Abstract
This study aimed to detect methicillin resistant and slime producing Staphylococcus aureus in cases of bovine mastitis. A triplex PCR was optimized targetting 16S rRNA, nuc and mecA genes for detection of Staphylococcus species, S. aureus and methicillin resistance, respectively. Furthermore, for detection of slime producing strains, a PCR assay targetting icaA and icaD genes was performed. In this study, 59 strains were detected as S. aureus by both conventional tests and PCR, and 13 of them were found to be methicillin resistant and 4 (30.7%) were positive for mecA gene. Although 22 of 59 (37.2%) S. aureus isolates were slime-producing in Congo Red Agar, in PCR analysis only 15 were positive for both icaA and icaD genes. Sixteen and 38 out of 59 strains were positive for icaA and icaD gene, respectively. Only 2 of 59 strains were positive for both methicillin resistance and slime producing, phenotypically, suggesting lack of correlation between methicillin resistance and slime production in these isolates. In conclusion, the optimized triplex PCR in this study was useful for rapid and reliable detection of methicillin resistant S. aureus. Furthermore, only PCR targetting icaA and icaD may not sufficient to detect slime production and further studies targetting other ica genes should be conducted for accurate evaluation of slime production characters of S. aureus strains.
Keywords: Staphylococcus aureus, icaA, icaD, slime, mastitis
INTRODUCTION
S. aureus is an important etiologic agent of mastitis in ruminants and also has an economical importance in cattle industry (43,47). Several tecniques are used to characterize the bovine S. aureus strains in veterinary microbiology. In addition to the phenotypic methods, Polymerase Chain Reaction (PCR) is valuable in identification and genotypic characterization of the S. aureus strains (36,42). Several efforts to removig this pathogen from farms are hampered by some factors. One of these factors is antibiotic resistance (22). One of the major mechanisms of resistance to β-lactam antibiotics is β-lactamase producing by staphylococci. This enzyme hydrolyzes the β-lactam ring and causes inactivation of β-lactams. In the early 1950s, it has been aware of the effectiveness of penicilin in treatment of S. aureus infections because of β-lactamaseproducing plasmids. In 1959, methicillin, synthetic, penicillinase-resistant penicillin, was introduced and solved problem in clinical practice, for a time. However, by 1960, Staphylococcus aureus strains were found to be resistant to the new semisynthetic β-lactams (methicillin, oxacillin, flucloxacillin), and became known as methicillin-resistant S. aureus (MRSA). This type of resistance was termed “intrinsic resistance” because it was not due to destruction of the antibiotic by β-lactamase (12). Methicillin resistance in S. aureus is mediated by the production of an altered penicilin-binding protein (PBP2a), a transpeptidase. mecA encodes this enzyme involved in cell-wall peptidoglycan synthesis. Unlike conventional PBPs of S. aureus, PBP2a does not bind to β-lactam antibiotics with high affinity (13,28,32,38,41). A distinctive feature of methicillin resistance is its heterogenous nature, with the level of resistance varying according to the culture conditions and β-lactam antibiotic being used. Heterogeneous strains can be considered to be composed of two populations of cells: relatively susceptible cells and highly resistant cells. Another type of methicillin resistance is borderline (or low level) resistance. Borderline strains characterized by methicillin MICs at or just above the susceptibility breakpoint (e.g., oxacillin MICs of 4-8 μg/ml) and may divided into two categories on the basis of presence of mecA gene (12). Strains not contain mecA differ from the others that contain mecA in the absence of highly resistant clones. The reasons for mecA-negative borderline resistance may be the modifications of normal PBP genes, the overproduction of β-lactamase and meticillinase (40).
It is noted that several studies concerning the evaluation of rapid methods for diagnosis of intrinsic resistance of oxacillin have been reported. Among them, PCR has succesfully used for the detection of mecA gene (3). Most of the clinical isolates show hetereogenous resistance in routine culture conditions and therefore detection of the presence of mecA gene by PCR is accepted as “gold standard”. Detection of methicillin resistance is influenced by several factors as mec regulatory genes, β-lactamase regulatory genes and fem genes (12,40). Detection of mecA gene is the most reliable and fundamental method of identifying methicillin-resistant Staphylococcus aureus (3).
Several factors such as exotoxins, surface proteins (44) and extracellular polysaccharides (1) having important roles in virulence of S. aureus isolated from mastitis cases have been reported. Furthermore, it has been determined that production of slime factor in S. aureus strains causing mastitis was an important virulence factor affecting pathogenesis (44). It is considered that the first step in mastitis progress is adhesion of S. aureus to mammary epithelial cells (14) and slime factor plays an important role for adhesion and colonization (10,47). Production of slime factor also plays an important role in antibiotic resistance and it has been reported that slime producing strains are more resistant to antibiotics than non-slime producing strains (4). Intracellular adhesin is encoded in the ica locus containing icaA, icaB, icaC, icaD genes in S. aureus strains (17,34). icaA gene encodes N-acetylglucosaminyltransferase. Further, icaD plays an important role in expression of this enzyme. icaA and icaD were found to be in high prevalance among S. aureus mastitis isolates and this finding confirms that ica locus has a potential role as a virulence factor in the pathogenesis of mastitis in ruminants (47). Although production of slime factor has been well characterized in other staphylococci isolated from different infections, there is a little information in literature about the formation of slime factor and detection of ica locus in S. aureus mastitis isolates.
