Abstract
Chronic inflammation is an important risk factor for the development of colorectal cancer; however, the mechanism of tumorigenesis especially tumor progression to malignancy in the inflamed colon is still unclear. Our study shows that epithelial signal transducer and activator of transcription 3 (STAT3), persistently activated in inflamed colon, is not required for inflammation-induced epithelial overproliferation and the development of early-stage tumors; however, it is essential for tumor progression to advanced malignancy. We found that one of the mechanisms that epithelial STAT3 regulates in tumor progression might be to modify leukocytic infiltration in the large intestine. Activation of epithelial STAT3 promotes the infiltration of the CD8+ lymphocyte population but inhibits the recruitment of regulatory T (Treg) lymphocytes. The loss of Stat3 in epithelial cells promoted the expression of cytokines/chemokines including CCL19, CCL28, and RANTES, which are known to be able to recruit Treg lymphocytes. Linked to these changes was the pathway mediated by sphingosine 1-phosphate receptor 1 and sphingosine 1-phosphate kinases, which is activated in colonic epithelial cells in inflamed colon with functional STAT3 but not in epithelial cells deleted of STAT3. Our data suggest that epithelial STAT3 plays a critical role in inflammation-induced tumor progression through regulation of leukocytic recruitment especially the infiltration of Treg cells in the large intestine.
Introduction
Tumorigenesis is a multistage process often initiated by mutations that activate oncogenes or inhibit tumor suppressor genes. However, neoplastic cells often require additional factors from the microenvironment to support their survival, growth, and angiogenesis [1]. Clinical data and experimental mouse models have provided a definitive link between inflammation and cancer [2,3]. Recent evidence has shown that the activity of most of the inflammatory cytokines converges on the nuclear factor κ-beta and signal transducer and activator of transcription 3 (STAT3) [4,5]. However, the functions of STAT3 in tumor and stromal cells have not been clearly defined.
Transcriptional factor STAT3 is known to mediate inflammation acting downstream of a number of cytokines including interleukin-6 (IL-6), IL-10, IL-17, IL-21, IL-23, and vascular endothelial growth factor [4]. STAT3 functions in stromal cells have been shown to stimulate CD8+ cell production of interferon-γ [6], stimulate regulatory T (Treg) cells infiltrating into the tumor site [6], inhibit the maturation of functional dendritic cells [7], and inhibit immune stimulation in macrophages and neutrophils [8]. The anti-inflammatory effect of IL-10 on macrophage, for example, required STAT3 for its inhibition. Thus, knockout of either IL-10 or Stat3 causes severe inflammation [9,10]. Persistent activation of STAT3 has also been reported in many solid tumors [5]. Depending on tumor type, STAT3 has diverse functions. In head and neck tumors, for example, it has been shown to regulate cell cycling in conjunction with cyclin D1 (or CCND1) [12], and it inhibits cell growth by upregulating kinase inhibitor CDKN1B (or p27Kip1) [13] in melanoma cells and inhibits apoptosis by suppressing proapoptotic gene expression in breast, skin, and colon neoplastic cells [14–18]. Several reports have further shown that STAT3 activation in tumors is associated with poor prognosis [12,19–21], suggesting that STAT3 promotes tumor progression and/or metastasis. However, the mechanism by which STAT3 promotes tumor progression is unknown.
In the pathogenesis of inflammatory bowel disease and its associated colorectal cancer, an aberrant signaling cascade involving IL-6, IL-10, and STAT3 has been reported [3]. Recent genome-wide analysis of Crohn disease has identified the Stat3 gene as one of the susceptibility loci in this form of inflammatory bowel disease [22]. Furthermore, somatic mutations of Stat3 linked to persistent activation of STAT3 and colorectal cancer have been shown [16]. A knock-in mouse model in which constitutive active GP130 (gp130Y757F) leads to hyperactivation of JAK1/2-STAT3 signaling cascade on IL-6 binding develops tumors during the inductive state of chronic inflammation [23]. Recently, a link between chronic inflammation and sphingosine 1-phosphate kinase 1 (SPHK1) in the development of colitis-associated cancer was reported [24–26]. In all these models, the essential function of STAT3/S1P in tumor growth was examined extensively, but the role of STAT3 and its relationship with S1P in tumor progression has not been addressed sufficiently.
We previously established a mouse model of spontaneous inflammation-associated colonic cancer, conditional knockout mice with Stat3 deletion in hematopoietic cells (Stat3-IKO), by inactivating Stat3 in hematopoietic cells using colony-stimulating factor 1 receptor promoter driving Cre recombinase in a control mouse with floxed P sites in the introns of Stat3 gene (Stat3flox/flox mouse) [10]. This model has significant advantages for the study of this disease, because a single myeloid mutation is used to generate chronic inflammation that leads to tumor development at frequencies similar to human colon cancer in the setting of inflammatory bowel disease (IBD) [10]. Importantly, no germ line mutation is introduced into the colonic epithelium, and consequently, the model is appropriate for the study of early genetic and phenotypic changes within the critical epithelial subsets. In this model, we found that STAT3 in epithelial cells was persistently activated through early stage of inflammation development, tumor formation, and tumor progression to malignancy. To determine the role of STAT3 activation in epithelial/tumor cells in inflamed colon, we have inactivated Stat3 specifically in the intestinal epithelium of Stat3-IKO mice. In the Stat3 double knockout mice [conditional knockout mice with Stat3 deletion in hematopoietic and intestinal epithelial cells (Stat3-EIKO)], the colonic epithelium still exhibited hyperproliferation and formation of early-stage tumors in response to the chronic inflammation in the colons, but the rate of tumorigenesis and progression to advanced malignancy was significantly reduced. The delayed tumor progression is associated with a decreased CD8+ cell and a reduced activation of sphingosine 1-phosphate receptor 1 (S1PR1)-SPHK1 pathway in epithelial cells but an enhanced Treg lymphocyte in the large intestine, suggesting that epithelial STAT3 regulates tumor progression by coordinating immune cell recruitment using the S1PR1 pathway.
Materials and Methods
Mice and Human Samples
Mice were housed in the pathogen-free barrier facility at Albert Einstein College of Medicine (AECOM, Bronx, NY) and were handled in accordance with National Institutes of Health regulations concerning the usage and care of experimental animals. All procedures involving animal usage were approved by the Institutional Animal Care and Use Committee (IACUC) committee at AECOM. Residual human colonic tissues from biopsies or colectomy taken from patients with inflammatory bowel disease and colorectal carcinoma, which would otherwise be discarded, with Institutional Review Board (IRB)-approved protocol 09-03-096X, were used. We have examined three patient samples. One was a 71-year-old patient with a history of ulcerative colitis (UC) and adenocarcinoma in a background of chronic active colitis pathology. A second case was a 49-year-old male with UC for 7 years with low-grade dysplasia. A third case was a 66-year-old male with UC and poorly differentiated carcinoma with signet ring morphology and multifocal dysplasia. Negative samples were taken from patients with diverticulitis or normal region of the colon from patients with UC.
Stat3-IKO and Stat3-EIKO Mice
The establishment of Stat3-IKO mice was described previously [10]. To delete Stat3 in intestinal epithelial cells in Stat3-IKO mice, the transgene vCRE (Cre regulated by the villin promoter), kindly provided by Dr Sylvie Robine (Institut Curie, Paris, France) [27], was introduced into Stat3-IKO mice (in a C57BL/6 background) to obtain mice with both hematopoietic and epithelial inactivation of Stat3 (Stat3-EIKO mice).
