontaminação do solo.
Palavras-chave: biodegradação, Stenotrophomonas, compostos aromáticos, dioxigenases
REFERENCES
- 1.Adebusoye S.A., Picardal F.W., Ilori M.O., Amund O.O., Fuqua C., Grindle N. Aerobic degradation of di- and trichlorobenzenes by two bacteria isolated from polluted tropical soils. Chemosphere. 2007;66:1939–1946. doi: 10.1016/j.chemosphere.2006.07.074. [DOI] [PubMed] [Google Scholar]
- 2.Almeida M.T.G., Rubio F.G., Garcia D.O., Pavarino-Bertelli E.C., Rossit A.R.B., Bando S.Y., Silbert S., Goloni-Bertollo E.M., Soares M.M.C.N., Martinez M.B. Genetic relatedness among clinical strains of Stenotrophomonas maltophilia in tertiary care hospital settings in Sao Paulo State, Brazil. Braz. J. Microbiol. 2007;38:278–284. [Google Scholar]
- 3.Assih E.A., Ouattara A.S., Thierry S., Cayol J-L., Labat L., Macarie H. Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int. J. Syst. Evol. Microbiol. 2002;52:559–568. doi: 10.1099/00207713-52-2-559. [DOI] [PubMed] [Google Scholar]
- 4.Boonchan S., Britz M.L., Stanley G.A. Surfactant-enhanced biodegradation of high molecular weight polycyclic aromatic hydrocarbons by Stenotrophomonas maltophilia. Biotechnol. Bioengin. 1998;59:482–494. doi: 10.1002/(sici)1097-0290(19980820)59:4<482::aid-bit11>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 5.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–258. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Bugg T.D.H. Dioxygenase enzymes: catalytic mechanisms and chemical models. Tetrahedron. 2003;59:7075–7101. [Google Scholar]
- 7.Deveryshetty J., Suvekbala V., Varadamshetty G., Phale P.S. Metabolism of 2-, 3- and 4-hydroxyenzoate by soil isolates Alcaligenes sp. strain PPH and Pseudomonas sp. strain PPD. FEMS Microbiol. Lett. 2007;268:59–66. doi: 10.1111/j.1574-6968.2006.00561.x. [DOI] [PubMed] [Google Scholar]
- 8.Finkmann W., Altendorf K., Stackebrandt E., Lipski A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000;50:273–282. doi: 10.1099/00207713-50-1-273. [DOI] [PubMed] [Google Scholar]
- 9.Glucksman A.M., Skipper H.D., Brigmon R.L., Santo Domingo J.W. Use of the MIDI-FAME technique to characterize groundwater communities. J. Appl. Microbiol. 2000;88:711–719. doi: 10.1046/j.1365-2672.2000.01058.x. [DOI] [PubMed] [Google Scholar]
- 10.Hauben L., Vauterin L., Moore E.R.B., Hoste B., Swings J. Genomic diversity of the genus Stenotrophomonas. Int. J. Syst. Bacteriol. 1999;49:1749–1760. doi: 10.1099/00207713-49-4-1749. [DOI] [PubMed] [Google Scholar]
- 11.Hegeman G.D. Synthesis of enzymes of the mandelate pathways by Pseudomonas putida. Synthesis of enzyme by the wild type. J. Bacteriol. 1966;91:1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holt J.G., Krieg N.R., Sneath P.H.A., Staley J.T., Williams S.T. USA: Williams and Wilkins; 1994. Bergey’s manual of determinative bacteriology. [Google Scholar]
- 13.Kahng H-Y., Cho K., Song S-Y., Kim S-J., Leem S-H., Kim S.I. Enhanced detection and characterization of protocatechuate 3,4-dioxygenase in Acinetobacter lwoffii K24 by proteomics using a column separation. Biochem. Biophys. Res. Commun. 2002;295:903–909. doi: 10.1016/s0006-291x(02)00778-7. [DOI] [PubMed] [Google Scholar]