In this study, we aimed to determine the methicillin resistance and slime factor production of S. aureus in bovine mastitis phenotypically and genotypically.
MATERIALS AND METHODS
Bacterial isolates and phenotypic identification
The bacteria used in this study consisted of 161 Staphylococcus spp. isolated from subclinic mastitic milk samples. The staphylococcal isolates were identified morphologically and biochemically by standart laboratory procedures (35). For discrimination of S. aureus from coagulase-negative stahylococci (CoNS), the coagulase test was performed.
DNA extraction
Staphylococcus strains were inoculated on Triptycase Soy Agar. After incubation period, fresh colonies were suspended in 500 μl of DEPC-treated water (DNase-RNase free). The suspension was held in a 100ºC of water bath for 10 min. After centrifugation at 10 000 rpm for 5 min, the supernatant containing bacterial DNA was used as a template for subsequent PCR mixture (50).
Triplex PCR
A triplex PCR assay was performed to discriminate the S. aureus from other staphylococci and determinate the methicillin resistance, genotypically. For the detection of 16S rRNA (Staphylococcus spp. specific) 5’-AAC TCT GTT ATT AGG GAA GAA CA-3’ was used as the forward primer and 5’-CCA CCT TCC TCC GGT TTG TCA CC-3’ was used as the reverse primer. For the detection of nuc (S. aureus specific) gene 5’-GCG ATT GAT GGT GAT ACG GTT-3’ was used as forward primer and 5’-AGC CAA GCC TTG ACG AAC TAA AGC-3’) was used as reverse primer. Two primer, Forward: 5’-GTA GAA ATG AC GAA CGT CCG ATA-3’and Reverse: 5’-CCA ATT CCA CAT TGT TTC GGT CTA A -3’ were used to detect mecA (methicillin resistance specific) gene.
Five microliter of the rapid extracted DNA was used as a template in a 25 μl PCR mixture containing 1XPCR buffer (50 mm KCl, 20 mM Tris HCl), 5 μl of 25 mM MgCl2, 3 μl of 10 mM deoxynucleoside triphosphate (dNTP) mix, 1 μl of 20 μM each 16S rRNA and mecA primers, 0,4 μl of 20 μM each nuc primer and 2U of Taq DNA polymerase. The buffers and enzymes used in this assay were obtained from Fermentas Inc. (Canada). The oligonucleotide primers were synthesized from Bio Basic Inc. (Canada). The amplification of DNA was performed as follows: 94ºC for 5 min of initial denaturation; 30 cycles of 94ºC for 45 s, 68ºC for 45 s and 72ºC for 90 s; and a final extantion at 72ºC for 10 min. Amplicons were loaded onto 1.5% Agarose Gel containing 1 μg/ml ethidium bromide. The 756-bp (16S rRNA), 310-bp (mecA) and 279-bp (nuc) amplified DNA fragments were seperated by agarose gel electrophoresis and visualized under UV-light.
Phenotypic detection of methicillin resistance
Disc diffusion sensitivity testing of S. aureus isolates was performed with 5 μg oxacillin discs. On Mueller Hinton Agar, according to NCCLS recommendation, oxacillin complete inhibition zone diameter of ≤12 mm were considered resistant, those with inhibition zone of ≥13 mm were susceptible.
Slime production assay
Slime production assay was performed by cultivation of S. aureus strains on Congo Red Agar (CRA) plates containing 0.8 g of Congo Red dye and 36 g saccharose (49). Strains were inoculated on CRA plates and incubated for 24-72 h at 37ºC. Slime producing strains and non-slime producing strains constitutes rough black colonies and red colonies on CRA, respectively.
PCR detection of icaA and icaD genes
For genotypic determination of slime production, the PCR targetting icaA and icaD genes were performed (47). The primers for icaA and icaD genes were designated from published sequence of the ica locus in GenBank and synthesized from Bio Basic Inc. For the amplifying of icaA, AF (5’-CCT AAC TAA CGA AAG GTA G-3’) and AR (5’-AAG ATA TAG CGA TAA GTG C -3’) primers and of icaD gene, DF (5’-AAA CGT AAG AGA GGT GG-3’) and DR (5’-GGC AATATG ATC AAG ATA-3’) primers were used. The PCR with icaA and icaD genes were amplified a products of 1315-bp and 381-bp, respectively.
