Abstract
Little is known about the etiology of progressive macular hypomelanosis, although it has been suggested that Propionibacterium acnes plays an important role. While microbiological culture is commonly employed to identify Propionibacterium acnes, new identification methods have been under investigation, amongst them polymerase chain reaction. To determine the cut-off point for the number of genome copies of Propionibacterium acnes in the lesional skin of patients with progressive macular hypomelanosis as a positive marker, employing quantitative real-time polymerase chain reaction and anaerobic culture, considered gold standard. An observational study with a comparison group, included 35 patients with dermatosis, attended at the Oswaldo Cruz University Hospital, Pernambuco, Brazil, between March and May 2008. Lesional skin was compared to non-lesional skin through positive testing with real-time polymerase chain reaction and culture. The Statistical Package for Social Sciences, version 12.0, was employed for the association analysis with the McNemar test, and the cut-off point with the ROC curve for maximum values. Propionibacterium acnes was most frequently encountered in lesional areas (p<0,025). The cut-off point of Propionibacterium acnes in lesional skin was 1,333 genome copies, with a sensitivity of 87,9% and a specificity of 100,0%. Since Propionibacterium acnes is a saprophyte, identifying the cut-off point may assist in determining its positivity in lesional skin in patients suffering with this dermatosis.
Keywords: Hypopigmentation/progressive macular hypomelanosis, Propionibacterium acnes, Polymerase chain reaction, Biopsy/bacteriology, ROC curve
INTRODUCTION
Progressive macular hypomelanosis (PMH) is a dermatosis that affects young adults and is characterized by asymptomatic, nummular hypochromic macules, with well-defined borders, localized mainly on the trunk, abdomen (19) and upper parts of the limbs (18), tending to spread towards the middle of the back (10).
It is frequently confused with pityriasis versicolor, Hansen’s disease, hypopigmented mycosis fungoides (10), pityriasis alba, vitiligo and residual hypochromic stains (17). The histopathological examination is unspecific, and only demonstrates a reduction of epidermal melanin, with no alteration to the dermis (26).
Although its etiology is unknown, in 2004, Westerhof et al (26), using Wood’s lamp observed red follicular fluorescence in the lesional skin of eight patients, and after lesional skin cultures, confirmed the presence of Propionibacterium acnes (P. acnes) in seven of these eight patients. Since these same findings were not confirmed in non-lesional skin, the authors formulated the hypothesis that the bacteria could be associated to the development of PMH.
P. acnes is a gram-positive, non-motile, pleomorphic, saprophytic facultatively anaerobic bacilli (16), which under the microscope exhibits a coryneform appearance (8, 10). Under Wood’s lamp examination, it presents a red follicular fluorescence (15, 22), proportional to its concentration (22), due to the production of porphyrins, especially coproporphyrin III (22, 25).
Apart from PMH, the involvement of P. acnes, an opportunistic pathogen of low virulence (5), has also been cited in the development of several diseases (7). It has been investigated in acne (25), in the granuloma of patients with primary biliary cirrhosis (8), infections of the cerebral-spinal fluid (4), endophthalmitis (3, 9), prosthetic joint infections (28) and in bacteremia (11).
Microbiological cultures have generally been employed to identify P. acnes. However, due to the difficulties involved in cultivating this bacteria with regular aerobic techniques and because of the need for a long incubation period (16), new methods of identification have been investigated, amongst them polymerase chain reaction (PCR).
If compared to conventional PCR, real-time PCR demonstrates a greater sensitivity for identifying target DNA, since detection is carried out through the capture of fluorescence amplification. This technique also presents greater specificity due to the use of a probe specific to the target fragment of the reaction (13, 24) as well as greater reproducibility, thus permitting the quantification of the genome copy numbers (23).
PMH is an unsightly dermatosis, of an ill-defined etiopathogenesis, which causes a profound psychosocial impact, lowering the self-esteem of the patient, especially in severe cases and in individuals with darker skin. Since P. acnes is a saprophyte of the cutaneous microbiota, it is important to use methods that permit it to be quantified and compared to non-lesional skin.