- 14.Kim D., Kim S.W., Choi K.Y., Lee J.S., Kim E. Molecular cloning and functional characterization of the genes encoding benzoate and p-hydroxybenzoate degradation by the halophilic Chromohalobacter sp. strain HS-2. FEMS Microbiol. Lett. 2008;280:235–241. doi: 10.1111/j.1574-6968.2008.01067.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim S., Kim J.Y., Yun S-H., Kim J-Y., Leem S-H., Lee C. Proteome analysis of Pseudomonas sp. K82 biodegradation pathways. Proteomics. 2004;4:3610–3621. doi: 10.1002/pmic.200400977. [DOI] [PubMed] [Google Scholar]
- 16.Kim Y.H., Cho K., Yun S-H., Kim J.Y., Kwom K-H., Yoo J.S., Kim S.I. Analysis of aromatic catabolic pathways in Pseudomonas putida KT 2440 using a combined proteomic approach: 2-DE/MS and cleavable isotope-coded affinity tag analysis. Proteomics. 2006;6:13301–1318. doi: 10.1002/pmic.200500329. [DOI] [PubMed] [Google Scholar]
- 17.Liu Z., Yang C., Qiao C.H. Biodegradation of p-nitrophenol and 4-chlorophenol by Stenotrophomonas sp. FEMS Microbiol. Lett. 2004;277:150–156. doi: 10.1111/j.1574-6968.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- 18.Lonergan D.J., Jenter H.L., Coates J.D., Phillips E.J., Schmidt T.M., Lovley D.R. Phylogenetic analysis of dissimilatory Fe (III) – Reducing Bacteria. J. Bacteriol. 1996;178:2402–2408. doi: 10.1128/jb.178.8.2402-2408.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthew W., Vetting M.W., Douglas H., Ohlendorf D.H. The 1.8 Å crystal structure of catechol 1,2-dioxygenase reveals a novel hydrophobic helical zipper as a subunit linker. Structure. 2008;8:429–440. doi: 10.1016/s0969-2126(00)00122-2. [DOI] [PubMed] [Google Scholar]
- 20.Orville A.M., Lipscomb J.D., Ohlendorf D.H. Crystal structures of substrate and substrate analog complexes of protocatechuate 3,4-dioxygenase: endogenous Fe3+ ligand displacement in response to substrate binding. Biochemistry. 1997;36:10052–10066.. doi: 10.1021/bi970469f. [DOI] [PubMed] [Google Scholar]
- 21.Park S-H., Kim J-W., Yun S-H., Lem S.H., Kahng H-Y., Kim S.I. Characterization of β-ketoadipate pathway from multi-drug resistance bacterium, Acinetobacter baumannii DU202 by proteomic approach. J. Microbiol. 2006;44:632–640. [PubMed] [Google Scholar]
- 22.Radianingtyas H., Robinson G.K., Bull A.T. Characterization of a soil-derived bacterial consortium degrading 4-chloroaniline. Microbiology. 2003;149:3279–3287. doi: 10.1099/mic.0.26303-0. [DOI] [PubMed] [Google Scholar]
- 23.Sasser M. Technical note #. 1990:101–110. [Google Scholar]
- 24.Song J., Sung J., Kim Y.M., Zylstra G.J., Kim E. Roles of the meta- and the ortho-cleavage pathways for the efficient utilization of aromatic hydrocarbons by Sphingomonas yanoikuyae B1. J. Microbiol. 2000;38:245–249. [Google Scholar]
- 25.Stanier R.Y., Ingraham J.L. Protocatechuic acid oxidase. J. Biol. Chem. 1954;210:799–808. [PubMed] [Google Scholar]
- 26.Vaillancourt F.H., Bolin J.T., Eltis L.D. The Ins and Outs of ring-cleaving dioxygenases. Crit. Rev. Biochem. Mol. Biol. 2006;41:241–267. doi: 10.1080/10409230600817422. [DOI] [PubMed] [Google Scholar]
- 27.Yoon Y-H., Yun S-H., Park S-H., Seol S-Y., Leem S-H., Kim S.I. Characterization of a new catechol branch of the β-ketoadipate pathway induced for benzoate degradation in Acinetobacter lwoffii K24. Biochem. Biophys. Res. Commun. 2007;360:513–519. doi: 10.1016/j.bbrc.2007.05.132. [DOI] [PubMed] [Google Scholar]
- 28.Zaki S. Detection of meta- and orto-cleavage dioxygenases in bacterial phenol-degraders. J. Appl. Sci. Environ. Manage. 2006;10:75–81. [Google Scholar]