Ten microliters of the rapid extracted DNA was used as a template in a 50 μl PCR mixture, containing 1X PCR buffer (50 mm KCl, 20 mM Tris HCl), 5 μl of 25 mM MgCl2, 5 μl of 10 mM deoxynucleoside triphosphate (dNTP) mix, 1 μl of 20 μM each primers and 1U of Taq DNA polymerase. The buffers and enzymes used in the assay were obtained from Fermentas Inc. The amplification of DNA was performed as follows: 92ºC for 5 min of initial denaturation; 30 cycles of 92ºC for 1 min, 49ºC for 1 min and 72ºC for 1 min; and a final extention at 72ºC for 7 min. Amplicons were loaded onto 1.5% Agarose Gel containing 1 μg/ml ethidium bromide. The presence and molecular weight of the amplified DNA fragments were confirmed by agarose gel electrophoresis and visualized under UV-light.
RESULTS
Initial characterization of bacterial strains
The isolated bacteria from bovine mastitis were identified by conventional methods. All of the 161 strains were found gram positive and catalase positive cocci. They were identified as staphylococci. Among these staphylococci, 59 strains were rabbit plasma-coagulase positive and they were considered as S. aureus. Confirmations of the strains were done using triplex-PCR.
Agar disc diffusion test for detection MRSA
Thirteen of 59 (22.0%) S. aureus isolates tested in this study gave a zone of 12 mm or more and were considered as methicillin resistant (Table 1).
Table 1.
Cross-evaluation of slime factor production and methicillin resistance of S.aureus strains analyzed phenotypically and genotypically.
| Slime factor production on CRA (n) | icaA (n) | icaD (n) | icaA and icaD (n) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| P | N | P | N | P | N | P | N | ||
| Methicillin resistance with oxacillin disk (n) | R | 3 | 10 | 4 | 9 | 7 | 6 | 4 | 9 |
| S | 19 | 27 | 12 | 34 | 31 | 15 | 11 | 35 | |
| mecA (n) | P | 0 | 4 | 2 | 2 | 2 | 2 | 2 | 2 |
| N | 22 | 33 | 14 | 41 | 36 | 19 | 13 | 42 | |
P: Positive, N: Negative, R: Resistant, S: Sensitive.
Slime production assay
Rough-black colonies on CRA plates were considered to be slime producing strains and red colonies were evaluated as non-slime producing strains. Among the 59 S. aureus strains tested, 22 (37.2%) were found to produce black colonies within 24-48 h. After 72 h, no more strains produced black colonies (Fig. 1).
Figure 1.
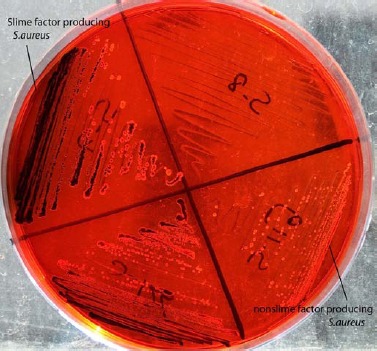
Slime Factor Production of S.aureus on Congo Red Agar.
Triplex PCR
Triplex PCR was used to discriminate S. aureus strains among all our mastitis isolates and to simultaneously detect methicillin resistant strains. This assay was targetted the 16S rRNA, nuc and mecA genes. Fragments of expected sizes were 756, 279 and 310 bp for the 16S rRNA, mecA and nuc genes, respectively (Fig. 2). All the 59 S. aureus strains identified phenotypically were found to possess both 16S rRNA and nuc genes and were confirmed as S. aureus. Strains were characterized in this PCR analysis targetting mecA gene simultaneously for methicillin resistance. Among the 13 S. aureus strains found as methicillin resistant phenotypically, 4 (30.7%) were positive for mecA gene. Also; the phenotypically methicillin sensitive 46 S. aureus strains were found as negative for mecA gene.
Figure 2.
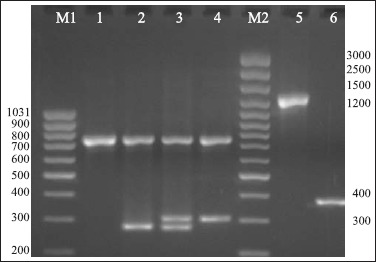
The results of triplex-PCR for confirmation of the identification of S. aureus and determination of methicillin resistance; single PCR for icaA and icaD genes.
M1: Marker (80-1031 bp); M2: Marker (100-3000 bp); 1: Staphylococcus spp., 16S rRNA (756 bp) pozitive; 2: S. aureus, 16S rRNA (756 bp) and nuc (279 bp) pozitive; 3: MRSA, 16S rRNA (756 bp), nuc (279 bp) and mecA (310 bp) pozitive; 4:Methicilline resistant Staphylococcus spp., 16S rRNA (756 bp) and mecA (310 bp) pozitive; 5: icaA (1315 bp); 6: icaD(381 bp).
PCR detection of icaA and icaD genes
Among all S. aureus isolates, 15 of 59 (25.4%) were positive for both icaA and icaD genes and only 8 of them (53.3%) were slime positive on CRA. 16 of 59 (27.1%) strains were positive for on icaA and 38 of them (64.4%) were positive for icaD gene (Fig. 1, Table 1 and 2).
Table 2.