Isolation of Colonic Epithelial Cells
Colonic epithelial cells were prepared as described previously [28]. Briefly, colons from the anal cavity to the cecum were removed from the mice, cut into halves, and washed with Hank's balanced salt solution (HBSS; Sigma, St Louis, MO) to remove fecal material. The colons were treated with 1 mM DTT in HBSS for 10 minutes to remove the mucus. They were further treated with 25 ml of HBSS with 1 mM EDTA at 37°C with shaking for 20 minutes, and colonic epithelial cells were then scraped off the basal lamina. The scraped cells were washed three times in HBSS and once in phosphate-buffered saline (PBS) and then filtered through cell strainer with 70-µm nylon mesh (BD Falcon; Fisher Scientific, Pittsburgh, PA). Epithelial cells were isolated from 5 x 106 scraped cells using the biotinylated anti-mouse CD326 epithelial cell adhesion molecule (CAM) antibody (Biolegend, San Diego, CA) and CELLection Biotin Binder Kit (Invitrogen, Life Technologies, Grand Island, NY) following the manufacturer's protocol.
Quantitative Polymerase Chain Reaction
RNA from colonic epithelial cells was extracted using TRIzol (Invitrogen). RNA (50–100 ng) was used in reverse transcription with the Omniscript RT Kit (Qiagen, Valencia, CA). cDNA was subjected to quantitative polymerase chain reaction (qPCR) using 0.1 µMprimers from commercially available Mouse Cytokine Primer Library II (RealtimePrimers.com) or from synthesized sequences (Sigma) and subjected to 50 cycles of denaturation at 95°C for 10 seconds and annealing at 58°C for 45 seconds.
Histologic Analysis
The large intestine from mice were fixed in 10% formalin and then paraffin embedded [28]. Sections were stained with hematoxylin and eosin. The pathology of tissue sections were evaluated in blinded fashion by Dr Qiang Liu, and disease stages were classified as follows: Hyperplasia is characterized by thickened mucosa with elongated crypts; low-grade dysplasia is classified by thickened mucosa with elongated, irregularly branched glands that contain epithelium with cytologic and nuclear atypia including loss of differentiation (goblet cells), polarity, and enlarged nuclei; high-grade dysplasia has stratified nuclei that are more extensive than low-grade dysplasia with loss of cell polarity and presence of cribriform architecture or gland within gland; carcinoma is characterized by tumor cell invasion into the submucosa as the key distinguishing feature for this lesion. Specific features also include the presence of atypical and irregular glands with desmoplastic reaction.
Immunohistochemistry
Standard immunohistochemical (IHC) procedures were used according to a protocol from Cell Signaling Technology (Danvers, MA). All tissues stained for immune cells and intracellular molecules were fixed with 10% formalin except for those stained for macrophages with F4/80 antibody, which were fixed in zinc fixative (BD Biosciences, San Jose, CA). Antibodies used were 0.1 µg/ml anti-F4/80 (Invitrogen), 3 µg/ml anti-CD3 (Dako Cytomation, Carpinteria, CA), 1:200 of anti-MKI67 (or anti-Ki67), 4 µg/ml anti-pSTAT3 (Tyr705), 1:50 for anti-pSTAT3 (Ser727), 1:400 for anti-BIRC5 (or anti-Survivin; Cell Signaling Technology), 1:100 for anti-Forkhead box P3 (anti-FOXP3; eBioscience, San Diego, CA), and 1:200 for anti-EDG-1 (S1PR1; Abcam, Cambridge, MA).
Ex Vivo Organ Culture
Colons were isolated from mice, cut longitudinal, removed of fecal material, and washed with HBSS. The colons were cut into 2- to 3-mm pieces and placed in serum-free RPMI media with antibiotic-antimycotic (Gibco; Life Technologies, Grand Island, NY) before incubation at 37°C. Supernatants from overnight culture were collected, and chemokine and cytokine production were measured using MILLIPLEX MAP Mouse Cytokine/Chemokine Panel Kit (EMD Millipore, Billerica, MA) following the manufacturer's protocol.
Flow Cytometry
Cells from scraped colons were washed in HBSS and passed through a 70-µm filter. The cells were washed with FACS buffer (0.5% BSA in PBS) and stained for the following surface proteins: CD3+ T cells [CD3-fluorescein isothiocyanate (FITC)], CD4+ T cell (CD4-FITC), CD8+ T cells [CD8-phycoerythrin (PE)], CD25+ T cells (CD25-PE) for 20 to 30 minutes. All fluorescent antibodies were purchased from eBioscience and compared to recommended isotype controls. The cells were washed once in FACS buffer and then in PBS before fixation in 4% paraformaldehyde buffer for 20 to 30 minutes. Cells were washed three times in FACS buffer before analysis using a Becton Dickinson FACScan Flow Cytometer.
Results
Inflammation-Induced Epithelial Overproliferation in Colon Is Independent from STAT3
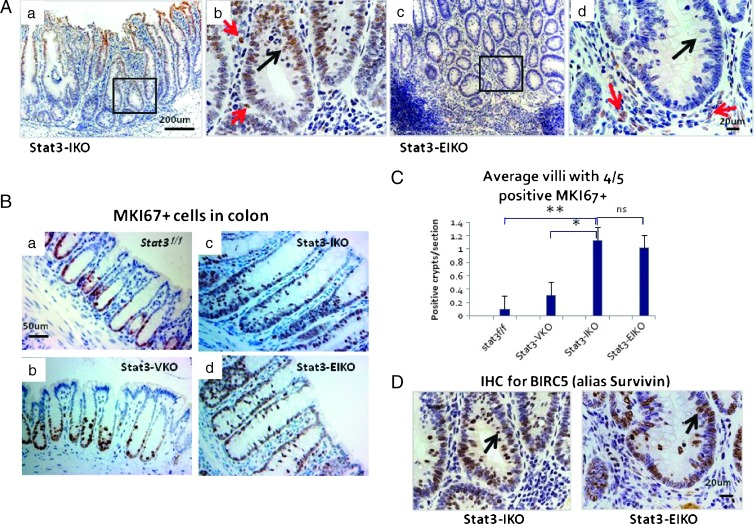
In a published study [10], we used the Stat3-IKO model to demonstrate the role of mammalian target of rapamycin (mTOR), in putative early stages of colitis-associated cancer, specifically the hyperproliferation of colonic epithelium and also the activation in these epithelial cells of STAT3, which has been implicated in colon tumorigenesis by other studies. To test the hypothesis that inflammation may promote tumorigenesis through STAT3-mediated epithelial hyperproliferation, we targeted inactivated Stat3 specifically in intestinal epithelial cells in this model by crossing the transgene villin-Cre [27] into the Stat3-IKO mice. The inactivation of epithelial Stat3 was efficient as shown by the much lower density of the active form of STAT3 (pSTAT3) in colonic epithelial cells in the double knockout mice, Stat3-EIKO (Figure 1A, a and b vs c and d), compared to the parental strain, Stat3-IKO.
Figure 1.
