Abstract
The present investigation deals with the kinetics of submerged extracellular lipases fermentation by both wild and mutant strains of Rhizopus oligosporus var.microsporus in a laboratory scale stirred fermentor. Other parameters studied were inoculum size, pH, agitation and rate of aeration. It was found that the growth and lipases production was increased gradually and reached its maximum 9.07± 0.42a U mL-1 (W) and 42.49 ± 3.91a U mL-1 (M) after 30h of fermentation for both wild and mutant strain. There is overall increase of 109% (W) and 124% (M) in the production of extracellular lipases as compared to shake flask. Another significant finding of the present study is that the fermentation period is reduced to 30 h in case of wild and 23 h in case of mutant from 48 h in shake flask studies. The specific productivity of mutant strain (qp = 377.3 U/g cells/h) was several folds higher than wild strain. The specific production rate and growth coefficient revealed the hyperproducibility of extracellular lipases using mutant IIB-63NTG-7.
Keywords: Extracellular lipases; Rhizopus oligosporus,; fermentor; kinetics
INTRODUCTION
Enzymes are considered as nature’s catalysts. Lipase (triacyl glycerol acyl-hydrolases, EC 3.1.1.3) catalyses hydrolysis of long chain acyl glycerol at an oil water interface. These are enzymes belonging to the group of hydrolases that present as main biological function to catalyze the hydrolysis of insoluble triacylglycerols to generate free fatty acids, mono and diacylglycerols and glycerol (20, 26). Ester synthesis is carried out in aqueous media in the presence of various lipases (23).
Interest in lipase enzyme has been greatly developed in the last few years, due to their potential applications. They are extensively used in fat splitting as well as in synthesis of glycerides. Lipases are also used in various reactions due to their high specificity, prevention of product and substrate deterioration and decreased energy consumption. The advantage of the enzymatic hydrolysis over the chemical process is less energy requirements and higher quality of the obtained products (19). Lipases can be classified in several groups according to their specificity as non-specific lipases, 1, 3-specific lipases, 2-specific lipases and fatty acyl lipases (2, 30).
Numerous species of fungi, yeast and bacteria produce lipases. Different microorganisms have been used for the production of lipases such as Rhizopus species,Rhizomucor mehei, Aspergillus niger, Penicillium species, Bacillus subtilus (12, 13, 14, 25). Gopinath et al. (9) reported about 34 wild fungal species associated with edible oil mill wastes which were isolated by the serial dilution technique. This study also confirmed that the isolated fungi present on a wide range of substrates in the ambient environment, and these results could also provide basic data for further investigations on fungal extracellular enzymes (6). Rhizopus species is among the most well known lipase producers and its enzyme is suitable for use in many industrial applications (3, 31). Many microorganisms produced both intra- and extracellular enzymes (18). The spectacular success examples of strain improvement in industry are mostly attributed to the extensive application of mutation and selection of microorganisms (15). There is also no cell disruption in case of extracellular enzymes, which will create heat that could be detrimental to the target proteins. Extracellular enzymes are also more stable as compared to intracellular enzymes due to the presence of sulfide bonds (4).
Batch culture has great influence on the enhanced production of lipases. Optimization of physico-chemical parameters such as the composition of production medium, aeration and agitation is an important step for its commercial usage (10, 27). It is reported that an increased lipase production in a batch culture system have been determined by the aeration (2.0, 2.5, 3.0, 4.0 L/min) during the bioprocess time. The lipolytic activities into the broth reached maximum levels within 20 - 22 h after inoculation, when the microorganisms were at the beginning of the stationary growth phase (27). Effect of the gradual increase in impeller speed has also been investigated that ranges between 400–900 rpm (5). For this purpose, scale up studies in a laboratory scale stirred fermenter for extracellular lipase production from selected mutant strain was subsequently evaluated in parallel with wild strain.
MATERIAL AND METHODS
Microorganism
Lipase producing Rhizopus oligosporus strains having Accession Nos IIB-63 (W) and IIB-63NTG-7 (M) was obtained from the culture collection of laboratory of Biotechnology, Department of Botany, GC University Faisalabad. The strains were maintained on Potato dextrose agar media (4%).