Distribution of S.aureus strains according to their phenotypic and genotypic slime factor production characteristics.
| icaA | icaD | icaA and icaD | |||||
|---|---|---|---|---|---|---|---|
| Positive (n) | Negative (n) | Positive (n) | Negative (n) | Positive (n) | Negative (n) | ||
| Slime production on CRA | Positive (n) | 8 | 14 | 15 | 7 | 8 | 7 |
| Negative (n) | 8 | 29 | 23 | 14 | 7 | 13 | |
DISCUSSION
Bovine mastitis is the most costly disease to the dairy industry worldwide as well as in Turkey. Staphylococcus aureus is a frequent cause of bovine mastitis and several tecniques are used to characterize the bovine S. aureus strains in veterinary microbiology. Rapid detection of methicillin resistance together with the identification of S. aureus is necessary for therapeutic and epidemiological purposes (16,36). S. aureus nuc gene encodes the TNase producing by these bacteria. S. aureus TNase has species-specific sequences and amplification of the nuc gene has potential for the rapid diagnosis of S. aureus infections (11,16). The primary tool for controlling staphylococcal mastitis is antimicrobial therapy. Therefore antimicrobial susceptibility tests are so important that the results of these help guide the veterinarian in selecting the most appropriate antimicrobial agent for treatment of mastitis caused by S. aureus (19). Many studies are avaliable on antibiotic resistance of Staphylococcus aureus (24,29,33,39). Methicillin-resistant Staphylococcus aureus (MRSA) and multi-resistant S. aureus strains has also been reported in some cases in veterinary medicine (9,31). MRSA show an intrinsic resistance to penicillinase-resistant beta lactam antibiotics. This resistance is based on “mecA” gene encoding PBP2a, an inducible and an altered PBP that has low affinity for binding β-lactam antibiotics. mecA gene is located on the chromosome of MRSA and methicillin resistance does not due to destruction of the antibiotic by beta-lactamase. MRSA strains show also a heterogeneous character with the level of resistance varying according to the culture conditions and β-lactam antibiotic being used. Because of this heterogeneous resistance, the detection of MRSA by phenotypic methods becomes problematic. Although laboratory conditions has been changed (e.g. additional NaCl to medium, prolonged incubation, incubation at low temperatures, higher inoculum) to enhance the phenotypic expression, all strains can not be classified. Furthermore, the conditions that are used to enhance expression of methicillin resistance also can cause the susceptibility test results for susceptible strains to shift toward or above the breakpoint for resistance (25). The multidrug-resistant (methicillin and other more common antibiotics such as oxacillin, penicillin and amoxicillin) phenotype of MRSA strains and their intrinsic betalactam resistance make them difficult and costly to treat (12,28,41). Methicillin and oxacillin are not used in veterinary medicine in Turkey except for cloxacillin used in mastitis cases (45). Considering the multiple antibiotic resistance, the rapid and correct detection of MRSA strains must be performed to select appropriate antibiotic regimens. However selected methods detecting MRSA strains must be useful, reliable, simple and rapid (27). Several phenotypic methods such as oxacillin disk diffusion test, agar plate screen, the microbroth dilution and the E-tests have been used to detect the MRSA isolates (5). Phenotypic-based identification and susceptibility testing methods are time consuming and most have inherent limitations (16). However, polymerase chain reaction (PCR)-based methods have shown to be a rapid and reliable approach for the identification and genotypic characterization of these organisms. mecA-based PCR methods has accepted as “gold standard” (5,12,40).
In the present study, S. aureus strains originated from bovine mastitis were identified phenotypically by some conventional tests and also were characterized genotypically by a triplex PCR targetting 16srRNA (staphylococcus genus-spesific), nuc (S. aureus species-spesific) and mecA (a determinant of methicillin resistance). In this study, all 59 S. aureus isolates identified by conventional tests were confirmed as S. aureus by this PCR genotypically and at the same time, methicillin resistance of these strains were detected. While 13 of 59 (22.0%) S. aureus isolates were found to be methicillin resistant in oxacillin disc diffusion method, only 4 of 59 (6.7%) S. aureus isolates were found to be MRSA in triplex PCR. Similarly, some investigators (5,46) have reported the discrepant results between disc diffusion methods and PCR for detection of methicillin resistance. It is reported that conventional susceptibility tests such as agar disc diffusion and broth dilution methods may not give reliable results in detecting MRSA because of heterogenic expression of resistance (46). In this study, MRSA lacking mecA gene are classified as false resistant by the oxacillin disc diffusion method and it was considered that it may due to another resistance mechanism such as hyperproduction of beta-lactamase.