The aim of the study was to conduct a quantitative investigation into the presence of P. acnes in lesional and non-lesional skin of patients suffering with PMH, and to determine the cut-off point for the genome copy numbers of P. acnes as a positive marker in lesional skin, through quantitative real-time PCR, considering culture as the gold standard test.
MATERIALS AND METHODS
An observational study with a comparison group was undertaken, involving 38 patients, of 18 years or over, and diagnosed with PMH, regardless of sex. The patients sought medical attention at the dermatology outpatients clinic at the Oswaldo Cruz University Hospital (OCUH), Recife, Pernambuco, Brazil, either by spontaneous need or by referral from other services, in the period from March to May, 2008.
Exclusion criteria were: presence of acne on the back, lactation, presence of dermatosis on the trunk or taking medication for P. acnes, during the three months prior to the consultation with the researcher.
Due to the lack of epidemiologic data concerning the frequency of PMH in Brazil, on identifying that a sample of 35 patients provided sufficient laboratory evidence, because of ethical questions and owing to the use of an invasive procedure to collect biological material for laboratory examinations, it was decided to finish data collection.
PMH was characterized by the presence of asymptomatic hypochromic macules on the back, with a confluence on the midline (Figures 1A and B), ruling out the differential diagnoses of pityriasis versicolor, atopic dermatitis, seborrheic dermatitis, hypopigmented mycosis fungoides, vitiligo, residual hypochromic spots and indeterminate Hansen’s disease.
Figures 1.
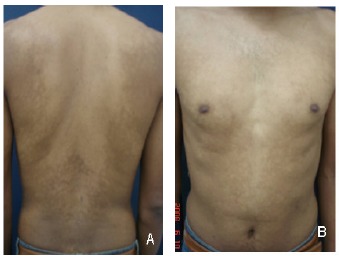
A and B- Multiple coalescent hypochromic macules on the trunk of a patient with PMH
All patients were submitted to a dermatological naked eye examination by three dermatologists and a Wood’s lamp examination, in a dark room. The follicular coral-red fluorescence suggested the presence of P. acnes.
All patients with a clinical diagnosis of PMH underwent a mycological examination of the lesional areas with tape lift-off method, to exclude pityriasis versicolor (12). These patients were submitted to a biopsy of lesional and non-lesional skin in order to carry out histopathological, molecular biological and microbiological studies.
Fragments of lesional and non-lesional skin were removed, with a minimum distance of 1 cm from the lesional skin, identified with naked eye examination and Wood’s lamp, from the same anatomic site of the trunk, using a 4 mm skin punch.
A histopathological examination was conducted on one part of the biopsy fragment of the lesional skin in order to exclude the differential diagnosis of PMH. Routine staining was employed (hematoxylin-eosin) and a specific stain for melanin (Fontana Masson). The other part, as well as the fragment of non-lesional skin, were taken in refrigerated containers for microbiological and molecular biological studies. The professionals conducting the studies were unaware of the origins of the skin fragments.
A homogenized portion of the each skin fragment was seeded using Casman medium base (Becton Dickinson®, MD, USA), enriched with 5% defibrinated rabbit blood. Isolation was conducted by seeding the collected material in Muller-Hinton medium and incubated under anaerobic conditions. An anaerobic atmosphere was generated in BBL GasPak Plus jars with palladium catalyst (Becton Dickinson, MD, USA). Plates were incubated at 37 ° C for 4 days. The suspected colonies of P. acnes were re-isolated and submitted to Gram staining and biochemical examinations to identify the species of bacteria (production of indole, nitrate, catalase and esculin degradation) (12). The results of the microbiological culture were regarded as gold standard to identify the bacteria.
The DNA of P. acnes was extracted by following the instructions for tissues in the QIAamp® Mini Kit (Qiagen®; Chattlesworth, CA, EUA). Amplification reactions of the genomic material were performed with a iCycler® iQ5 (BioRad®) thermocycler, and the TaqMan® (Applied Biosystems®, Foster City, CA) system was used to detect the amplification.
Specific primers were used for the detection and amplification of 131 base pairs of the 16S rRNA gene of P. acnes and a TaqMan ® probe labeled with fluorophore.