Inoculum Preparation (Seed medium)
Vegetative inoculum was used in the present studies. Hundred milliliter of seed medium (%W/V: g 100mL-1; soybean meal 10; olive oil 20; glucose 10; K2HPO4 2; NaNO3 0.5; MgSO4.7H2O 0.5; pH 7, Iftikhar et al. 2010) containing glass bead in 1 liter cotton wool plugged conical flask was sterilized at 15 lb in2 pressure (121°C) for 15 min. One milliliter of spore suspension (containing 4.63 x 107 spores) was aseptically transferred to the flask. The flask was incubated at 30°C in an incubator shaker at 200 rpm for 24 h.
Fermenter Study
A fermentor (KoBiotech. Co., Ltd, KB 250, Korea), of 5L capacity with a working volume of 3 L was used for lipase production. Seed Medium and culture medium (g 100mL-1; soybean meal 10; olive oil 20; glucose 10; K2HPO4 2; NaNO3 0.5; MgSO4.7H2O 0.5; pH 7, Iftikhar et al. 2007) used were the same in composition. Culture media were sterilized at 121 °C at 15 lb / inch2pressure for 15 min. The incubation temperature was kept at 30°C throughout the fermentation period. Agitation speed of the stirred reactor was 200 rpm unless mentioned otherwise. Sterilized silicone oil was used to control the foaming. After every 8 h the samples were withdrawn from fermentor and subjected to different assays.
Spectrophotometric Assay of lipases
After specific time interval lipase activity was assayed spectrophotometrically using p-nitrophenyl palmitate (p-NPP) (Sigma) as substrate according to the method described by Krieger et al., (1999) (18). Enzyme solution (50 µL) was added to 950 µL of the substrate solution consisting of one part solution A (3.0 mM p-NPP in 2-propanol) and nine parts solution B [100 mM potassium phosphate buffer (pH 7), 0.4% Triton X 100 and 0.1% gum arabic], which was freshly prepared before use. The reaction mixture was incubated at 37oC for 20 min. The reaction was stopped by boiling for 10 min, followed by centrifugation at 8000 x g for 10 min. The p-nitrophenol released was measured at 410 nm against a blank containing only buffer and carried out under the conditions described above. One unit of enzyme activity is defined as the amount of enzyme that released 1 µmole p-nitrophenol per minute.
Glucose Estimation
Glucose was estimated by glucose kit (BMI 30092, Korea). Twenty µL of enzyme solution was transferred to 3 mL of the buffer provided with the kit. Parallel to it 20 µL of control solution (provided) was added in the kit in 3 mL of the same buffer solution used in the first step. After vortexing placed in water bath at 37oC for 5 min and then measure the absorbance at 505 nm by using buffer as blank.
Dry Cell Mass Determination
The mycelium was filtered through Whatman GF/C circles 47mmФ (Cat#1822 047), England. Ten mL of the sample was poured through filter paper placed on filter assembly attached with diaphragm vacuum pump (ULVA Sinku kiko DA-60D, Japan) and placed in an oven at 80oC. After 12 h the filter papers were re-weighed and dry cell mass was determined.
Statistical analysis
Treatment effects were compared after Snedecor & Cochran (1980). Each value is an average of three replicates. Y error bars indicate the standard error among replicates.
Kinetic Studies
Kinetic parameters for batch fermentation experiments were determined according to the methods described by Pirt (1975) and Lawford and Roseau (1993). The following parameters of kinetics were studied:
µ(h-1) max Specific growth rate;
qp Unit product produced/g cells/h.
qs g substrate consumed/g cells/h.
Yp/s Unit product produced/g substrate consumed.