Extracellular polysaccharides, slime factor, are considered to be significant virulence factors for some staphylococci (20). Slime layer surrounding the S. aureus strains help in adherence and colonization of these microorganisms on the mammary gland epithelium. It is reported that slime factor production in S. aureus isolates from mastitis cause antibiotic resistance which is due to the decreased diffusion of antibiotics through the biofilm matrix and decreased metabolic activity of bacteria (4,36). Several phenotypic methods such as standard tube method, Christensen’s method, Congo Red Agar and microdilution methods are used to detect slime production (2). In the present study, slime producing S. aureus isolates were detected on Congo Red Agar (CRA) plates in vitro. CRA plate test has been reported as a simple, rapid, sensitive, and reproducible and advantageous in those colonies remain viable on the medium and also more specific than standard tube test (23,37,48). We found that only 22 of 59 (37.2%) S. aureus isolates were slime-producing on Congo Red Agar. Slime-producing S. aureus isolates from different clinical origins such as wound infection (49), catheter-associated infections (6), bovine mastitis (47) has been detected in vitro by using Congo Red Agar plates as 52%, 60.8% and 91.4%, respectively. Knobloch et al (30) have reported that the phenotype on CRA was found to be an unreliable indicator of slime-forming capacity among clinical isolates of S. aureus. Therefore, although CRA methods may be easier to perform than a molecular analysis of the genes implicated in biofilm production and could be performed easily in a diagnostic laboratory, it may be a poor method for determining the slime-producing capacity of clinical isolates in the diagnostic laboratory (21).
Recently developed molecular methods provided a direct evidence of the genetic basis of slime production complementary to the CRA test. Slime synthesis is controlled by the ica (intercellular adhesion) operon (7). The ica locus consists ica A, D, B, C genes which encode the proteins mediating the synthesis polysaccharide intercellular adhesin and capsular polysaccharide/adhesin (PS/A) in S. epidermidis and S. aureus, respectively (49). N-acetylglucosaminyltransferase that synthesizes the polysaccharide intercellular adhesin from UDP-N-acetylglucosamine encoded by the icaA particularly. However sole expression of icaA induces only low enzymatic activity, coexpression of icaA with icaD leads to a significant increase in activity and is related to phenotypic expression of the capsular polysaccharide (6). In the operon, coexpression of icaA and icaD is required for full slime synthesis. In this study slime production of S. aureus isolates were detected by PCR targetting icaA and icaD. In PCR analysis, not all isolates, only 15 of 59 were positive for both icaA and icaD genes and only 8 of them were slime positive in CRA. 16 of 59 strains were positive for icaA and 38 of them were positive for icaD gene. This result was contrast to Arciola et al. (6) who have reported that all strains which were positive for icaA were also positive for icaD. Further Vasudevan et al. (47) have reported that although only 24 of 35 S. aureus mastitis isolates produced slime factor in vitro, all of them were found to possess the ica locus as well as the icaA and icaD. Arciola et al. (8) have been found that the genes of the ica locus appear, in all the clinical isolates analyzed in their study, strictly linked each other, so they are either all present or all absent. Our discrepant findings that icaA and icaD genes were not be together in some isolates may due to some mutations on icaA. Although coexpression of icaA and icaD is necessary for slime production, it was considered that other genes in ica locus play role in controlling slime expression. Fitzpatrick et al (21) have reported that there are ica-independent mechanisms of biofilm formation in S. aureus. In this study, among the 37 strains which did not produce slime factor on CRA plate in vitro, 7 strains were positive for both icaA and icaD genes. This suggests the possibility that some environmental conditions or the presence of accessory genes can influence the phenotypic behavior on the Congo red agar plate, giving colonies which did not fully express the ica genes.
Slime factor may also play a role in antibiotic resistance (18,44) and it is reported that slime-producing strains are more resistant to antibiotics than non slime producing strains (4). Günaydin et al. (26) are also reported that there is a correlation between slime producing and multiple antibiotic resistances. In contrast to these studies, Ciftci et al. (15) have not found any effect of slime producing on antibiotic resistance in their study and furthermore they have reported that slime producing strains are more susceptible to antibiotics than nonslime producing strains. In this study, we found that only 2 of 59 strains were positive for both methicillin resistance and slime producing, phenotypically. Thus, we suggested that there is no correlation between methicillin resistance and slime producing in our isolates.
In conclusion, it was showed that several cases of mastitis in cows in Turkey were caused by MRSA despite of low percentage. It was considered that the triplex PCR optimized in this study is useful for diagnostic laboratory because of its rapid and reliable characters for detection of methicillin resistance of S. aureus. Using the phenotypic and genotypic methods together is required for accurate identification of MRSA. Thus it was found that not all isolates phenotypically slime positive were positive both icaA and icaD and there were differences in the presence of these two genes. We considered that only PCR targetting icaA and icaD was not sufficient to detect slime production. Further studies targetting other genes that are present in ica locus and studies for investigation of mutations on these genes should be conducted to accurate evaluation of slime production characters of S. aureus strains.
Acknowledgments
This work was supported by OMU-BAP (VET-013).