Colonic hyperproliferation does not require functional STAT3. (A) IHC analysis of phosphorylated STAT3 (STAT3-P) in Stat3-IKO (a and b) and Stat3-EIKO (c and d) colons. Boxed regions in a and c are shown at higher magnification in b and d. Red arrows in b and d point to positive STAT3-P stromal cells, black arrow in b points to positive Stat3-IKO colonic epithelial cells, and black arrow in d points to cell loss of STAT3-P. (B) IHC analysis for cellular proliferation. Representative sections show MKI67+ (or anti-Ki67+) cells extended up to 1/2 of the crypt of control Stat3flox/flox (a) and Stat3-VKO (b) and to 4/5 of crypts of Stat3-IKO (c) and Stat3-EIKO (d) colons. (C) Significantly more villi with MKI67+ extended up to 4/5 of the crypt are seen in both Stat3-IKO (c) and Stat3-EIKO (d) compared to control Stat3flox/flox (a) and Stat3-VKO (b) [unpaired t test Stat3flox/flox vs Stat3-IKO or Stat3-EIKO. *P = .001 and **P = .0001 and no significant (ns) difference between Stat3-IKO and Stat3-EIKO]. (D) IHC analysis shows that BIRC5 (or Survivin) is localized to the nucleus (black arrows) of epithelial cells in Stat3-IKO and Stat3-EIKO colons taken from the same sections as in A (b and d).
No abnormality was observed in mice carrying villin-Cre and Stat3flox/flox [conditional knockout mice with Stat3 deletion in intestinal epithelial cells (Stat3-VKO)], which are phenotypically similar to Stat3flox/flox control mice, suggesting that the activation of STAT3 in colonic epithelial cells in Stat3-IKO mice is specifically induced by inflammation and it is not required for the normal development of the large intestine. Unlike Stat3-IKO mice, which exhibited colitis symptoms including persistent diarrhea, blood feces, and weight loss by 18 to 20 weeks of age [10], age-matched littermates of Stat3EIKO mice appeared healthy and developed much milder symptoms even in older mice (35–40 weeks of age). Examining the histology of the large intestine, we found that, in a similar fashion to Stat3-IKO mice, hyperproliferation of epithelial cells and an expanded proliferative compartment was observed in the large intestine of Stat3-EIKO mice (Figure 1B, c and d vs a and b). There were many regions in both Stat3-IKO and Stat3-EIKO colons with MKI67+ (or Ki67+) cells extending up to 4/5 (80%) of the length of the crypt, whereas most control (Stat3flox/flox or Stat3-VKO) colons showed MKI67+ (or Ki67+) cells only in the lower 1/2 (50%) of the crypt (Figure 1B, a and b vs c and d). A quantitative analysis of crypts with extended MKI67+ (or Ki67+) epithelial cell proliferation demonstrated a significant higher density of such crypts in the large intestines of both Stat3-IKO and Stat3-EIKO mice compared to normal controls (Figure 1C) with no significant difference between Stat3-IKO and Stat3-EIKO colons, suggesting that epithelial cell overproliferation can occur without functional epithelial Stat3. BIRC5 (or Survivin) is a major downstream target of STAT3 and is known to have antiapoptotic functions [11,29]. Analysis of BIRC5 (or Survivin) production indicated that the functional BIRC5 (or Survivin) in Stat3-EIKO colons was comparable to Stat3-IKO colons (Figure 1D). These data support the idea that Survivin in colonic epithelial cells can be regulated by intracellular signaling molecules other than STAT3 in inflamed colon, as a deletion of intestinal Stat3 has little effect on the Stat3-EIKO epithelial overproliferation.
Inactivation of Epithelial Stat3 Delayed Tumor Progression in Inflamed Colon
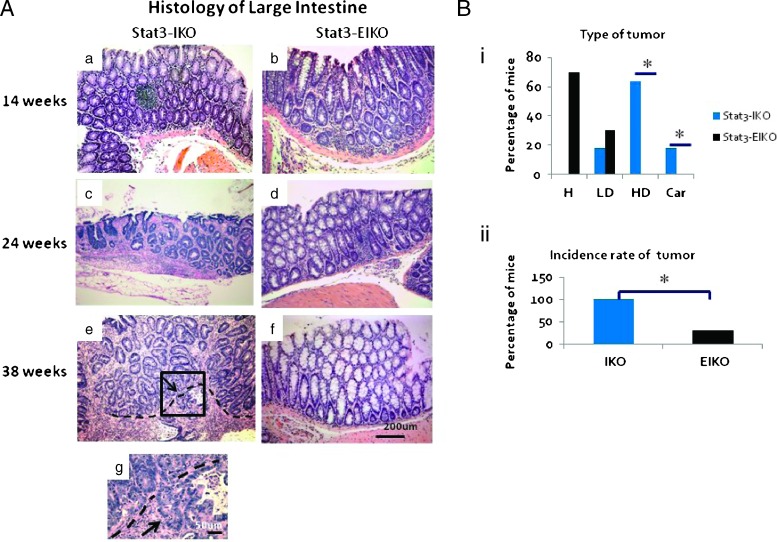
There were significant differences in colonic histopathology between age-matched Stat3-IKO and Stat3-EIKO littermates. Our previous study classified tumor progression in the large intestine of Stat3-IKO mice into three stages: low-grade dysplasia, high-grade dysplasia, and carcinoma. Although both Stat3-IKO and Stat3-EIKO mice developed hyperplasia in the large intestine at 14 weeks of age, most Stat3-IKO mice exhibited high-grade dysplasia in colon by 24 weeks, while the majority of age-matched Stat3-EIKO littermates had hyperplasia and a few developed low-grade dysplasia in colon (Figure 2, A, c vs d, and Bi). By 38 weeks of age, a large percentage of the Stat3-IKO mice had tumors progressed to carcinoma with tumor invasion into the submucosa or muscular layer, whereas none of the eight age-matched Stat3-EIKO littermates exhibited progression to carcinoma (Figure 2A, e and g vs f). Further analysis of tumor stages revealed that a similar percentage of both Stat3-IKO and Stat3-EIKO mice developed early-stage tumor, low-grade dysplasia (Figure 2Bi, LD); a significant reduction of tumors at advanced stages, including high-grade dysplasia and carcinoma, classified with severe irregularly branched glands, cytologic and nuclear atypia, as well as tumor invasion (Materials and Methods section), was observed in Stat3-EIKO mice compared to Stat3-IKO mice (Figure 2Bi). Histologic analysis also demonstrated a significant reduction of tumor incidence observed in the Stat3-EIKO mice compared to Stat3-IKO mice (Figure 2Bii). Neither inflammation nor tumors were found in control (Stat3flox/flox) or Stat3-VKO mice. Taken together, our data indicated that activation of epithelial STAT3, induced by inflammation in Stat3-IKO colon, is required for tumor progression to malignancy.
Figure 2.
Loss of colonic epithelial Stat3 does not affect inflammation-induced hyperplasia. (A) Hematoxylin and eosin examination of representative sections of large intestines at different ages. Early carcinoma is seen by 24 weeks of Stat3-IKO (c) and invasive carcinoma by 38 weeks (e and higher magnification in g), whereas aged-matched Stat3-EIKO mice develop hyperplasia (d and f). Arrow in e points to the invading front, and arrow in g points to dysplastic cells in the muscularis layer. (B) Graphical analysis of histologic differences between Stat3-IKO and Stat3-EIKO colons taken from 14 to 38 weeks (n > 20). H, hyperplasia; LD, low-grade dysplasia; HD, high-grade dysplasia; Car, carcinoma. (i) Significant difference is seen at the high-grade dysplasia stage and carcinoma between Stat3-IKO and Stat3-EIKO large intestines [Fisher exact test (two-tail); *P < .05]. (ii) Significant difference is observed in the incidence of tumor development in the Stat3-IKO compared to Stat3-EIKO mice (Fisher exact test, *P = .001).