Yp/x Unit product produced/g cells formed
Yx/s g cells/g substrate utilized
Qp g cells produced/L/h
Qs g substrate consumed/L/h
RESULTS AND DISCUSSION
The rate of fermentation of both wild (IIB-63) and mutant (IIB-63NTG-7) strains of R. oligosporus for the production of extracellular lipases was investigated in stirred fermenter (Figure 1). Time course aliquots were withdrawn after every 8 hours aseptically and subjected to enzyme estimation (UmL-1), dry cell mass determination (gL-1) and glucose consumption (g L-1) up to 40 hours of fermentation period. It was found that the growth and lipases production was increased gradually and reached its maximum 9.07± 0.42a U mL-1 (W) and 42.49 ± 3.91a U mL-1 (M) after 30 h for wild and 23 h of fermentation for mutant strain. The dry cell mass (DCM) was found maximum after 16 h of fermentation by both wild (5.0 ± 0.4a gL-1) and mutant (5.5± 0.5a gL-1) strains that remain stable up to 30 h. There is overall increase of 109% (W) and 124% (M) in the production of extracellular lipases as compared to shake flask (Data not shown). Another significant finding of the present experiment is that the fermentation period is reduced to 30 h in case of wild and 23 h in case of mutant from 48 h in shake flask studies (17). Rapid decline in the enzyme production was seen in case of mutant strain when the incubation period was increased from optimum time period. It might be due to that fact that the important factors like pH, temperature, aeration and agitation were more precisely controlled by the fermenter which made the environment more suitable for the growth of organism. The carbon source (glucose) was exhausted after 15 h of cultivation, corresponding to the maximum growth rate value of the microorganism (19). Data obtained from above experiment was subjected to kinetic analysis for the calculations of µ(h-1)max (specific growth rate), qp(unit product produced/g cells/h), qs(g substrate consumed/g cells/h), Yp/s(unit product produced/g substrate consumed), Yp/x(unit product produced/g cells formed), Yx/s(g cells/g substrate utilized), Qp (g cells produced/L/h) and Qs(g substrate consumed/L/h). The kinetic evaluation of results also revealed that the optimum fermentation period for extracellular lipases production was 30 h in case of wild and 23 h in case of mutant strains of R. oligosporus (IIB-63NTG-7) (Table 1).
Figure 1.
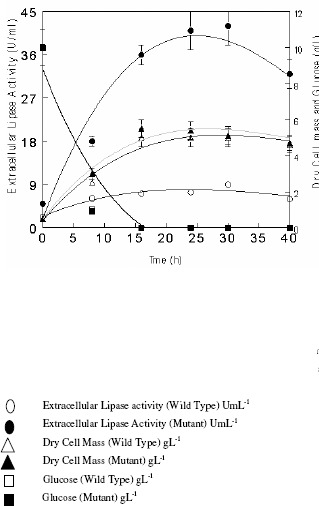
Rate of fermentation for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
Initial pH 8.0, incubation temperature 30oC, agitation speed 200rpm, aeration rate 1.0vvm, working volume 80%, fermentation medium M5, inoculum size 5%.
Table 1.
Kinetic evaluation of the rate of fermentation for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
| Kinetic Parameters | Wild | Mutant |
|---|---|---|
| µ (h-1) | 0.1 | 0.11 |
| Qx(g cells/L. h) | 0.40 | 0.44 |
| Qs(g /L. h) | 0.89 | 0.89 |
| Qp(U/L. h) | 540 | 3430 |
| qs(g/g. h) | 0.2 | 0.22 |
| qp(U/g. h) | 180 | 377.3 |
| Yp/x(U/g. cells) | 1800 | 8400 |
| Yx/s(g/g. S) | 0.5 | 0.51 |
| Yp/s(U/g. S) | 900 | 4200 |
Effect of different sizes of inoculum was investigated for extracellular lipases production by both wild (IIB-63) and mutant (IIB-63NTG-7) strains of R. oligosporus in stirred fermenter (Figure 2). The size of the vegetative inoculum was varied from 1–5% and fermentation was carried out. It was observed that extracellular lipases activity of the both wild (13.76 ± 0.99a U mL-1) and mutant (46.34 ± 3.05a U mL-1) strains was gradually increased with the increase of inoculum size and reached its maximum at 3% of inoculum (Figure 2). It is clear from the figure that a lower level of enzyme resulted in poor growth of organism which in turn caused a lesser production of enzyme while a higher concentration of inoculum caused a rapid increase in the rate of growth and consequently a lower enzyme production (32). However, Gutarra et al. (11) found that the more inoculum concentration is insignificant in case of Penicillium simplicissimum lipase production by solid-state fermentation. All kinetic parameters Qp1706 U/L.h (W) and 13646 U/L.h (M), Yp/s1300 U/g.S (W) and 4600 U/g.S (M), Yp/x 2600 U/g.h (W) and 9200 U/g.h for product formation showed 3% vegetative inoculum to be adequate for optimal production of extracellular lipase (Table 2). Thus the inoculum size of 3% was used in the further studies for the production of extracellular lipases by microorganism in the stirred fermenter.