RESUMO
Detecção de resistência a meticilina e produção do fator slime por Staphylococcus aureus em mastite bovina
Este estudo objetivou a detecção de Staphylococcus aureus resistente a meticilina e produtor do fator slime em casos de mastite bovina. Um PCR triplex foi otimizado, com alvo no genes 16SrRNA, nuc e mecA para detecção de Staphylococcus spp, S. aureus e resistencia a meticilina, respectivamente. Para detecção das cepas produtoras do fator slime, empregou-se um PCR com alvo nos genes icaA e icaD. No estudo, 59 cepas foram identificadas como S. aureus por testes convencionais e PCR, sendo 13 resistentes a meticilina e quatro positivas para o gene mecA. Embora 22 das 59 cepas tenham sido produtoras do fator slime em Agar Vermelho Congo, no teste PCR somente 15 foram positivas para os genes icaA e icaD. Dezesseis e 38 das 59 cepas foram positivas para os genes icaA e icaD, respectivamente. Somente duas das 59 cepas foram positivas simultaneamente para resistência a meticilina e produção do fator slime, sugerindo falta de correlação entre estas características. Em conclusão, o PCR triplex otimizado neste trabalho mostrou-se ser um método rápido e confiável para detecção de S.aureus meticilina resistente. Por outro lado, somente PCR para os genes icaA e icaD pode não ser suficiente para detectar produção de fator slime e outros estudos com alvo em outros genes ica são necessários para um avaliação correta da produção do fator slime por S. aureus.
Palavras-chave: Staphylococcus aureus, icaA, icaD, mecA, slime, mastite
REFERENCES
- 1.Aguilar B., Amorena B., Iturralde M. Effect of slime on adherence of Staphylococcus aureus isolated from bovine and ovine mastitis. Vet. Microbiol. 2001;78:183–191. doi: 10.1016/s0378-1135(00)00287-x. [DOI] [PubMed] [Google Scholar]
- 2.Alcaraz L.E., Satorres S.E., Lucero R.M., Centorbi O.N.P. Species identification, slime production and oxacillin susceptibility in coagulase-negative staphylococci isolated from nosocomial specimens. Braz. J. Microbiol. 2003;34:45–51. [Google Scholar]
- 3.Allaouchiche B., Jaumain H., Zambardi G., Chassard D., Freney J. Clinical impact of rapid oxacillin susceptibility testing using a PCR assay in Staphylococcus aureus bactaeremia. J. Infect. 1999;39:198–204. doi: 10.1016/s0163-4453(99)90049-x. [DOI] [PubMed] [Google Scholar]
- 4.Amorena B., Gracia E., Monzon M., Leiva J., Oteiza C., Perez M., Alabart J.L., Hernandez-Yago J. Antibiotic susceptibility assay for Staphylococcus aureus in biofilms developed in vitro. J. Antimicrob. Chemother. 1999;44:43–55. doi: 10.1093/jac/44.1.43. [DOI] [PubMed] [Google Scholar]
- 5.Araj G.F., Talhouk R.S., Siman C.J., Maasad M.J. Discrepancies between mecA PCR and conventional tests used for detection of methicillin resistant Staphylococcus aureus. Int. J. Antimicrob. Agent. 1998;11:47–52. doi: 10.1016/s0924-8579(98)00047-8. [DOI] [PubMed] [Google Scholar]
- 6.Arciola C.R., Baldassarri L., Montanaro L. Presence of icaA and icaD genes and slime production in a collection of staphylococcal strains from catheter-associated infections. J. Clin. Microbiol. 2001;39(6):2151–2156. doi: 10.1128/JCM.39.6.2151-2156.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arciola C.R., Campocciaa D., Gamberinia S., Cervellatia M., Donatia E., Montanar L. Detection of slime production by means of an optimised Congo red agar plate test based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterial. 2002;23:4233–4239. doi: 10.1016/s0142-9612(02)00171-0. [DOI] [PubMed] [Google Scholar]
- 8.Arciola C.R., Gamberini S., Campoccia D., Visai L., Speziale P., Baldassarri L., Montanaro L. A multiplex PCR method for the detection of all five individual genes of ica locus in Staphylococcus epidermidis. A survey on 400 clinical isolates from prosthesis-associated infections. J. Biomed. Material. Res. 2005;75(2):408–413. doi: 10.1002/jbm.a.30445. [DOI] [PubMed] [Google Scholar]
- 9.Baptiste K.E., Williams K., Williams J., Wattret A., Clegg P.D., Dawson S., Corkill J.E., O’Neill T., Hart C.A. Methicillimresistant staphylococci in companion animals. Emerg. Infect. Dis. 2005;11(12):1942–1944. doi: 10.3201/eid1112.050241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baselga R., Albizu I., De La Cruz M., Del Cacho E., Barberan M., Amorena B. Phase variation of slime production in Staphylococcus aureus: implications in colonization and virulence. Infect. Immun. 1993;61(11):4857–4862. doi: 10.1128/iai.61.11.4857-4862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brakstad O.G., Aasbakk K., Maeland J.A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J. Clin. Microbiol. 1992;30(7):1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chambers H.F. Methicillin resistance in staphylococci: Molecular and Biochemical Basis and Clinical Implications. Clin. Microbiol. Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chambers H.F. Methicillin resistance in staphylococci. Clin. Microbiol. Rev. 1998;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cifrian E., Guidry A.J., O’Brien C.N., Nickerson S.C., Marquardt W.W. Adherence of Staphylococcus aureus to cultured bovine mammary epithelial cells. J. Dairy Sci. 1994;77:970–983. doi: 10.3168/jds.S0022-0302(94)77033-8. [DOI] [PubMed] [Google Scholar]