The primer sequences were: PA-F: 5´- GCGTGAGTGAC GGTAATGGGTA-3´ e PA-R: 5´-TTCCGACGCGATCAACC A-3´. The TaqMan® probe was FAM-5´- AGCGTTGTCCGGA TTTATTGGGCG 3´-TAMRA (6).
A cycle protocol was employed for the following conditions: step at 50°C for 2 min; step at 95°C for 10 min; 40 cycles at 95°C for 15 sec and 60°C for 1 min.
As a positive control, DNA was used after being extracted from a culture of P. acnes, and was previously isolated and identified. A negative control was carried out using water. The action of the eventual inhibitors of the DNA polymerase was verified by being tested alongside the success of the amplification of the human beta-globin gene.
The variables studied were the results of the culture and the quantitative real-time PCR to identify the bacteria in lesional (study group) and non-lesional (comparison group) skin.
The programs employed were: Microsoft Office Excel, version 2003, to administrate the database, Statistical Package for Social Science for Windows (SPSS), version 12.0, to calculate statistics, and Microsoft Office Word, version 2003, to produce the tables.
The McNemar test was applied to evaluate the results of the PCR and the culture. The receiver operating characteristic curve (ROC curve) was used to determine the cut-off point of the number of P. acnes genomes with maximum sensitivity and specificity.
The present study was analyzed and approved by the Ethics Commission at the OCUH, n°. 147/2007.
This research did not involve any conflict of interests.
RESULTS
Of the 38 patients who initiated the study, three were excluded. In one of these excluded patients, the non-lesional skin biopsy included part of the lesional skin, which may have interfered with the quantitative real-time PCR results. Another patient refused to undergo the skin biopsy and for the third patient it was not possible to complete the culture. The sample was therefore composed of 35 patients.
To guarantee reproducibility, the PCR was carried out in triplicate with similar results and the mean numbers of genome copies were calculated.
The DNA quantification for the positive control used in the assays presented a value of Ct=27, background 2,5x104 fg, i.e. an equivalent of 9.260 bacterial genomes per reaction.
The purified DNA extracted from the culture of the strain control was quantified with a spectrophotometer and used in dilutions to determine the standard curve.
Using the ROC curve, using the culture as gold standard, it was possible to determine that the best cut-off point for the quantitative real-time PCR was 1,333 genome copies of P. acnes, which corresponded to a sensitivity of 87.9%, a specificity of 100.0%, a positive predictive value of 100.0%, and a negative predictive value of 33.3%. The concordance between this test and the culture was 88.6%.
P. acnes was identified in all samples of lesional and non-lesional skin when the PCR was qualitatively analyzed, i.e. regardless of the cut-off point. By adopting the proposed cutoff point, in the lesional skin, the two diagnostic methods present statistically significant (p=0.025) concordant positive results (Table 1).
Table 1.
Results of the culture and the quantitative real-time PCR to identify P. acnes in the lesion and non-lesional skin biopsies of 35 patients with PMH, adopting the proposed cut-off point – OCUH – March – May/2008
| Biopsied skin | Culture | Result of PCR adopting cut-off point | p-Value | |
|---|---|---|---|---|
| Positive (n;%) | Negative (n;%) | |||
| Lesional | Positive | 29 (100.0) | 4 (66.7) | 0.025 |
| Negative | - | 2 (33.3) | ||
| Non-lesional | Positive | - | 4 (12.1) | 0.782 |
| Negative | 2 (100.0) | 29 (87.9) | ||
NOTE: McNemar test
DISCUSSION
In this study, there was a significant predominance of P. acnes in the lesional skin of patients with PMH, demonstrated through culture and quantitative real-time PCR. Similar results were described by Westerhof et al.(26), who demonstrated the presence of the bacteria through cultures from the lesional skin in seven out of eight patients with PMH, and in no patients from the non-lesional skin. Since this is a saprophytic bacteria of the pilosebaceous unit, the study not only identified it through culture, but also determined a cut-off point for the number of P. acnes genome copies as a positive marker on lesional skin.
This sample size of 35 patients was also accepted because, having established a cut-off point for the number of genome copies greater than or equal to 1,333 and accepting a error of 11%, within this total of patients, sensitivity was 87.9% and specificity was 100%.