Figure 2.
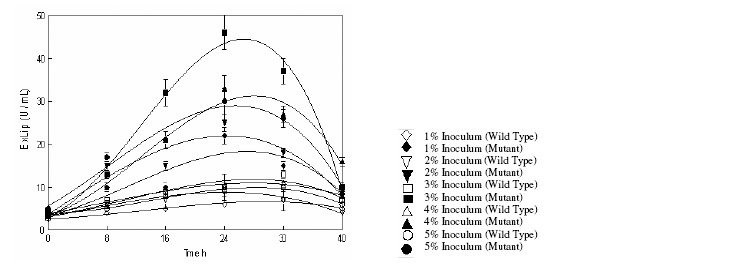
Effect of the size of inoculum on the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter*
* Initial pH 8.0, incubation temperature 30oC, agitation speed 200rpm, aeration rate 1.0vvm, working volume 80%, fermentation medium M5.
Table 2.
Kinetic evaluation of different inoculum sizes for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
| Kinetic Parameters/ Size of Inoculum | 1% | 2% | 3% | 4% | 5% | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | |
| Qp(U/L.h) | 902 | 3492 | 539 | 7940 | 1706 | 13646 | 1115 | 7670 | 2087 | 4123 |
| Yp/s(U/g.S) | 700 | 2200 | 1000 | 2500 | 1300 | 4600 | 1100 | 3300 | 1000 | 3000 |
| Yp/x(U/g. cells) | 1400 | 4400 | 2000 | 5000 | 2600 | 9200 | 2200 | 6600 | 2000 | 6000 |
Effect of the control of the pH of the fermentation medium by both wild and mutant strain of R. oligosporus was investigated in stirred fermenter. The initial pH of the fermentation medium was varied from 7.0 to 9.0 and was controlled throughout the fermentation process despite the experiment with uncontrolled pH was also carried out. The pH was controlled by the automatic addition of 0.1 N HCl/NaOH with the help of peristaltic pump attached with the fermenter. The production of extracellular lipases was found maximum 30.39 ± 2.58a U mL-1 (W) and 50.42 ± 4.37a U mL-1 (M) when the pH of the medium was maintained at 8.0 (Figure 3A). This finding is of utmost importance that reveals that the production of enzyme is optimal in the slightly alkaline conditions which made it useful in detergent industry (29). However, in the experiment with uncontrolled pH there is no remarkable increase in the production of enzyme and growth of fungus does not alter the culture conditions which show its maintained uniform growth conditions in terms of pH (Figure 3B).
Figure 3.
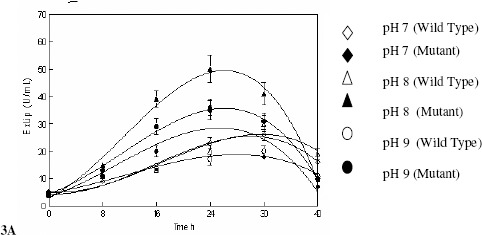
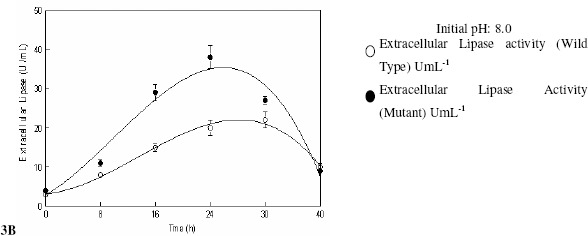
Effect of the controlled and uncontrolled pH of the medium on the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
* Incubation temperature 30oC, agitation 200 rpm, aeration rate 1.0 vvm, working volume 80%, fermentation medium M5, inoculum size 3%
Furthermore, the metabolite profile of the fungus seems to be non-polar as in case of any electrophilic or nucleophilic extrusion from the fungus would definitely interfere with pH. It was also verified that lipase from Rhizopus sp. showed a better stability in the range of pH 5.0 to 8.5 (10). Comparison of kinetic relations, i.e., Qp(g cells produced/L/h), Yp/s(unit product produced/g substrate consumed) and Yp/x(unit product produced/g cells formed) was also made (Table 3) and it was found that in both pH controlled and variant cases there is significant difference in product formation. It shows that pH of the medium is an important parameter for enzyme production. So the pH of the medium was maintained at 8.0 for the production of extracellular lipases by R. oligosporus (IIB-63NTG-7) throughout the subsequent studies.