- 15.Ciftci A., Ica T., Onuk E.E., Baş B., Tosun G. çeşitli klinik örneklerden izole edilen Staphylococcus aureus suşlarinda slime faktor üretimi ve antibiyotik dirençliliği. Vet. Hek. Mikrobiyol. Derg. 2003;3(1-2):51–55. [Google Scholar]
- 16.Costa A., Kay I., Palladino S. Rapid detection of mecA and nuc genes in staphylococci by real-time multiplex polymerase chain reaction. Diag. Microbiol. Infect. Dis. 2004;51(1):13–17. doi: 10.1016/j.diagmicrobio.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 17.Cramton S.E., Gerke C., Schnell N.F., Nichols W.W., Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect. Immun. 1999;67(10):542–5433. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davenport D.S., Massanari R.M., Pfaller M.A., Bale M.J., Streed S.A., Hierholzer W.J. Usefulness of a test for slime production as a marker for clinically significant infections with coagulase-negative staphylococci. J. Infect. Dis. 1986;153(2):332–339. doi: 10.1093/infdis/153.2.332. [DOI] [PubMed] [Google Scholar]
- 19.De Oliveira A.P., Watts J.L., Salmon S.A., Aarestrup F.M. Antimicrobial susceptibility of Staphylococcus aureus isolated from bovine mastitis in Europe and United States. J. Dairy Sci. 2000;83(4):855–862. doi: 10.3168/jds.S0022-0302(00)74949-6. [DOI] [PubMed] [Google Scholar]
- 20.Drewry D.T., Galbraith L., Wilkinson B.J., Wilkinson S.G. Staphylococcal slime: A cautionary tale. J. Clin. Microbiol. 1990;28(6):1292–1296. doi: 10.1128/jcm.28.6.1292-1296.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzpatrick F., Humphreys H., O’Gara J.P. The genetics of staphylococcal biofilm formation—will a greater understanding of pathogenesis lead to better management of device-related infection? Clin. Microbiol. Infect. 2005;11:967–973. doi: 10.1111/j.1469-0691.2005.01274.x. [DOI] [PubMed] [Google Scholar]
- 22.Fox L.K., Bayles K.W., Bohach G.A. Staphylococcus aureus Mastitis. In: Honeyman A.L., Friedman H., Bendinelli M., editors. Staphylococcus aureus Infection and Disease. New York: Kluwer Academic/Plenum Publishers; 2001. pp. 271–293. [Google Scholar]
- 23.Freeman D.J., Falkiner F.R., Keane C.T. New method for detecting slime production by coagulase negative staphylococci. J. Clin. Pathol. 1989;42:872–874. doi: 10.1136/jcp.42.8.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fthenakis G.G. Susceptibility to antibiotics of staphylococcal isolates from cases of ovine or bovine mastitis in Greece. Small Ruminant Res. 1998;28:9–13. [Google Scholar]
- 25.Gerberding J.L., Miick C., Liu H.H., Chambers H.F. Comparison of conventional susceptibility tests with direct detection of penicillin-binding protein 2a in borderline oxacillin-resistant strains of Staphylococcus aureus. Antimicrob. Agent. Chemother. 1991;35(12):2574–2579. doi: 10.1128/aac.35.12.2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Günaydin M., Leblebicioğlu H., Saniç A., Pirinççiler M. Koagulaz negatif stafilokoklarda slime yapimi ve antibiyotik direnci ile iliğkisi. Mikrobiyol. Bult. 1995;105:493–500. [Google Scholar]
- 27.Hasbek M., Hakgüdener Y., Kaya S., Bakici M.Z. Stafilokoklarda metisilin direncinin farkli yöntemlerle belirlenmesi ve çoğul antibiyotik direnci. Mikrobiyol. Bult. 1994;25:227–234. [Google Scholar]
- 28.Hiramatsu K., Katayama Y., Yuzawa H., Ito T. Molecular genetics of methicillin resistant Staphylococcus aureus. Int. J. Med. Microbiol. 2002;292:67–74. doi: 10.1078/1438-4221-00192. [DOI] [PubMed] [Google Scholar]
- 29.Kaszanyitzky E.J., Janosi S.Z., Egyed Z., Agost G., Semjen G. Antibiotic resistance of staphylococci from humans, food and different animal species according to data of the Hungarian resistance monitoring system in 2001. Acta Vet. Hung. 2003;51:451–464. doi: 10.1556/AVet.51.2003.4.3. [DOI] [PubMed] [Google Scholar]
- 30.Knobloch J.K.M., Horstkotte M.A., Rohde H., Mack D. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Med. Microbiol. Immunol. 2002;191(2):101–106. doi: 10.1007/s00430-002-0124-3. [DOI] [PubMed] [Google Scholar]