Epithelial STAT3 Regulates Leukocytic Infiltration in Colon
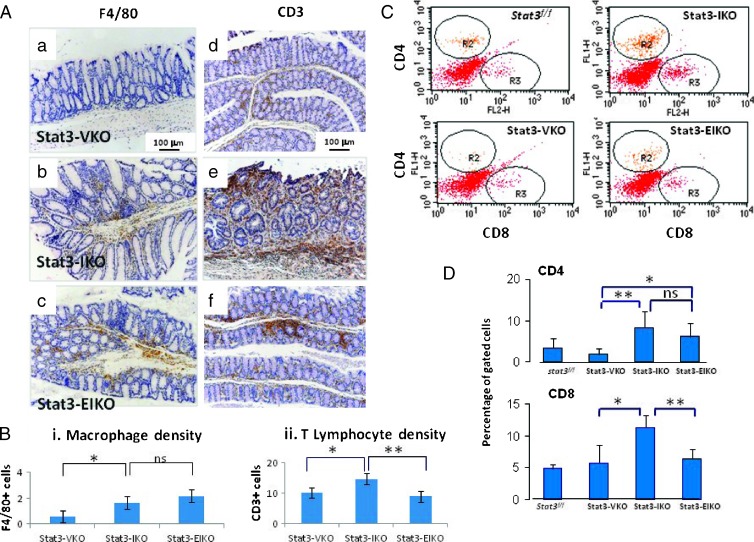
Our previous study has demonstrated a marked increase of macrophage infiltration, determined by macrophage-specific marker F4/80, in the large intestine of Stat3-IKO mice compared to the normal controls [10]. This was not reduced when epithelial Stat3 was inactivated (Stat3-EIKO; Figure 3, A, a–c, and Bi). Similarly, a significant increase of CD3+ lymphocytes was observed in Stat3-IKO colon compared to normal control (Figure 3A, d vs e). In this case, however, inactivation of epithelial Stat3 significantly reduced CD3+ T cell infiltration in the colon of Stat3-EIKO compared to Stat3-IKO mice (Figure 3, A, e vs f, and Bii). To further analyze T lymphocyte infiltration in the colon of Stat3-EIKO mice, we examined the subpopulations of T lymphocytes and found that inactivation of Stat3 in epithelial cells significantly reduced the infiltration of CD8+ but not CD4+ lymphocytes in the inflamed intestine of Stat3-EIKO compared to Stat3-IKO mice (Figure 3, C and D). Consistent with the reduced infiltration of CD8+ lymphocytes, a significant reduction of secretory proinflammatory cytokine IL-1α and a trend toward lower IL-6, tumor necrosis factor-alpha (TNF-α), and IL-2 were found in colons of Stat3-EIKO compared to Stat3-IKO mice, in which the production of these cytokines was markedly increased relative to normal colon (Figure 4A). These data suggest that inflammation-induced activation of STAT3 in colonic epithelial cells promote inflammation in colon by facilitating proinflammatory cytokine expression.
Figure 3.
Epithelial STAT3 regulates inflammation. (A) IHC analysis of F4/80+ macrophages (a–c) and CD3+ T cells (d–f): control Stat3-VKO (a and d), Stat3-IKO (b and e), and Stat3-EIKO (c and f). (B) Macrophage and CD3+ densities were quantified using National Institutes of Health ImageJ: (i) A significant higher macrophage population is observed in either Stat3-IKO or Stat3-EIKO colons compared to control (unpaired t test, *P = .0001) but no significant difference between Stat3-IKO and Stat3-EIKO. (ii) Significantly higher CD3+ T cell density is observed in the Stat3-IKO compared to control or Stat3-EIKO (Stat3-IKO vs control, unpaired t test, * P = .02 and Stat3-IKO vs Stat3-EIKO, unpaired t test, **P = .003). (C) FACS analysis of stromal cells in colons using CD4-FITC and CD8-PE. (D) Gated cells were used to calculate the percentage of CD4+ and CD8+ T cells. CD4: Stat3-EIKO versus Stat3-VKO, unpaired t test, *P = .049; Stat3-IKO versus Stat3-VKO, **P = .02. No significant difference in CD4+ T cell population between Stat3-IKO and Stat3-EIKO colons. CD8: Stat3-IKO versus Stat3-VKO, unpaired t test, *P = .007 and Stat3-IKO versus Stat3-EIKO colons, **P = .0176.
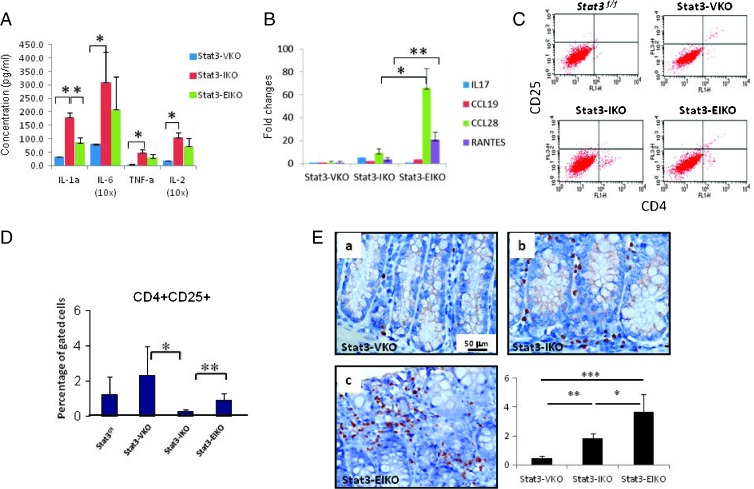
Figure 4.
Colonic epithelial STAT3 regulates cytokine, chemokine production, and Treg recruitment. (A) Cytokine production (pg/ml) from overnight organ culture. Stat3-IKO versus control Stat3-VKO, unpaired t test, *P = .0001 for IL-1α,*P = .02 for IL-6, *P = .0164 for TNF-α, and *P = .0044 for IL-2. No significant (ns) difference in IL-6, TNF-α, and IL-2 production between Stat3-IKO and Stat3-EIKO culture, but a significant difference is seen with IL-1α production, *P = .017. (B) Relative gene expression of chemokines from isolated colonic epithelial cells. A higher trend of IL-17 expression is seen in Stat3-IKO compared to Stat3-EIKO and CCL19 in Stat3-EIKO compared to Sta3-IKO, but a significant higher expression of CCL28 and RANTES is seen in Stat3-EIKO versus Stat3-IKO, unpaired t test, *P = .018 for CCL28 and **P = .04 for RANTES. (C) FACS analysis of CD4+ and CD25+ Treg cells. (D) A significantly higher population of CD4+CD25+ in Stat3-EIKO colons and Stat3-VKO compared to Stat3-IKO colons, unpaired t test, *P = .048 and **P = .02. (E) IHC analysis of FOXP3+ cells in representative section from controls, Stat3-VKO (a; similar to Stat3flox/flox), Stat3-IKO (b), and Stat3-EIKO mice (c). Graphical analysis of FOXP3+ cells/CD3+ T cells. Stat3-EIKO versus Stat3-IKO colons (unpaired t test, *P = .0412), Stat3-IKO versus Stat3-VKO (unpaired t test, **P = .0149), and Stat3-EIKO versus Stat3-VKO (unpaired t test, ***P = .0033).