Table 3.
Kinetic evaluation of controlled and uncontrolled pH values of media for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
| pH of Medium/KineticParameters | 7.0 | 8.0 | 9.0 | Uncontrolled pH | ||||
|---|---|---|---|---|---|---|---|---|
| Wild | Mutant | Wild | Mutant | Mutant | Wild | Wild | Mutant | |
| Qp(U/L.h) | 7753 | 5992 | 7325 | 8134 | 12948 | 2920 | 6194 | 7654 |
| Yp/s(U/g. S) | 2600 | 3600 | 3000 | 3500 | 5000 | 2000 | 2200 | 3800 |
| Yp/x(U/g. cells) | 5200 | 7200 | 6000 | 7000 | 10,000 | 4000 | 4400 | 7600 |
The effect of the rate of agitation for the production of extracellular lipases by wild and mutant strain of R. oligosporus was investigated in the stirred fermenter. The rate of the agitation was varied from 150–300 rpm (Figure 4). Maximum enzyme production by both wild (27.30 ± 1.98 U mL-1) and mutant (54.01 ± 4.54 U mL-1) strains was obtained when the agitation speed was maintained at 250 rpm. Change in the rate of agitation resulted in decreased enzyme production. Higher stirring speeds than 250 rpm resulted in mechanical and/or oxidative stress, excessive foaming, disruption and physiological disturbance of the cells, while lower stirring speeds seemed to limit oxygen levels along with the lacking of homogeneous suspension of the fermentation medium and breaking of the clumps of the cells Elibol and Ozer (2000) also reported that main reason for the negative effect of agitation speed may be due to the perturbation of protein structure during the biosynthesis of lipase. Alonso et al. (1) have also reported 200 rpm, stirring speeds as optimum while Freire et al., (8) have reported maximum production of lipase at stirring speed of 300rpm with Pencillium sp.
Figure 4.
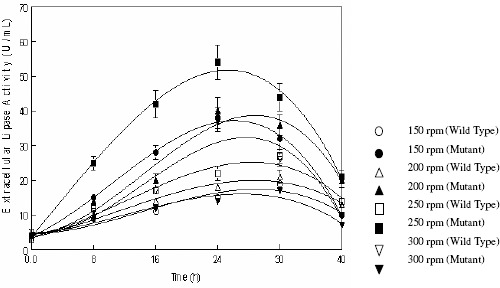
Effect of agitation rate on the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter*
* Medium pH 8.0, incubation temperature 30oC, aeration rate 1.0vvm, working volume 80%, fermentation medium M5, inoculum size 3%
Evaluation of the kinetic parameters i.e., Qp, Yp/s,andYp/xrevealed that production yield by both wild and mutant strains was found optimum when the agitation speed of the impeller was set at 250 rpm (Table 4). Thus the agitation speed of 250 rpm was selected for further studies. Figure 5 and Table 5 show the effect of rate of aeration on the production of extracellular lipases by both wild (IIB-63) and mutant (IIB-63NTG-7) strain of Rhizopus oligosporus. The rate of aeration was varied from 0.2–1.0 vvm. Production of enzyme by mutant strain was found maximum i.e., 59.0 ± 4.88a U mL-1 when the aeration rate was set at 0.8 vvm while wild strain gave maximum lipase units (28.0 ± 1.97a U mL-1) at 1.0 vvm. The lipase production was increased with the increase of aeration and reached its maximum at 1.0 vvm (wild) and 0.8 vvm (mutant) but further increase in air supply resulted in decreased product yield. Lower rates of aeration seems to be insufficient to fulfill the organisms demand for oxygen while higher concentration rates also have some detrimental effects on the growth of the microorganism and subsequently enzyme production during the bioprocess time (19). Foam formation also affects the enzyme production by making the volume of the vessel uncontrollable (28). Alonso et al., (1) also reported 0.8 vvm aeration for the best lipase production by Yarrowia lipolytica.