If the cut-off point is disregarded, P. acnes was identified in all skin samples by real-time PCR, and it was not possible to differentiate between lesional skin and non-lesional skin. However, the culture itself managed to identify, without quantifying, a significant difference and was positive in 33/35 (94.3%) in lesional skin and in only 4/35 (11.4%) in non-lesional skin.
When PCR was analyzed quantitatively, considering the cut-off point of 1,333 P. acnes genome copies, there was significant concordance in relation to the positive values, between the PCR results and those of the culture. These results demonstrate an abnormal accumulation of the bacteria in the PMH lesions and indicate that, since this is a saprophyte bacteria is, it is important to use diagnostic methods that permit a comparison with non-lesional tissue. An established cut-off point for the number of P. acnes genome copies helps to provide a better evaluation of a positive real-time PCR result, when identifying this bacteria in lesional skin.
Further to the cut-off point, the fact that 4 (11.4%) patients were identified with positive cultures and negative real-time PCR, in lesional skin, and another 4 (11.4%) patients, in non-lesional skin, would seem to be due to the primers employed. The primers, adopted at a time when it had been generally accepted that there were of two strains of P. acnes, had been previously analyzed by other researchers (6). In 2008, P. acnes type III was discovered, amplifying the recA gene, by using the PAR-1 and PAR-2 primers (14). It is possible that the primers used in the present study did not identify all types of P. acnes and if a higher number of sequences of the 16S rRNA gene had been analyzed, it is possible that a greater number of copies of P. acnes would have been observed in these samples.
It may be observed that unexpectedly, the results of 2 (5.7%) non-lesional skin biopsies presented negative cultures and PCRs with higher cut-off values. The number of genome copies identified by PCR are independent of microorganism viability (1), unlike the culture, which requires viable P. acnes.
It is probable that the P. acnes was not viable in those particular skin samples, which therefore impeded growth in the culture medium and consequently, the appearance of hypomelanosis.
While PMH does not actually cause any functional incapacity, it causes a profound psycho-social impact, especially in extreme cases and in patients with darker skins, in whom the lesions are much more evident. It is of the utmost importance therefore, to clarify the etiology and thus, encounter the best treatment for this dermatosis.
Although it is not possible to draw any definitive conclusions regarding the participation of P. acnes in the etiology of PMH, it is evident that the bacteria accumulates in lesional skin of patients with this dermatosis. The hypothesis that P. acnes may be related to the development of PMH has led several authors to employ medications active against this microorganism. Repigmentation of the lesions has been observed after using doxycycline (19), benzoyl peroxide associated to topical clindamycin (1) , minocycline (2) and lymecycline associated to benzoyl peroxide, by Cavalcanti et al., in 2009 (data currently unpublished).
There are no studies that indicate whether the strains of P. acnes encountered in the lesional skin of patients with PMH are genotypically or phenotypically different from those presented in non-lesional skin. It is of great importance that further studies be carried out in order to determine if specific subtypes of P. acnes are involved in PMH.
CONCLUSION
The results of this study suggest that quantitative real-time PCR is a quick, sensitive, specific technique for detecting P. acnes. However, even while this technique permits an established cut-off point for the number of genome copies as a positive marker for P. acnes in the lesional skin of patients with PMH, its positive results are independent of the viability of the microorganism, and may differ according to the primers used. It is the belief of this study that culture still remains the best method to identify P. acnes in PMH lesions.
ACKNOWLEDGEMENTS
We are grateful to Dr. Eliane Ruth Alencar for carrying out the histopathological studies on the skin fragments of the patients taking part in the research.
And also to all our dermatological colleagues who have contributed to this work.