To determine whether epithelial STAT3 regulated leukocytic infiltration in the colon, we next analyzed the expression of cytokines/chemokines in isolated colonic epithelial cells using qPCR. Among 30 different chemokines in the CC and CXC families screened, an increased production of IL-17 was observed in epithelial cells from Stat3-IKO mice compared to normal controls, whereas inactivation of Stat3 in the colonic epithelial cells reduced their expression (Figure 4B). In contrast, the expression of CCL19, CCL28, and RANTES was mildly increased in epithelial cells isolated from Stat3-IKO colon compared to normal control but highly expressed in epithelial cells from Stat3-EIKO colon (Figure 4B), indicating that this set of cytokines may be negatively regulated by functional STAT3.
Because cytokines with higher expression in the colonic epithelial cells of Stat3-EIKO compared to Stat3-IKO mice, including CCL19, CCL28, and RANTES, were reported as Treg chemoattractants, we next tested whether epithelial STAT3 regulates Treg cell recruitment in the colon. FACS analysis of cells isolated from scraped colons and gated for CD4+ and CD25+ population showed a reduced infiltration of this population in Stat3-IKO colon compared to normal control but a significant increase in the colons of Stat3-EIKO compared to Stat3-IKO (Figure 4, C and D). Interestingly, a significantly higher population of CD4+ CD25+ was also observed in Stat3-VKO colon compared to Stat3-IKO or control colons. These observations suggested that activation of epithelial STAT3 may have an inhibitory effect on Treg infiltration in colon. We then verified the result by IHC using an antibody that recognizes the Treg-specific marker, FOXP3 [30]. Although a three-fold increase of FOXP3+ T cells was found in Stat3-IKO colon compared to the controls (Figure 4E, a vs b), an additional two-fold increase of FOXP3+ T cells was observed in Stat3-EIKO compared to Stat3-IKO colon (Figure 4E, b vs c; unpaired t test, P = .0412). A totally eight-fold increase of FOXP3+ T cells was found in the large intestine of Stat3-EIKO compared to Stat3-VKO (unpaired t test, P < .0033). Taken together, consistent to the expression of pattern of cytokine/chemokine expression in colonic epithelial cells in Stat3-IKO and Stat3-EIKO mice, the data suggest that inflammation-induced activation of epithelial STAT3 plays a critical role in regulating leukocytic infiltration in inflamed intestine and that it promotes CD8+ but inhibits Treg infiltration in the large intestine.
Epithelial STAT3 Regulates Leukocytic Infiltration in Colon Associated with S1P Pathway
In tumors with persistent activation of STAT3, the S1P signaling pathway has been shown to act as a positive feedback loop that supports tumor cell growth involving IL-6 [31]. Because the ex vivo culture of both Stat3-IKO and Stat3-EIKO colons showed a robust IL-6 production (Figure 4A), we determined whether the S1P signaling pathway was upregulated in these mice. IHC analysis for the S1PR1 receptor indicated that, in the inflamed areas of Stat3-IKO, there was more intense staining of S1PR1 in colonic epithelial cells and stromal cells compared to control colons (Stat3flox/flox or Stat3-VKO; Figure 5A, a, b vs c). Fewer S1PR1+ cells were seen in the colon of Stat3-EIKO mice compared to Stat3-IKO (Figure 5A, c vs d). Interestingly, in Stat3-IKO colon, S1PR1 appeared to be expressed on the cell surface of the epithelial cells (Figure 5Ac, arrows), whereas in Stat3-EIKO colon, S1PR1-positive staining appeared to be in enclosed vesicles in epithelial cells (Figure 5Ad, arrow). The S1PR1 expression in isolated colonic epithelial cells was verified by qPCR, which showed that Stat3-IKO epithelial cells had more than 10-fold higher expression than controls and that inactivation of epithelial Stat3 significantly reduced the expression (Figure 5Bi). Higher expression of S1PR1 suggested that this pathway may be activated in the colonic epithelial cells in Stat3-IKO colon. In a positive feedback loop, S1P, the substrate that binds S1PR1, is phosphorylated by two kinases that are downstream targets of S1PR1 signaling [31,32]. Using qPCR analysis, we detected significantly increased expression of SPHK2 in the epithelial cells of Stat3-IKO mice compared to controls and Stat3-EIKO epithelial cells (Figure 5Bii). A similar pattern was seen with SPHK1, but the expression did not show a significant difference between Stat3-IKO and Stat3-EIKO. Consistent with the observations in our mouse models, overexpression of S1PR1 in the colonic epithelial cells of human IBD colon was verified by IHC. All colon samples from three inflammatory bowel disease patients showed intense S1PR1 staining in the colonic epithelium (Figure 5C). Taken together, these data indicate that the S1P pathway is active in human inflammatory bowel disease and that the high expression of S1PR1 in colonic epithelial cells in the inflamed sites is dependent on functional epithelial STAT3 as a positive feedback mechanism.
Figure 5.
Persistent activation of epithelial STAT3 in inflammatory bowel diseases regulates the expression of S1PR1 receptor and SPHKs. (A) Representative IHC analysis of S1PR1 receptor or EDG+ cells in Stat3flox/flox (a), Stat3-VKO (b), Stat3-IKO (c), and Stat3-EIKO colon (d). Red arrows point to S1PR1+ cells in stromal cells and in epithelial cells of Stat3-IKO and Stat3-EIKO colons (c and d). (B) qPCR analysis of (i) S1PR1 and (ii) SPHK's expression in the isolated colonic epithelial cells. Significantly higher expression of S1PR1 is detected in Stat3-IKO colonic epithelial cells (Stat3-IKO vs control Stat3-VKO, unpaired t test, *P = .02 and Stat3-IKO vs Stat3-EIKO, **P = .05), and significantly higher expression of SPHK2 is seen in Stat3-IKO colonic epithelial cells compared to control and Stat3-EIKO (Stat3-IKO vs control Stat3-VKO, unpaired t test, *P = .028 and Stat3-IKO vs Stat3-EIKO, unpaired t test, **P = .039). (C) Representative IHC section shows S1PR1-positive cells in biopsied colon taken from an inflammatory bowel disease patient (b) and normal colon (a).
Discussion
This study identified a novel role of epithelial STAT3 in inflammation-induced tumor development in the large intestine. We demonstrated that, in the large intestine with chronic inflammation, epithelial STAT3 is not required for inflammation-induced epithelial overproliferation but is critical for tumor progression from early-stage malignancy to invasive carcinoma. Because inactivation of Stat3 in epithelium alone (Stat3-VKO) did not have any noticeable effect on colonic epithelium, our study supports the hypothesis that the activation of epithelial STAT3 in inflamed intestine is specifically linked to inflammation-induced tumor development.
Our study highlights the function of epithelial STAT3 in tumor development that is different from the commonly used models of azoxymethane/dextran sodium sulfate (DSS)-induced colorectal cancers, in which epithelial STAT3 is believed to contribute to tumorigenesis in inflamed colon by promoting tumor cell growth and inhibiting cell death [23,33,34]. It is noteworthy that mice with functional epithelial STAT3 under azoxymethane and DSS treatment developed tubular adenomas as a consequence of compensatory hyperproliferation [23] because of the physical damage of mucosal layer. Our model has significant advantages for the study of tumorigenesis in chronic inflammation such as inflammatory bowel diseases, because a single myeloid mutation is used to generate chronic inflammation that leads to tumor development at frequencies similar to human colon cancer in the setting of IBD [10]. In contrast, in the DSS model, inflammation is secondary to acute mucosal injury that has overwhelming normal mucosal immunity/functions.