- 31.Lee J.H. Methicillin (oxacillin)-resistant Staphylococcus aureus strains isolates from major food animals and theirpotential transmission to humans. Appl. Environ. Microbiol. 2003;69(11):6489–6494. doi: 10.1128/AEM.69.11.6489-6494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louie L., Matsumura S.O., Choi E., Louie M., Simor A.E. Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J. Clin. Microbiol. 2000;38(6):2170–2173. doi: 10.1128/jcm.38.6.2170-2173.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malinowski E., Klossowska A., Kaczmarowski M., Lassa H., Kuzma K. Antimicrobial susceptibility of staphylococci isolated from affected with mastitis cows. Bull. Vet. Inst. Pulawy. 2002;46:289–294. [Google Scholar]
- 34.McKenney D., Hubner J., Muller E., Wang Y., Goldmann D.A., Pier G.B. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infect. Immun. 1998;66(10):4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murray P.R. Manuel of clinical microbiology. 8th ed. Washington D.C: ASM Pres; 2003. [Google Scholar]
- 36.Nak D. Subklinik mastitislerin teþhis yöntemleri üzerine çalişmalar. U.Ü. Vet. Fak. Derg. 1999;3(18):15–27. [Google Scholar]
- 37.Nourizadeh E., Sultan N. Stafilokoklarda slime faktör yapiminin çeşitli yöntemlerle gösterilmesi. Infeks. Derg. 1993;7:31–36. [Google Scholar]
- 38.Ontengco D.C., Baltazar L.A., Santiago R.S., Matias R.R., Isaac C.A., Tuazon A.O. Methicillin-resistant Staphylococcus aureus isolates from Filipino patients (1999-2003). Phil. J. Microbiol. Infect. Dis. 2004;33(3):105–110. [Google Scholar]
- 39.Roesch M., Perreten V., Doherr M.G., Scheren W., Schalllbaum M., Blum J.W. Comparison of antibiotic resistance of udder pathogens in dairy cows kept on organic and on conventional farms. J. Dairy Sci. 2006;89:989–997. doi: 10.3168/jds.S0022-0302(06)72164-6. [DOI] [PubMed] [Google Scholar]
- 40.Sancak B. S. aureus’ta metisilin direnç mekanizmalari. Mikrobiyol. Bult. 2000;34:381–389. [Google Scholar]
- 41.Shopsin B., Kreiswirth B.N. Molecular epidemology of meticillin-resistant Staphylococcus aureus. Emerg. Infect. Dis. 2001;7(2):323–326. doi: 10.3201/eid0702.010236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Silva W.P., Silva J.A., Macedo M.R.P., Araujo M.R., Mata M.M., Gandra E.A. Identification of Staphylococcus aureus, S. intermedius and S. hyicus by PCR amplification of coa and nuc genes. Braz. J. Microbiol. 2003;34:125–127. [Google Scholar]
- 43.Takeuchia S., Maedaa T., Hashimotoa N., Imaizumia K., Kaidoha T., Hayakawab Y. Variation of the agr locus in Staphylococcus aureus isolates from cows with mastitis. Vet. Microbiol. 2001;79:267–274. doi: 10.1016/s0378-1135(00)00354-0. [DOI] [PubMed] [Google Scholar]
- 44.Türkyilmaz S., Eskiizmirliler S. Detection of slime factor production and antibiotic resistance in staphylococcus strains isolated from various animal clinical samples. Turk. J. Vet. Anim. Sci. 2006;30:201–206. [Google Scholar]
- 45.Türütoğlu H., Erçelik S., Öztürk D. Antibiotic resistance of Staphylococcus aureus and coagulase-negative staphylococci isolated from bovine mastitis. Bull. Vet. Inst. Pulawy. 2006;50:41–45. [Google Scholar]
- 46.Unal S., Werner K., Degirolami P., Barsanti F., Eliopoulos G. Comparison of tests for detection of methicillin-resistant Staphylococcus aureus in a clinical microbiology laboratory. Antimicrob. Agent Chemother. 1994;38(2):345–347. doi: 10.1128/aac.38.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vasudevan P., Nair M.K.M., Annamalai T., Venkitanarayanan K.S. Phenotypic and genotyping characterization of bovine mastitis isolates of Staphylococcus aureus for biofilm formation. Vet. Microbiol. 2003;92:179–185. doi: 10.1016/s0378-1135(02)00360-7. [DOI] [PubMed] [Google Scholar]
- 48.Woznicova V., Votava M., Skalka B. Comparison of two methods of detecting slime production by coagulase-negative staphylococci. Cesk. Epidemiol. Microbiol. Imunol. 1993;42:51–53. [PubMed] [Google Scholar]
- 49.Yazdani R., Oshaghi M., Havayi A., Pishva E., Salehi R., Sadeghizadeh M., Foroohesh H. Detection of icaAD gene and biofilm formation in Staphylococcus aureus isolates from wound infections. Iranian J. Publ. Health. 2006;35(2):25–28. [Google Scholar]
- 50.Zhang K., Sarling J., Chow B.L., Elsayed S., Hussain Z., Church D.L., Gregson D.B., Louie T., Conly J.M. New quadriplex PCR assay for detection of methicillin resistance and simultaneous discrimination of S. aureus from coagulase-negative staphylococci. J. Clin. Microbiol. 2004;42:4947–4955. doi: 10.1128/JCM.42.11.4947-4955.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