REFERENCES
- 1.Alexeyev O., Olsson J., Elgh F. Is there evidence for a role of Propionibacterium acnes in prostatic disease? Urology. 2009;3(2):220–224. doi: 10.1016/j.urology.2008.02.052. [DOI] [PubMed] [Google Scholar]
- 2.Almeida A.R.T., Bedani T.P., Debs E.A.F., Ferreira J.A.D. Estudo piloto para avaliar a eficácia da minociclina no tratamento da hipomelanose macular progressiva (HMP) Surg Cosmetic Dermatol. 2009;1(1):25–35. [Google Scholar]
- 3.Bagvalakshunt R., Madhavan H.N., Threse K.L. Development and application of multiplex polymerase chain reaction for the etiological diagnosis of infectious endophthalmitis. J Postgrad Med. 2006;52(3):179–182. [PubMed] [Google Scholar]
- 4.Banks J.T., Bharara S., Tubbs R.S., Wolff C.L., Gillespie G.Y., Markert J.M., Blount J.P. Polymerase chain reaction for the rapid detection of cerebrospinal fluid shunt or ventriculostomy infections. Neurosurgery. 2005;57(6):1237–1243. doi: 10.1227/01.neu.0000186038.98817.72. [DOI] [PubMed] [Google Scholar]
- 5.Cogen A.L., Nizet V., Gallo R.L. Skin microbiota: a source of disease or defence? Brit J Dermatol. 2008;158:442–455. doi: 10.1111/j.1365-2133.2008.08437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eishi Y., Suga M., Ishige I., Kobayashi D., Yamada T., Takemura T., Takizawa T., Koike M., Kudoh S., Costabel U., Guzman J., Rizzato G., Gambacorta M., du Bois R., Nicholson A.G., Sharma O.P., Ando M. Quantitative analysis of mycobacterium and propionibacterial DNA in lymph nodes of Japanese and European patients with sarcoidosis. J Clin Microbi. 2002;40(1):198–204. doi: 10.1128/JCM.40.1.198-204.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa A., Uchida K., Ishige Y., Ishige I., Kobayayshi I., Takemura T., Yokoyama T., Iwai K., Watanabe K., Shimizu S., Ishida N., Suzuki Y., Suzuki T., Yamada T., Ito T., Eishi Y. Characterization of Propionibacterium acnes isolates from sarcoid and non-sarcoid tissues with special reference to cell invasiveness, serotype, and trigger factor gene polymorphism. Microb Pathog. 2009;46:80–87. doi: 10.1016/j.micpath.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Harada K., Tsuneyama K., Sudo Y., Masuda S., Nakanuma Y. Molecular identification of bacterial 16S ribossomal RNA gene in liver tissue of primary biliary cirrhosis: Is Propionibacterium acnes involved in granuloma formation? Hepatology. 2001;33(3):530–536. doi: 10.1053/jhep.2001.22653. [DOI] [PubMed] [Google Scholar]
- 9.Hollander D.A., Dodds E.M., Wood I.S., Alvarado J.A. Propionibacterium acnes Endophthalmitis with bacterial sequestration in a Molteno’s implant after cataract extraction. Am J Ophthalmol. 2004;138(5):878–879. doi: 10.1016/j.ajo.2004.05.065. [DOI] [PubMed] [Google Scholar]
- 10.Kumarasinghe S.P.W., Tan S.H., Thng S., Thamboo T.P., Liang S., Lee Y.S. Progressive macular hypomelanosis in Singapore: a clinic-pathological study. Int J Dermatol. 2006;45:737–742. doi: 10.1111/j.1365-4632.2006.02755.x. [DOI] [PubMed] [Google Scholar]
- 11.Lau S.K., Woo P.C., Fung A.M., Chan K.M., Woo G.K., Yuen K.Y. Anaerobic, non–sporulating, Gram-positive bacilli bacteraemia characterized by 16S RNA gene sequencing. J Med Microbiol. 2004;53(12):1247–1253. doi: 10.1099/jmm.0.45803-0. [DOI] [PubMed] [Google Scholar]
- 12.Macfaddin J.F. Biochemical tests for identification of medical bacteria. Philadelphia: Lippincott Williams & Wilkins; 2000. [Google Scholar]
- 13.Mackay I.M. Real-time PCR in the microbiology laboratory. Clin Microbiol Inf. 2004;10:190–212. doi: 10.1111/j.1198-743x.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 14.McDowell A., Perry A.L., Lambert P.A., Patrick S. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol. 2008;57:218–224. doi: 10.1099/jmm.0.47489-0. [DOI] [PubMed] [Google Scholar]