Our data demonstrate a potential mechanistic link between epithelial STAT3 and tumor progression. Activation of epithelial STAT3 in the inflamed region is associated with overproduction of proinflammatory cytokines in colon including IL-6, TNF-α, and IL-1α. Our data also showed that an increased infiltration of CD8+ lymphocytes in colon is linked to the activation of Stat3 in epithelial cells and tumor progression. The role of CD8+ lymphocytes in colitis-associated colorectal cancer is still controversial in that CD8+ T cells is considered to play a central role in cancer immunosurveillance, but these cells were also found in intestinal mucosa of active IBD including both Crohn disease and UC [35]. The infiltration of CD8+ T cells was reported to have a direct cause of tissue damage and destruction of colonic epithelial cells [36], which have the potential to favor tumor development. Our data demonstrated that, consistent with the observation in human IBD [35], an increased infiltration of CD8+ T cells is associated with more severe inflammation and the development of advanced malignancy in colon, whereas reduced tumor progression is correlated with a decreased infiltration of CD8+ T cells, suggesting a protumorigenic function of this population. We also demonstrated that the infiltration of CD8+ is potentially regulated by epithelial STAT3.
Our data further suggest that inflammation-activated STAT3 in epithelial cells may promote tumor progression through inhibiting Treg recruitment in colon. Treg function primarily suppresses immune responses by mediating tolerance against self-antigen or preventing excessive inflammation against intestinal microbes [37]. The role of Treg in inflammation-associated cancer, however, is still unclear. Some studies have shown that Treg cells suppress the anticancer immune reactions [38,39]; however, others have shown that Treg cells can repress inflammation and therefore reduce inflammation-promoted tumor risk [37]. Our finding that lower density of Treg cells is associated with more advanced tumor progression, but intensive infiltration is associated with reduced tumor development, suggests the tumor inhibitory role of Treg cells in colitis-associated colorectal cancer. This observation is supported by clinical studies that high density of FOXP3+ Treg infiltration in colon cancer tissues was associated with improved survival and more favorable prognosis [40,41]. Our study demonstrates a novel regulation of Treg infiltration by epithelial STAT3 that may inhibit the recruitment of this population through regulating the expression of a set of cytokines and chemokines including CCL28, CCL19, and RANTES. Several reports have shown that Treg expressed CCR4 and CCR10 receptors for CCL22 and CCL28 [42,43] and, more recently, CCR5 receptor for RANTES [44].
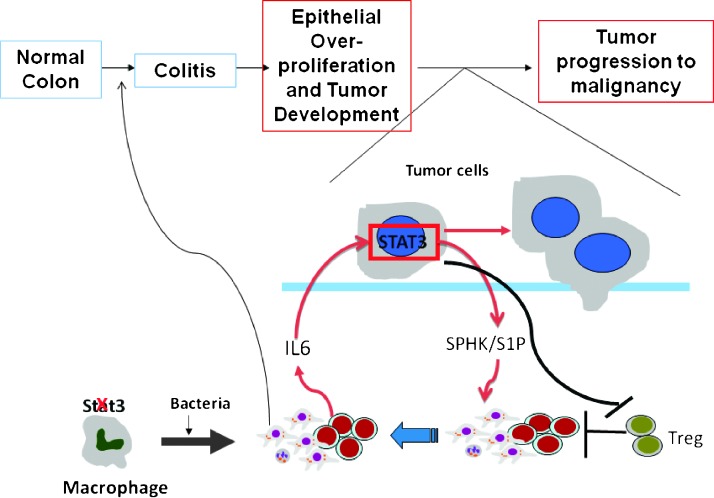
Activated STAT3 by inflammation in the colon may act as a key component of a positive feedback cross talk between inflammatory and epithelial cells. For example, IL-6, STAT3, and S1PR1 mediate an autocrine loop that favors cancer cell growth [31,34,45]. In this context, recruited stromal cells such as macrophages and T cells would produce IL-6 that stimulates cancer cell growth by acting on the STAT3 pathway. Our data would support this loop in tumor progression, as intense expression of S1PR1 is seen in areas with dysplastic cell region of Stat3-IKO colons and moderate expression in hyperplastic cell region of the Stat3-EIKO colons. Our observation that epithelial cells with inflammation-activated STAT3 had higher expression of SPHKs suggests further that STAT3 regulates the key enzymes responsible for phosphorylation of sphingosine to S1P. Significantly high CD4+ and CD8+ T cells seen in Stat3-IKO mice may be due to an increase of S1P-regulating lymphocytes exiting from secondary lymphoid organs consistent with others showing S1PR1 and STAT3 induction in inflammatory responses [46]. Loss of epithelial Stat3 in Stat3-EIKO showed that reduced expression of SPHK2 suggests epithelial STAT3-mediating immune cell recruitment through S1P pathway. Collectively, our data have established the linkage between STAT3 regulation of SPHKs in the inflamed environment and tumor progression. To our knowledge, this is the first time in which a link between STAT3 and S1PR1 expression can be associated with a defined stage of tumor progression (Figure 6).
Figure 6.
Model of inflammatory bowel disease-associated colorectal cancer. During chronic inflammation, IL-6 activates epithelial STAT3, which regulates S1P production through action of S1P/SPHK1 pathway. Recruited immune cells stimulated tumor progression from hyperplasia to carcinoma.
In summary, we observed that epithelial STAT3 regulates tumor progression but not the growth of the cells in the inflamed colon and that epithelial STAT3 regulates leukocytic infiltration that may play a crucial role in tumorigenesis and tumor progression. Epithelial STAT3 may promote tumor progression through regulating the infiltration of proinflammatory cells and inhibiting Treg infiltration. Recruited inflammatory cells may accelerate tumor progression through the activation of the S1PR1 pathway in tumor cells. The up-regulation of the S1P pathway is associated with advanced stage of tumor progression, suggesting a possible target for therapeutic intervention. A recent retrospective study of more than 900 patients shows a significant association between colorectal cancer patients with phosphatidylinositol 3-kinase mutation (PIK3CA) on regular regimen of aspirin and reduced mortality rate [47]. This study demonstrated the crucial role of inflammation, especially the interaction between inflammation and tumor markers such a PI3K and MT-CO2 (or Cox2), in tumor progression in colorectal cancer. Our finding is consistent with this study and further demonstrates that inflammation regulated by epithelial STAT3 is critical for promoting tumor progression to advanced stage in colon. Interestingly, overexpression of SPHK1 in cancer progression has been recently reviewed by Alshaker et al. [48], and SPHK1 activation in cancer cells mediated by IL-6 through PI3K and mitogen-activated protein kinase (MAPK) pathways has also been reported [49]. Collectively, these data suggest that SPHK1/S1P pathway regulates PI3K activity and inhibiting either the upstream signaling cascade with S1P analog (dihydrosphingosine or dimethylsphingosine), SPHK1 inhibitors (F-12509a, B-5354c, and SKI-I-V) [48], or downstream through PI3K activity (aspirin) would improve the clinical outcome of colorectal cancer. We are currently evaluating the effectiveness of blocking the S1P signaling pathway with immunosuppressive drug, such as fingolimod (FTY720) [50–52], in our mouse model. The usage of FTY720 would offer additional strategy that could be used to block inflammation-induced tumorigenesis.
Acknowledgments
We thank Sylvie Robine (Institut Curie) for providing the villin-Cre mice. We thank Ernestina Middleton, Joseph Albanese, Wa Shen, Wai Ba, and Michele Houston for technical support and Neva Morales-Grajales for administrative support. We thank the Pathology Laboratory at Montefiore Medical Center and facilities at AECOM, including Flow Cytometry, Analytical Imaging, Histology and Comparative Pathology, and Animal Housing, for the support.