Table 4.
Kinetic evaluation of different agitation speeds for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter
| Agitation Speed/Kinetic Parameters | 150 rpm | 200 rpm | 250 rpm | 300 rpm | ||||
|---|---|---|---|---|---|---|---|---|
| Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | |
| Qp(U/L.h) | 4337 | 8238 | 2769 | 11936 | 4841 | 17222 | 3424 | 12376 |
| Yp/s(U/g.S) | 1900 | 3800 | 2100 | 4000 | 2700 | 5400 | 1700 | 3700 |
| Yp/x(U/g. cells) | 3800 | 7600 | 4200 | 8000 | 5400 | 10800 | 3400 | 7400 |
Kinetic parameters
Qp g cells produced/L/h
Yp/s Unit product produced/g substrate consumed:
Yp/x Unit product produced/g cells formed
Figure 5.
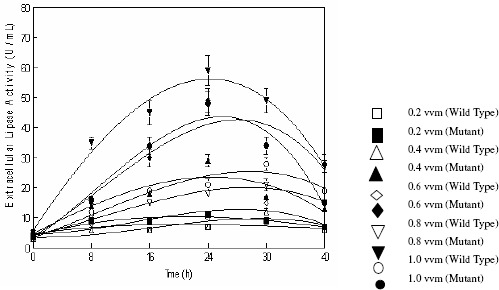
Effect of the aeration rate on the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter*
* Medium pH 8.0, incubation temperature 30oC, agitation speed 250rpm, working volume 80%, fermentation medium M5, inoculum size 3%
Table 5.
Kinetic evaluation of the aeration rate for the production of extracellular lipases by Rhizopus oligosporus IIB-63 and its mutant derivative in stirred fermenter*
| Aeration rate (vvm)/Kinetic Parameters | 0.2 | 0.4 | 0.6 | 0.8 | 1.0 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | Wild | Mutant | |
| Qp(U/L.h) | 4575 | 7915 | 2170 | 10501 | 2908 | 5567 | 2091 | 34328 | 2281 | 7293 |
| Yp/s(U/g.S) | 900 | 1100 | 1200 | 2900 | 1500 | 4900 | 2100 | 5900 | 2800 | 4800 |
| Yp/x(U/g. cells) | 1800 | 2200 | 2400 | 5800 | 3000 | 9800 | 4200 | 11,800 | 5600 | 9600 |
Kinetic parameters
Qp g cells produced/L/h
Yp/s Unit product produced/g substrate consumed
Yp/x Unit product produced/g cells formed
The kinetic analysis of the above parameters revealed that the values of Qp, Yp/s and Yp/x were significant at an air supply of 1.0 vvm (W) and 0.8 vvm (M). Therefore 0.8 vvm was optimized for further studies for enhanced production of extracellular lipase.
CONCLUSION
Scale up studies showed an overall increase of 109% (W) and 124% (M) in the production of extracellular lipases as compared to shake flask. Another significant finding of the present experiment is that the fermentation period is reduced to 30 h in case of wild and 23 h in case of mutant from 48 h in shake flask studies which was ultimately helpful to economize the process. There is further increase in the lipolytic potential of the fungus after optimization of inoculum, controlled and uncontrolled pH, rate of aeration and rate of agitation.
ACKNOWLEDEGMENT
Special thanks to Prof. Dr. Kye Joon, Seoul National University, South Korea and Dr. M. Ibrahim Rajoka, National Institute of Biotechnology and Genetic Engineering, Faisalabad, Pakistan.