- 15.McGinley K.J., Webster G.F., Leyden J.J. Facial follicular porphyrin fluorescence; correlation with age and density of Propionibacterium acnes. Br J Dermatol. 1980;102(4):437. doi: 10.1111/j.1365-2133.1980.tb06557.x. [DOI] [PubMed] [Google Scholar]
- 16.Mohammadi T., Pietersz R.N., Scholtalbers L.A., Vandenbroucke-Grauls C.M., Savelkoul P.H., Reesink H.W. Optimal sampling time after preparation of platelet concentrates for detection of bacterial contamination by quantitative real-time polymerase chain reaction. Vox Sang. 2005;89(4):208–214. doi: 10.1111/j.1423-0410.2005.00707.x. [DOI] [PubMed] [Google Scholar]
- 17.Mollet I., Ongenae K., Naeyaert J.M. Origin, Clinical Presentation, and Diagnosis of Hypomelanotic Skin Disorders. Dermatologic clinics. 2007;25(3):363–371. doi: 10.1016/j.det.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 18.Ortone J.P., Bahadoran P., Fitzpatrick T.B., Mosher D., Hori Y. Hypomelanosis and hypermelanosis. In: Freedberg I.M., Eisen A.Z., Wolff K., Austen K.F., Goldsmith L.A., Katz S.I., editors. Fitzpatrick`s dermatology in general medicine. New York: McGraw-Hill; 2003. pp. 836–881. [Google Scholar]
- 19.Perman M., Sheth P., Lucky A. Progressive macular hypomelanosis in a 16 years old. Pediatr Dermatol. 2008;25:63–65. doi: 10.1111/j.1525-1470.2007.00585.x. [DOI] [PubMed] [Google Scholar]
- 20.Perry A.L., Lambert P.A. Propionibacterium acnes. Lett Appl Microbiol. 2006;42(3):185–188. doi: 10.1111/j.1472-765X.2006.01866.x. [DOI] [PubMed] [Google Scholar]
- 21.Relyveld G.N., Menkie H.E., Westerhof W. Benzoyl peroxide/clindamycin/UVA is more effective than fluticasone/UVA in progressive macular hypomelanosis: A randomized study. Am J Clin Dermatol. 2006;55(8):836–843. doi: 10.1016/j.jaad.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 22.Ross E.V. Optical treatments for acne. Dermatol Ther. 2005;18(3):253–266. doi: 10.1111/j.1529-8019.2005.05024.x. [DOI] [PubMed] [Google Scholar]
- 23.Rothman R.E., Yang S. PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet. 2004;4(6):337–347. doi: 10.1016/S1473-3099(04)01044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coordenadoria de Controle de Doenças (CCD). Introdução da PCR convencional e em tempo real para o diagnóstico laboratorial das meningites bacterianas no instituto Adolfo Lutz. Bol Epidemiol Paulista. 2007;4(40):1–29. [Google Scholar]
- 25.Schaller M., Loewenstein M., Borelli C., Jacob K., Vogeser M., Burgdorf W.H., Plewig G. Induction of a chemoattractive proinflammatory cytokine response after stimulation of keratinocytes with Propionibacterium acnes and coproporphyrin III. Br J Dermatol. 2005;153(1):66–71. doi: 10.1111/j.1365-2133.2005.06530.x. [DOI] [PubMed] [Google Scholar]
- 26.Westerhof W., Relyveld G., Kingswijk M.M., Man P. Propionibacterium acnes and the pathogenesis of progressive macular hypomelanosis. Arch Dermatol. 2004;140(2):210–214. doi: 10.1001/archderm.140.2.210. [DOI] [PubMed] [Google Scholar]
- 27.Zaitz C., Ruiz L.R.B., Souza V.M. Infecções causadas por agentes do reino Fungi (micoses) In: Zaitz C., Ruiz L.R.B., Souza V.M., editors. Atlas de micologia médica – diagnóstico laboratorial. Rio de Janeiro, Brazil: Medsi; 2004. pp. 37–38. [Google Scholar]
- 28.Zeller V., Ghorbani A., Strady C., Leonard P., Mamoudy P., Desplaces N. Propionibacterium acnes: an agent of prosthetic joint infection and colonization. J Infec. 2007;55:119–124. doi: 10.1016/j.jinf.2007.02.006. [DOI] [PubMed] [Google Scholar]