Abbreviations
- DSS
dextran sodium sulfate
- S1P
sphingosine 1-phosphate
- S1PR1
sphingosine 1-phosphate receptor 1
- STAT3
signal transducer and activator of transcription 3
- Stat3flox/flox mice
control mice with floxed P sites in the introns of Stat3 gene
- Stat3-EIKO
conditional knockout mice with Stat3 deletion in hematopoietic and intestinal epithelial cells
- Stat3-IKO
conditional knockout mice with Stat3 deletion in hematopoietic cells
- Stat3-VKO
conditional knockout mice with Stat3 deletion in intestinal epithelial cells
Footnotes
This work was supported in part by Montefiore Medical Center New Research Initiative Award, Miriam Mandel Faculty Scholar Award, the Albert Einstein Cancer Center Core grant P30 CA13330, Montefiore Medical Center Pathology Department Faculty Research Fund, and Professional Staff Congress of the City University of New York to A.V.N. The authors declare no competing financial interests and no conflict of interests between the authors and reviewers.
References
- 1.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 3.Karin M, Greten FR. NF-κB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 4.Yu H, Kortylewski M, Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 5.Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. 2009;9:798–809. doi: 10.1038/nrc2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kortylewski M, Kujawski M, Wang T, Wei S, Zhang S, Pilon-Thomas S, Niu G, Kay H, Mulé J, Kerr WG, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–1321. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 7.Wang T, Niu G, Kortylewski M, Burdelya L, Shain K, Zhang S, Bhattacharya R, Gabrilovich D, Heller R, Coppola D, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 8.Takeda K, Clausen BE, Kaisho T, Tsujimura T, Terada N, Forster I, Akira S. Enhanced Th1 activity and development of chronic enterocolitis in mice devoid of Stat3 in macrophages and neutrophils. Immunity. 1999;10:39–49. doi: 10.1016/s1074-7613(00)80005-9. [DOI] [PubMed] [Google Scholar]
- 9.Rennick DM, Fort MM, Davidson NJ. Studies with IL-10-/- mice: an overview. J Leukoc Biol. 1997;61:389–396. doi: 10.1002/jlb.61.4.389. [DOI] [PubMed] [Google Scholar]
- 10.Deng L, Zhou JF, Sellers RS, Li JF, Nguyen AV, Wang Y, Orlofsky A, Liu Q, Hume DA, Pollard JW, et al. A novel mouse model of inflammatory bowel disease links mammalian target of rapamycin-dependent hyperproliferation of colonic epithelium to inflammation-associated tumorigenesis. Am J Pathol. 2010;176:952–967. doi: 10.2353/ajpath.2010.090622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ernst M, Putoczki TL. Stat3: linking inflammation to (gastrointestinal) tumourigenesis. Clin Exp Pharmacol Physiol. 2012;39:711–718. doi: 10.1111/j.1440-1681.2011.05659.x. [DOI] [PubMed] [Google Scholar]
- 12.Masuda M, Suzui M, Yasumatu R, Nakashima T, Kuratomi Y, Azuma K, Tomita K, Komiyama S, Weinstein IB. Constitutive activation of signal transducers and activators of transcription 3 correlates with cyclin D1 over-expression and may provide a novel prognostic marker in head and neck squamous cell carcinoma. Cancer Res. 2002;62:3351–3355. [PubMed] [Google Scholar]
- 13.Kortylewski M, Heinrich PC, Mackiewicz A, Schniertshauer U, Klingmuller U, Nakajima K, Hirano T, Horn F, Behrmann I. Interleukin-6 and oncostatin M-induced growth inhibition of human A375 melanoma cells is STAT-dependent and involves upregulation of the cyclin-dependent kinase inhibitor p27/Kip1. Oncogene. 1999;18:3742–3753. doi: 10.1038/sj.onc.1202708. [DOI] [PubMed] [Google Scholar]
- 14.Stephanou A, Brar BK, Knight RA, Latchman DS. Opposing actions of STAT-1 and STAT-3 on the Bcl-2 and Bcl-x promoters. Cell Death Differ. 2000;7:329–330. doi: 10.1038/sj.cdd.4400656. [DOI] [PubMed] [Google Scholar]
- 15.Garcia R, Bowman TL, Niu G, Yu H, Minton S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R, Parsons S, et al. Constitutive activation of Stat3 by the Src and JAK tyrosine kinases participates in growth regulation of human breast carcinoma cells. Oncogene. 2001;20:2499–2513. doi: 10.1038/sj.onc.1204349. [DOI] [PubMed] [Google Scholar]
- 16.Corvinus FM, Orth C, Moriggl R, Tsareva SA, Wagner S, Pfitzner EB, Baus D, Kaufmann R, Huber LA, Zatloukal K, et al. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin Q, Lai R, Chirieac LR, Li C, Thomazy VA, Grammatikakis I, Rassidakis GZ, Zhang W, Fujio Y, Kunisada K, et al. Constitutive activation of JAK3/STAT3 in colon carcinoma tumors and cell lines: inhibition of JAK3/STAT3 signaling induces apoptosis and cell cycle arrest of colon carcinoma cells. Am J Pathol. 2005;167:969–980. doi: 10.1016/S0002-9440(10)61187-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Timofeeva OA, Tarasova NI, Zhang X, Chasovskikh S, Cheema AK, Wang H, Brown ML, Dritschilo A. STAT3 suppresses transcription of proapoptotic genes in cancer cells with the involvement of its N-terminal domain. Proc Natl Acad Sci USA. 2013;110:1267–1272. doi: 10.1073/pnas.1211805110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horiguchi A, Oya M, Shimada T, Uchida A, Marumo K, Murai M. Activation of signal transducer and activator of transcription 3 in renal cell carcinoma: a study of incidence and its association with pathological features and clinical outcome. J Urol. 2002;168:762–765. [PubMed] [Google Scholar]
- 20.Kawada M, Seno H, Uenoyama Y, Sawabu T, Kanda N, Fukui H, Shimahara Y, Chiba T. Signal transducers and activators of transcription 3 activation is involved in nuclear accumulation of β-catenin in colorectal cancer. Cancer Res. 2006;66:2913–2917. doi: 10.1158/0008-5472.CAN-05-3460. [DOI] [PubMed] [Google Scholar]
- 21.Morikawa T, Baba Y, Yamauchi M, Kuchiba A, Nosho K, Shima K, Tanaka N, Huttenhower C, Frank DA, Fuchs CS, et al. STAT3 expression, molecular features, inflammation patterns, and prognosis in a database of 724 colorectal cancers. Clin Cancer Res. 2011;17:1452–1462. doi: 10.1158/1078-0432.CCR-10-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmada MM, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn's disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bollrath J, Phesse TJ, von Burstin VA, Putoczki T, Bennecke M, Bateman T, Nebelsiek T, Lundgren-May T, Canli O, Schwitalla S, et al. gp130-mediated Stat3 activation in enterocytes regulates cell survival and cell-cycle progression during colitis-associated tumorigenesis. Cancer Cell. 2009;15:91–102. doi: 10.1016/j.ccr.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 24.Kawamori T, Kaneshiro T, Okumura M, Maalouf S, Uflacker A, Bielawski J, Hannun YA, Obeid LM. Role for sphingosine kinase 1 in colon carcinogenesis. FASEB J. 2009;23:405–414. doi: 10.1096/fj.08-117572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Snider AJ, Kawamori T, Bradshaw SG, Orr KA, Gilkeson GS, Hannun YA, Obeid LM. A role for sphingosine kinase 1 in dextran sulfate sodium-induced colitis. FASEB J. 2009;23:143–152. doi: 10.1096/fj.08-118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang J, Nagahashi M, Kim EY, Harikumar KB, Yamada A, Huang WC, Hait NC, Allegood JC, Price MM, Avni D, et al. Sphingosine-1-phosphate links persistent STAT3 activation, chronic intestinal inflammation, and development of colitis-associated cancer. Cancer Cell. 2013;23:107–120. doi: 10.1016/j.ccr.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.el Marjou F, Janssen KP, Chang BH, Li M, Hindie V, Chan L, Louvard D, Chambon P, Metzger D, Robine S. Tissue-specific and inducible Cre-mediated recombination in the gut epithelium. Genesis. 2004;39:186–193. doi: 10.1002/gene.20042. [DOI] [PubMed] [Google Scholar]