REFERENCES
- 1.Alonso F.O.M., Oliveira E.B.L., Ortiz G.M.D., Meirelles F.V.P. Improvement of lipase production at different stirring speeds and oxygen levels. Brazilian J. Chem. Eng. 2005;22(01):9–18. [Google Scholar]
- 2.Arroyo M., Sanchez-Montero J.M., Sinisterra J.V. Thermal stabilization of immobilized lipase B from Candida antarctica on different supports: effect of water activity on enzymatic activity in organic media. Enzyme Microb. Technol. 1999;24:3–12. [Google Scholar]
- 3.Belarbi E.H., Molina E., Chisti Y. A process for high yield and scaleable recovery of high purity eicosapentaenoic acid esters from microalgae and fish oil. Enzyme Microb. Technol. 2000;26:516–529. doi: 10.1016/s0141-0229(99)00191-x. [DOI] [PubMed] [Google Scholar]
- 4.Cheetham P.S.J. The application of enzymes in industry. In: Wiseman A., editor. Hand Book of Enzyme Biotechnology. UK: Ellis Horwood; 1995. pp. 419–522. [Google Scholar]
- 5.Dai D., Xia L. Enhanced production of Penicillium expansum PED-03 lipase through control of culture conditions and application of the crude enzyme in kinetic resolution of racemic Allethrolone. Biotechnol Prog. 2005;21(4):1165–1168. doi: 10.1021/bp0500563. [DOI] [PubMed] [Google Scholar]
- 6.Damaso M.C.T., Passianoto M.A., Freitas S.C., Lago R.C.A., Couri S. Utilization of agroindustrial residues for lipase production by solid-state fermentation. Utilization of agroindustrial residues for lipase production by solid-state fermentation. Braz. J. Microbiol. 2008;39(4) doi: 10.1590/S1517-838220080004000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elibol M., Ozer D. Lipase production by immobilized Rhizopus arrhizus. Process Biochem. 2000;36:219–223. [Google Scholar]
- 8.Freire D.M.G., Teles E.M.F., Bon E.P.S., Anna G.L.S. Lipase production by Penicillium restrictum in a Bench-scale fermenter: Effect of carbon and nitrogen nutrition, agitation and aeration. Appl. Biochem. Biotech. 1997;63:409–421. doi: 10.1007/978-1-4612-2312-2_36. [DOI] [PubMed] [Google Scholar]
- 9.Gopinath S.C.B., Hilda A., Priya T.L., Annadurai G., Anbu P. Purification of lipase from Geotrichum candidum: conditions optimized for enzyme production using Box-Behnken design. World J. Microbial. Biotechnol. 2003;19:681–689. [Google Scholar]
- 10.Gupta N., Sahai V., Gupta R. Alkaline lipase from a novel strain Burkholderia multivorans: Statistical medium optimization and production in a bioreactor. Process Biochem. 2007;42:518–526. [Google Scholar]
- 11.Gutarra M.L.E., Godoy M.G., Castilho L.R., Freire D.M.G. Inoculum strategies for Penicillium simplicissimum lipase production by solid-state fermentation using a residue from the babassu oil industry. J. Chem. Technol. Biotechnol. 2007;82:313–318. [Google Scholar]
- 12.Herrgard S., Gibas C.J., Subramaniam S. Role of electrostatic network of residues in enzymatic action of Rhizomucor mehei lipase family. Biochemistry. 2000;39:2921–2930. doi: 10.1021/bi9916980. [DOI] [PubMed] [Google Scholar]
- 13.Ibrik A., Chanian H., Rugani N., Sarda L., Comeau L.C. Biochemical and structural characterization of triacylglycerol lipase from Penicillium cyclopium. Lipids. 1998;33:377–384. doi: 10.1007/s11745-998-0218-6. [DOI] [PubMed] [Google Scholar]
- 14.Iftikhar T., Hussain A. Effect of nutrients on the extracellular lipase production by the mutant strain of R. oligosporous Tuv-31. Biotech. 2002;1(1):15–20. [Google Scholar]
- 15.Iftikhar T., Niaz M., Abbas S. Q., Zia M.A., Ashraf I., Lee K.J., Haq I.U. Mutation induced enhanced biosynthesis of lipases by rhizopus oligosporus var. microsporus. Pak. J. Bot. 2010;42(2):1235–1249. [Google Scholar]