- 28.Rigas B, Tsioulias GJ, Allan C, Wali RK, Brasitus TA. The effect of bile acids and piroxicam on MHC antigen expression in rat colonocytes during colon cancer development. Immunology. 1994;83:319–323. [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz N, Minton S, Cox C, Bowman T, Gritsko T, Garcia R, Eweis I, Wloch M, Livingston S, Seijo E, et al. Activation of Stat3 in primary tumors from high-risk breast cancer patients is associated with elevated levels of activated SRC and survivin expression. Clin Cancer Res. 2006;12:20–28. doi: 10.1158/1078-0432.CCR-04-1749. [DOI] [PubMed] [Google Scholar]
- 30.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. [PubMed] [Google Scholar]
- 31.Lee H, Deng J, Kujawski M, Yang C, Liu Y, Herrmann A, Kortylewski M, Horne D, Somlo G, Forman S, et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat Med. 2010;16:1421–1428. doi: 10.1038/nm.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olivera A, Kohama T, Tu Z, Milstien S, Spiegel S. Purification and characterization of rat kidney sphingosine kinase. J Biol Chem. 1998;273:12576–12583. doi: 10.1074/jbc.273.20.12576. [DOI] [PubMed] [Google Scholar]
- 33.Waldner MJ, Wirtz S, Jefremow A, Warntjen M, Neufert C, Atreya R, Becker C, Weigmann B, Vieth M, Rose-John S, et al. VEGF receptor signaling links inflammation and tumorigenesis in colitis-associated cancer. J Exp Med. 2010;207:2855–2868. doi: 10.1084/jem.20100438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muller S, Lory J, Corazza N, Griffiths GM, Z'Graggen K, Mazzucchelli L, Kappeler A, Mueller C. Activated CD4+ and CD8+ cytotoxic cells are present in increased numbers in the intestinal mucosa from patients with active inflammatory bowel disease. Am J Pathol. 1998;152:261–268. [PMC free article] [PubMed] [Google Scholar]
- 36.Okazaki K, Morita M, Nishimori I, Sano S, Toyonaga M, Nakazawa Y, Yamamoto Y. Major histocompatibility antigen-restricted cytotoxicity in inflammatory bowel disease. Gastroenterology. 1993;104:384–391. doi: 10.1016/0016-5085(93)90405-2. [DOI] [PubMed] [Google Scholar]
- 37.Erdman SE, Rao VP, Olipitz W, Taylor CL, Jackson EA, Levkovich T, Lee CW, Horwitz BH, Fox JG, Ge Z, et al. Unifying roles for regulatory T cells and inflammation in cancer. Int J Cancer. 2010;126:1651–1665. doi: 10.1002/ijc.24923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Atreya I, Neurath MF. Immune cells in colorectal cancer: prognostic relevance and therapeutic strategies. Expert Rev Anticancer Ther. 2008;8:561–572. doi: 10.1586/14737140.8.4.561. [DOI] [PubMed] [Google Scholar]
- 39.Deschoolmeester V, Baay M, Lardon F, Pauwels P, Peeters M. Immune cells in colorectal cancer: prognostic relevance and role of MSI. Cancer Microenviron. 2011;4:377–392. doi: 10.1007/s12307-011-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ladoire S, Martin F, Ghiringhelli F. Prognostic role of FOXP3+ regulatory T cells infiltrating human carcinomas: the paradox of colorectal cancer. Cancer Immunol Immunother. 2011;60:909–918. doi: 10.1007/s00262-011-1046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 42.Eksteen B, Miles A, Curbishley SM, Tselepis C, Grant AJ, Walker LS, Adams DH. Epithelial inflammation is associated with CCL28 production and the recruitment of regulatory T cells expressing CCR10. J Immunol. 2006;177:593–603. doi: 10.4049/jimmunol.177.1.593. [DOI] [PubMed] [Google Scholar]
- 43.Iellem A, Mariani M, Lang R, Recalde H, Panina-Bordignon P, Sinigaglia F, D'Ambrosio D. Unique chemotactic response profile and specific expression of chemokine receptors CCR4 and CCR8 by CD4+CD25+ regulatory T cells. J Exp Med. 2001;194:847–853. doi: 10.1084/jem.194.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tan MC, Goedegebuure PS, Belt BA, Flaherty B, Sankpal N, Gillanders WE, Eberlein TJ, Hsieh CS, Linehan DC. Disruption of CCR5-dependent homing of regulatory T cells inhibits tumor growth in a murine model of pancreatic cancer. J Immunol. 2009;182:1746–1755. doi: 10.4049/jimmunol.182.3.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Grivennikov S, Karin M. Autocrine IL-6 signaling: a key event in tumorigenesis? Cancer Cell. 2008;13:7–9. doi: 10.1016/j.ccr.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 46.Gurgui M, Broere R, Kalff JC, van Echten-Deckert G. Dual action of sphingosine 1-phosphate in eliciting proinflammatory responses in primary cultured rat intestinal smooth muscle cells. Cell Signal. 2010;22:1727–1733. doi: 10.1016/j.cellsig.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 47.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et al. Aspirin use, tumor PIK3-CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alshaker H, Sauer L, Monteil D, Ottaviani S, Srivats S, Bohler T, Pchejetski D. Therapeutic potential of targeting SK1 in human cancers. Adv Cancer Res. 2013;117:143–200. doi: 10.1016/B978-0-12-394274-6.00006-6. [DOI] [PubMed] [Google Scholar]
- 49.Pchejetski D, Nunes J, Coughlan K, Lall H, Pitson SM, Waxman J, Sumbayev VV. The involvement of sphingosine kinase 1 in LPS-induced Toll-like receptor 4-mediated accumulation of HIF-1α protein, activation of ASK1 and production of the pro-inflammatory cytokine IL-6. Immunol Cell Biol. 2011;89:268–274. doi: 10.1038/icb.2010.91. [DOI] [PubMed] [Google Scholar]
- 50.Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 2004;427:355–360. doi: 10.1038/nature02284. [DOI] [PubMed] [Google Scholar]
- 51.Azuma H, Takahara S, Ichimaru N, Wang JD, Itoh Y, Otsuki Y, Morimoto J, Fukui R, Hoshiga M, Ishihara T, et al. Marked prevention of tumor growth and metastasis by a novel immunosuppressive agent, FTY720, in mouse breast cancer models. Cancer Res. 2002;62:1410–1419. [PubMed] [Google Scholar]
- 52.Daniel C, Sartory N, Zahn N, Geisslinger G, Radeke HH, Stein JM. FTY720 ameliorates Th1-mediated colitis in mice by directly affecting the functional activity of CD4+CD25+ regulatory T cells. J Immunol. 2007;178:2458–2468. doi: 10.4049/jimmunol.178.4.2458. [DOI] [PubMed] [Google Scholar]