- 16.Iftikhar T., Niaz M., Zia M.A., Qadeer M.A., Abbas S.Q., Rajoka M.I., Haq I.U. Biotechnologies International Symposium “Which biotechnologies for south countries” Proceedings of Biotech World 2007. Oran Algerie: 2007. Nov, Screening of media and Kinetic studies for the production of lipase from a mutant strain of Rhizopus oligosporus”; pp. 24–25. [Google Scholar]
- 17.Iftikhar T., Haq I.U., Javed M.M. optimization of cultural conditions for the production of lipase by submerged culture of Rhizopus oligosporous tuv -31. Pak. J. Bot. 2003;35(4):519–525. [Google Scholar]
- 18.Iftikhar T., Niaz M., Afzal M., Haq I.U., Rajoka M.I. Maximization of intracellular Lipase Production in a Lipase-Overproducing Mutant Derivative of Rhizopus oligosporus DGM 31: A Kinetic Study. Food Technol. Biotechnol. 2008;46(4):402–412. [Google Scholar]
- 19.Ionita A., Moscovici M., Dragolici A., Eremia M., Buca C., Albulescu R., Pavel I., Vamanu C. Lipase Production in Discontinuous Operation System Using a Candida lipolytica Strain. Roum. Biotechnol. Lett. 2001;7(1):547–552. [Google Scholar]
- 20.Kempka A.P., Lipke N.L., Pinheiro T.L.F., Menoncin S., Treichel H., Freire D.M.G., Luccio M.D., Oliveira D. Response surface method to optimize the production and characterization of lipase from Penicillium verrucosum in solid-state fermentation. Biop. Biosys. Eng. 2008;31:119–125. doi: 10.1007/s00449-007-0154-8. [DOI] [PubMed] [Google Scholar]
- 21.Krieger N., Taipa M.A., Melo E.H.M., Lima J.L., Baros M.R.A., Cabral J.M.S. Purification of Penicilum citrinum lipase by chromatographic processes. Bioprocess Eng. 1999;20:59–65. [Google Scholar]
- 22.Lacointe C., Dubreucq E., Galzy P. Ester synthesis in aqueous media in the presence of various lipases. Biotechnol. Lett. 1996;18:869–874. [Google Scholar]
- 23.Lawford H.G., Roseau J.D. Mannose fermentation by ethanologenic recombinants of Escherichia coli and kinetical aspects. Biotechnol. Lett. 1993;15:615–620. [Google Scholar]
- 24.Lesuisse E., Schanck K., Colson C. Purification and preliminary characterization of the extracellular lipase of Bacillus subtilis 168, an extremely basic pH-tolerant enzyme. Eur. J. Biochem. 1993;216:155–160. doi: 10.1111/j.1432-1033.1993.tb18127.x. [DOI] [PubMed] [Google Scholar]
- 25.Lutz S. Engineering lipase B from Candida antarctica. Tetrahedron Asymmetry. 2004;15(18):2743–2748. [Google Scholar]
- 26.Nair C.S., Bone D.H. Production of lipase of Aspergillus foetidus in a batch stirred reactor. Biotechnol. Lett. 1987;9(8):601–604. [Google Scholar]
- 27.Pirt S.J. Blackwell Scientific. Oxford. 1975. Principles of cell and microbe and cell cultivation; pp. 4–22. [Google Scholar]
- 28.Singh S., Kaur G., Chakraborti A.K., Jain R.K., Banerjee U.C. Study of the experimental conditions for the lipase production by a newly isolated strain of Pseudomonas aeruginosa for the enantioselective hydrolysis of (±)-methyl trans-3(4-methoxyphenyl) glycidate. Bioprocess Biosyst. Eng. 2006;28:341–348. doi: 10.1007/s00449-005-0039-7. [DOI] [PubMed] [Google Scholar]
- 29.Snedecor G.W., Cochran W.G. Statistical Methods. Ames, Iowa State University Press. (7th) 1980 ISBN 0–81381560–6. [Google Scholar]
- 30.Sugihara A., Shimada Y., Nakamura M., Nagao T., Tominaga Y. Positional and fatty acid specificities of Geotrichum candidum lipase. Protein Eng. 1994;7:585–588. doi: 10.1093/protein/7.4.585. [DOI] [PubMed] [Google Scholar]
- 31.Toshihiko K., Mari T., Ishii T., Iloth Y., Kirimura K., Usami S. Production of lipase by Rhizopus oligosporous a newly isolated fungus. J. Hokoku Waseda. 1989;50(125):61–66. [Google Scholar]
- 32.Ushio K., Hirata T., Yoshida K., Sakaue M., Hirose C., Suzuki T., Ishizuk Super inducer for induction of thermostable lipase production by Pseudomonas species NT-163 and other Pseudomonas like bacteria. Biotechnol. Techniques. 1996;10(4):267–272. [Google Scholar]


